Plant Growth Promoting Abilities of Novel Burkholderia-Related Genera and Their Interactions With Some Economically Important Tree Species
- 1Biomaterials and Biocatalysts Group, Temasek Life Sciences Laboratory, 1 Research Link, National University of Singapore, Singapore, Singapore
- 2Division of Natural Resources, ICAR—Indian Institute of Horticultural Research, Hesarghatta Lake Post, Bengaluru, India
- 3School of Biological Sciences, Nanyang Technological University, Singapore, Singapore
A survey of bacterial endophytes associated with the leaves of oil palm and acacias resulted in the isolation of 19 bacterial strains belonging to the genera Paraburkholderia, Caballeronia, and Chitinasiproducens, which are now regarded as distinctively different from the parent genus Burkholderia. Most strains possessed one or more plant growth promotion (PGP) traits although nitrogenase activity was present in only a subset of the isolates. The diazotrophic Paraburkholderia tropica strain S39-2 with multiple PGP traits and the non-diazotrophic Chitinasiproducens palmae strain JS23T with a significant level of 1-aminocyclopropane-1-carboxylate (ACC) deaminase activity were selected to investigate the influence of bacterial inoculation on some economically important tree species. Microscopic examination revealed that P. tropica S39-2 was rhizospheric as well as endophytic while C. palmae JS23T was endophytic. P. tropica strain S39-2 significantly promoted the growth of oil palm, eucalyptus, and Jatropha curcas. Interestingly, the non-diazotrophic, non-auxin producing C. palmae JS23T strain also significantly promoted the growth of oil palm and eucalyptus although it showed negligible effect on J. curcas. Our results suggest that strains belonging to the novel Burkholderia-related genera widely promote plant growth via both N-independent and N-dependent mechanisms. Our results also suggest that the induction of defense response may prevent the colonization of an endophyte in plants.
Introduction
Plant-associated bacteria play important roles in plant development and differentiation (Hallmann et al., 1997; Reiter and Sessitsch, 2006). Rhizospheric bacterial communities are known to modulate the bio-availability of mineral nutrients, stimulate plant growth through phytohormone production, and enhance plant resistance to pathogens (Bringel and Couée, 2015). On the other hand, phyllospheric bacteria that inhabit the aerial parts of plants are lesser studied despite the fact that the global phyllosphere measures more than 108 km2 accommodating a diverse bacterial community of about 1026 bacterial cells (Lindow and Brandl, 2003). It is intriguing how the oxygen-sensitive nitrogen fixation system adapts itself to the photosynthetic O2-rich phyllosphere (Dixon and Kahn, 2004). How the tissue-specific localization of various endophytes is regulated by the plant system is also poorly understood.
Burkholderia is a fairly large genus with more than 100 validly described species. It has received little attention from the application point of view due to the fact that the B. cepacia complex (Bcc), comprising 20 species, cause opportunistic infections in cystic fibrosis patients while members of the B. pseudomallei group cause mellidiosis (Cheng and Currie, 2005). Burkholderia mallei is the causative agent of the Glanders disease in horses (Nierman et al., 2004). Recent data suggest that Paraburkholderia and Caballeronia are phylogenetically distinct from the Burkholderia cepacia complex and has therefore spawned an interest for their agrobiotechnology applications (Caballero-Mellado et al., 2007; Sawana et al., 2014; Dobritsa and Samadpour, 2016). Diazotrophic Paraburkholderia species have been isolated from the rhizosphere, nodules of legume plants, as well as root tissues of non-legume plants (Aizawa et al., 2010; Suárez-Moreno et al., 2012; De Meyer et al., 2013, 2014; Martínez-Aguilar et al., 2013; Sheu et al., 2013, 2015; Steenkamp et al., 2015), while a phosphate solubilizing Paraburkholderia tropica strain has been isolated from the rhizosphere of pomegranate (Kaur et al., 2016).
Paraburkholderia phytofirmans strain PsJN is a well-studied plant endophyte and has contributed greatly to the understanding of plant–endophyte interactions (Frommel et al., 1991; Conn et al., 1997; Pillay and Nowak, 1997; Barka et al., 2000; Kim et al., 2012b; Kost et al., 2013). Chitinasiproducens palmae isolated from the leaf tissues of oil palm is a newly validated genus genetically distinct from Burkholderia, Caballeronia, or Paraburkholderia (Madhaiyan et al., 2020).
Oil palm is one of the most economically important oil yielding crops in Southeast Asia (Koh and Wilcove, 2007). Acacia and eucalyptus are the dominant forestry species in tropical Southeast Asia, supplying raw material for paper and pulp, plywood, and wood composites. Jatropha curcas, on the other hand, is a shrub that grows in poor soils, produces oil-rich seeds, and is seen as an emerging biofuel crop (Madhaiyan et al., 2013b). The growth of such tree crops demands a high availability of soil nutrients, especially nitrogen since forest productivity is directly related to nutrient levels of the soil (Smethurst et al., 2004; Gobert and Plassard, 2008). Inoculation with N2-fixing endophytic bacteria is an attractive option to reduce the demand for nitrogen fertilizer and to enhance tree biomass production. This study was therefore undertaken to decipher the mechanisms of plant growth-promoting activities of the Burkholderia-related genera, which have not been studied in much detail in the past.
Materials and Methods
Isolation of Nitrogen-Fixing Endophytic Bacteria
Healthy oil palm leaves and phyllodes of Acacias were collected from various locations of Singapore. The collected samples were placed in plastic bags and processed within 2 h. Sample tissues (~5 g) were surface sterilized by sequential immersion in 90% (v/v) ethanol for 5 min and 10% (v/v) hydrogen peroxide solution for 10 min followed by repeated washing in sterile water. A 100-μl aliquot of the wash water from the third rinse was plated on a rich medium to confirm the efficiency of surface sterilization. Surface-sterilized tissues were macerated, enriched in an N-free medium, diluted serially with phosphate saline buffer, and plated on JNFb (Döbereiner et al., 1995), YEMA (Vincent, 1970), or BAc medium (Estrada-De Los Santos et al., 2001). An aliquot (100 μl) of the enriched suspension was simultaneously inoculated in a tube containing semi-solid N-free media. Sub-surface pellicles or colonies that appeared on agar plates after 72 h at 30°C were sub-cultured to obtain pure cultures. Isolates were routinely cultured on their respective agar plates at 30°C under aerobic conditions and stored at −80°C in 7% (v/v) DMSO.
DNA Amplification, Sequencing, and Phylogenetic Analysis
The phylogenetic positions of the isolated strains were identified by analyzing the complete 16S rRNA gene sequence as previously described (Wilson, 1997). The 16S rRNA gene was amplified from the genomic DNA of the isolated strains using the universal primers 27F and 1492R as described previously (DeLong, 1992; Madhaiyan et al., 2015). Sequencing was done with the BigDye Terminator Cycle Sequencing Kit (Perkin Elmer, Waltham, MA, USA) and samples were run on an Applied Biosystems 3730 Xl DNA sequencer (Applied Biosystems, Carlsbad, CA, USA). Sequences were analyzed against the EzTaxon-e Database (http://eztaxon-e.ezbiocloud.net/) (Kim et al., 2012a) and aligned using ClustalW tool in MEGA version 6 (Tamura et al., 2013). PCR amplification of the partial nifH fragment was performed as described (Pinto-Tomás et al., 2009). A 750-bp fragment of the ACC deaminase gene (acdS) was amplified using the primers F1937 and F1938 as described (Blaha et al., 2006). The nifH and acdS gene sequences were analyzed by BLAST searches against sequences in the GenBank at NCBI.
Screening for Plant Growth-Promoting Traits
The diazotophic trait of the isolates was confirmed by the presence of Acetylene Reduction Activity (ARA). The assay was performed using 125 ml serum bottles (Wheaton Industries Inc., Millville, NJ, USA) filled with the nitrogen-free medium (40 ml) of the following composition (per liter): 5 g Glucose, 5 g Mannitol, 0.1 g CaCl2·2H2O, 0.1 g MgSO4·7H2O, 5 mg Na2MoO4·2H2O, 0.9 g K2HPO4, 0.1 g KH2PO4, 0.01 g FeSO4·7H2O, 5 g CaCO3, and 1 ml trace element mixture. The trace element mixture comprised (per liter) of 0.1 g ZnSO4·7H2O, 0.03 g MnCl2·4H2O, 0.3 g H3BO3, 0.2 g CoCl2·6H2O, 0.01 g CuCl2·2H2O, and 0.02g NiCl2·6H2O in water. The assay was performed by injecting purified acetylene gas into the bottles that were sealed with gas-tight serum stoppers to achieve a yield of 15% acetylene (v/v), followed by incubation for 96 h at 30°C. For nitrogenase switch-off/switch-on assays, the isolates were cultured in N-free medium and supplemented with 1 mM NH4Cl or 10% O2. Protein concentration of the bacteria used for ARA assay was determined with the Bicinchoninic acid kit (B9643; Sigma-Aldrich, St. Louis, MO, USA) using bovine serum albumin (BSA) as the standard.
To determine the nitrogenase activity under in planta conditions, the ARA was performed using fresh plant tissues that were collected 30 and 60 days after inoculation of the individual bacteria strains. Root samples of the plants were separated from seedlings after carefully removing the adhering soil; placed in 250 ml glass bottles, and sealed with a rubber septum. After removing an equivalent volume of air, acetylene was injected into the bottles to achieve a final concentration of 15% (v/v) and incubated at 30°C for 24 h. Gas samples (0.8 ml for in planta samples) were removed at regular intervals with a PTFE-syringe (Hewlett-Packard, Palo Alto, CA, USA) and analyzed in a GCMS-QP2010 Ultra Gas Chromatograph (Shimadzu Corporation, Kyoto, Japan) with a flame ionization detector and GS-Alumina (30 m × 0.53 mm I.D.) column. The operational conditions were helium gas flow 30 ml/min; detector temperature: 200°C; pressure: 4.0 psi. The ethylene produced by the bacteria was quantified using a standard ethylene (C2H4, Product Number: 00489, Sigma-Aldrich) curve that was prepared in duplicate with concentrations ranging from 1 to 1000 nmol. All the obtained values were expressed after deducting the ethylene value of a blank treatment. The in planta ARA values were expressed as nmol C2H4 released day−1 seedlings−1 after deducting the plant's background C2H4 emission.
The ability of the strains to catabolize ACC was determined by assessing their growth in DF minimal salts medium (Dworkin and Foster, 1958) supplemented with 3 mM ACC. ACC deaminase activity was measured spectrophotometrically at 540 nm as described previously by Shah et al. (1998) and Honma and Shimomura (1978).
For the extraction of indole compounds, a single colony was used to inoculate 6 mL 2xYT medium supplemented with 100 μg ml−1 L-tryptophan and incubated at 30°C with agitation (200 rpm) for 4 days. A 2-ml aliquot of the cell-free supernatant was mixed with 100 μl of 10 mM orthophosphoric acid and 4 ml of Salkowski's reagent (1 ml of 0.5 M FeCl3 in 50 ml of 35% HClO4) and incubated at room temperature for 25 min. The concentration of indoles in the culture supernatant was determined spectrophotometrically at 530 nm (UV-1601, Shimadzu) by interpolating against the standard curve of indole-3-acetic acid prepared with concentrations ranging from 0 to 100 μg/ml (Sigma Chemical Co., St. Louis, MO, USA)
To evaluate the phosphate solubilization activity, strains were spot-inoculated on NBRIP agar plates (Nautiyal, 1999) containing 0.5% tricalcium phosphate [Ca3(PO4)2] as the inorganic phosphate source. The plates were incubated at 30°C for 7 days and solubilization of mineral phosphate was qualitatively estimated by measuring the diameter of the clear halo formed as a result of phosphate solubilization around the bacterial colonies.
The ability to produce siderophores by bacterial strains was assayed using the universal Chrome Azurol S (CAS; Sigma-Aldrich) dye assay (Schwyn and Neilands, 1987; Gao et al., 2006).
Screening for Cell Wall Degrading Endoglucanase Activity
The endoglucanase activity was determined as described previously (Reinhold-Hurek et al., 1993), with some modifications. A plate containing Kim–Wimpenny solid medium with 0.2% carboxymethyl cellulose (CMC) (Kim and Wimpenny, 1981), with or without 0.5% D-glucose, was spotted with 1 μl of an exponentially growing culture that was adjusted to an absorbance at 600 nm to 1.0; air-dried and incubated at 30°C for 3 days. Colonies were flushed off the plate with water and the plate was stained with a 0.1% Congo red solution for 30 min, followed by several washes with 1 M NaCl. The appearance of a clear yellow halo around the colony in a red background indicated a positive staining for endoglucanase activity.
Growth Inhibitory Activity
Rhodosporidium toruloides ATCC 10657 (Hu and Ji, 2016) and Kosakonia sacchari strain R4-368 (diazotrophic), the latter previously known as Enterobacter species (Madhaiyan et al., 2013a,b), were used as the fungal and bacterial test strains, respectively, for the determination of growth-inhibiting activity of the strains as described by Compant et al. (2005).
Construction of GFP-Tagged S39-2 and JS23T Strains
Escherichia coli strain S17-1λπ was used to mobilize the Tn5 transposon, constructed in pUT-tac-aph-sfGFP (Supplementary Figure 1), to the genome of S39-2 and JS23T strains by biparental mating. Mating was carried out overnight by mixing equal volume of an overnight culture of donor and acceptor cells on a 0.2-μm filter membrane placed on a 2xYT plate. Transformants were selected on succinate media (Nunn and Lidstrom, 1986) with 25 μg ml−1 kanamycin. The fluorescence was quantified using an Infinite® M200 microplate reader (TECAN, Männedorf, Switzerland). A single transformant that showed high fluorescence with no growth abnormality was selected for plant colonization studies.
The transformant's stability was determined by re-streaking a single colony from selective medium onto a 2xYT agar plate without any selection pressure. After 12 subcultures at a 5-day interval in 2xYT agar plates, colonies were scored for kanamycin resistance and GFP fluorescence. Fresh colonies that were scrapped off the agar plates were washed in sterile distilled water and starved at 4°C for 3 weeks. The suspensions were then plated onto succinate media with or without kanamycin. The colony, cell morphology, and growth of the GFP derivatives were compared with the wild type by culturing in 2xYT and succinate media, respectively.
Plant Inoculation and Endophytic Colonization of Seedlings
Lemon eucalyptus seeds were obtained from Richters Herbs (Goodwood, Canada). Seeds were surface sterilized by soaking in 70% alcohol (v/v) for 1 min and 15% H2O2 for 5 min followed by thorough rinsing with sterilized distilled water (5 × 15 min each) on a shaking platform. The surface-sterilized seeds were germinated in sterilized soil that had been autoclaved (121°C, 15 psi for 3 h). Strains JS23T and S39-2 were cultured in 2xYT medium for 36 h at 30°C, adjusted to a density of approximately 108 cfu ml−1, and applied near the root zone at a dose of 2 ml per seedling at 7 days after sowing (DAS). On day 14, the seedlings were transferred from germination trays to plastic pots (15 × 25 cm) containing 750 g non-sterilized red soil (Far East Flora, Singapore). Plants were maintained in a greenhouse with no artificial lighting and air-conditioning. Plant height, number of leaves, number of branches, chlorophyll content, and leaf area were determined at 30 and 60 DAI. Leaf chlorophyll levels were measured using the at LEAF chlorophyll meter (FT Green LLC, Wilmington, DE, USA). The relative chlorophyll concentration was recorded as the ratio of transmittance between the red (650 nm) and infrared (940 nm) emissions through the leaf. Leaf nitrogen (N) content was determined by the combustion method in an elemental analyzer (Vario EL Elemental Analyzer, Elementar, Germany) equipped with a thermal conductivity detector.
Oil palm seedlings derived from tissue culture of a hybrid variety were raised in plastic pots filled with 250 g potting mix (BVB substrates; Stealth Garden, Seacliff Park, Australia) and maintained in a greenhouse for 60 days. Selected healthy seedlings of uniform size were transferred to 45 × 60 cm pots containing ~5 kg non-sterilized red soil. The root zone of each seedling was inoculated with 5 ml of exponentially grown bacterial cells (108 cfu/ml). Plants were maintained in a greenhouse with no artificial lighting and air-conditioning. Plant growth data were recorded at 30, 60, and 90 DAI.
Seeds of Jatropha curcas were sown in germination trays filled with potting mixture and inoculated with 2 ml exponentially grown cells (108 cfu/ml). Root inoculation was done after germination, and the growth parameters were recorded at 60 DAI. For pot cultivation experiment under N-limiting condition, healthy seedlings derived from surface-sterilized seeds were transferred to pots containing a mixture of perlite, vermiculite, and sand in 1:1:1 ratio (v/v). Bacterial cultures (108 cfu ml−1; 50 ml pot−1) were applied to the root zone around the stem. Plants were maintained in a glasshouse. As a control, plants were watered as needed with a plant nutrient solution containing with 2 mM ammonium chloride (N source). For plants grown under N-limiting condition, a plant nutrient solution without the N source was used.
Plant Colonization and Monitoring Tissue Localizations by CLSM
To track plant colonization in lemon eucalyptus, seeds were germinated for 7 days under gnotobiotic conditions in multi-well plates containing MS medium (Murashige and Skoog, 1962) and transferred to PhytatrayTM II plant growth vessels (P5929 Sigma) each containing 200 g of sterilized sand (121°C, 15 psi, 3 h) filled with 40 ml of autoclaved nitrogen-free plant nutrient solution (Iniguez et al., 2004). The plants were cultured in a growth chamber with a temperature of 28°C and 16/8 h day–night cycles. The GFP labeled JS23T and S39-2 strains were cultured in succinate medium for 48 h, adjusted to 108 cfu ml−1, and inoculated at 5 ml per tray 3 days after seedling transplantation. Endophytic colonization was determined using surface-sterilized shoots (leaf + stem) and roots (30 DAI), which were homogenized with sterile pestle and mortar. Serially diluted samples were plated on R2A agar plates, incubated at 30°C for 3–5 days before colony forming units (CFU) were determined. The efficiency of surface sterilization was monitored by spreading an aliquot of the last wash on a 2xYT agar plate and incubated for 3 days at 30°C.
The localizations of GFP-labeled cells in eucalyptus tissues were examined with confocal laser scanning microscopy (CLSM) at 15, 30, and 45 days after inoculation (DAI). CLSM was carried out using a Zeiss LSM 510 upright confocal microscope with a motorized stage (Carl Zeiss Inc., Oberkochen, Germany) equipped with a multi-argon laser (458/477/488/514 nm), and a He/Ne laser (543 nm and 633 nm). Images were acquired using the Plan-Apochromat 63×/1.4 oil differential objective and analyzed using the Laser Scanning System LSM5 PASCAL software. Bacterial localization in eucalyptus root and shoot tissues was identified from 3-D confocal stacks.
Nucleotide Sequence Accessions
The 16S rRNA gene sequences of the strains have been submitted to NCBI under the accession numbers KU049648-KU049653, KU140660-KU140661, and KT337490-KT337503. The acdS gene and nifH gene sequences can be found under the accession numbers KX622566-KX622569 and KX622570-KX622579.
Statistical Analysis
Statistical analyses were carried out using the Statistical Analysis System (SAS) Version 9.4 (SAS Institute Inc., Cary, NC, USA). Analysis of variance (ANOVA) for the endophytic population was carried out using the General Linear Model (GLM) in SAS. The bacterial population data were log-transformed before being subjected to further analysis. The means of the treatment results were subjected to ANOVA and presented using Fisher's protected least significant difference (LSD). Endophytic bacterial inoculation data were subjected to analysis of variance and testing of means by Duncan's multiple range test (DMRT) at p ≤ 0.05 using SAS package.
Results
Isolation of Endophytic Bacteria From Leaves of Oil Palm and Phyllodes of Acacia
A survey of endophytic bacteria associated with the phyllosphere of oil palm and Acacia led to the isolation of 19 strains belonging to three Burkholderia related genera, Paraburkholderia, Caballeronia, and Chitinasiproducens (Figure 1). Recently, the genus Burkholderia has been split into three clades: Clade I retaining the genus name Burkholderia and consisting mainly of animal and plant pathogens; Clade II was named as genus Caballeronia which are mainly composed of strains of environmental origin (Dobritsa and Samadpour, 2016), while Clade III was named as genus Paraburkholderia, which are environmental bacteria including plant growth-promoting bacteria and nodule symbionts (Coenye and Vandamme, 2003; Suárez-Moreno et al., 2012; Sawana et al., 2014; Dobritsa and Samadpour, 2016; Dobritsa et al., 2016). 16S rRNA analysis of the Burkholderia-related genera from oil palm and acacias resulted in the identification of 16 distinct strains of Paraburkholderia, three strains of Caballeronia and one strain belonging to the novel genus, Chitinasiproducens. The isolates were closely related to Caballeronia udeis, B. anthina, B. vietnamiensis, B. ubonensis, P. tropica, P. sediminicola, P. unamae and P. phytofirmans (Table 1, Figure 1). Most of the strains from Paraburkholderia could be assigned to P. tropica. The endophytic nature of some of these Paraburkholderia species has already been demonstrated (Caballero-Mellado et al., 2004; Reis et al., 2004; Suárez-Moreno et al., 2012).
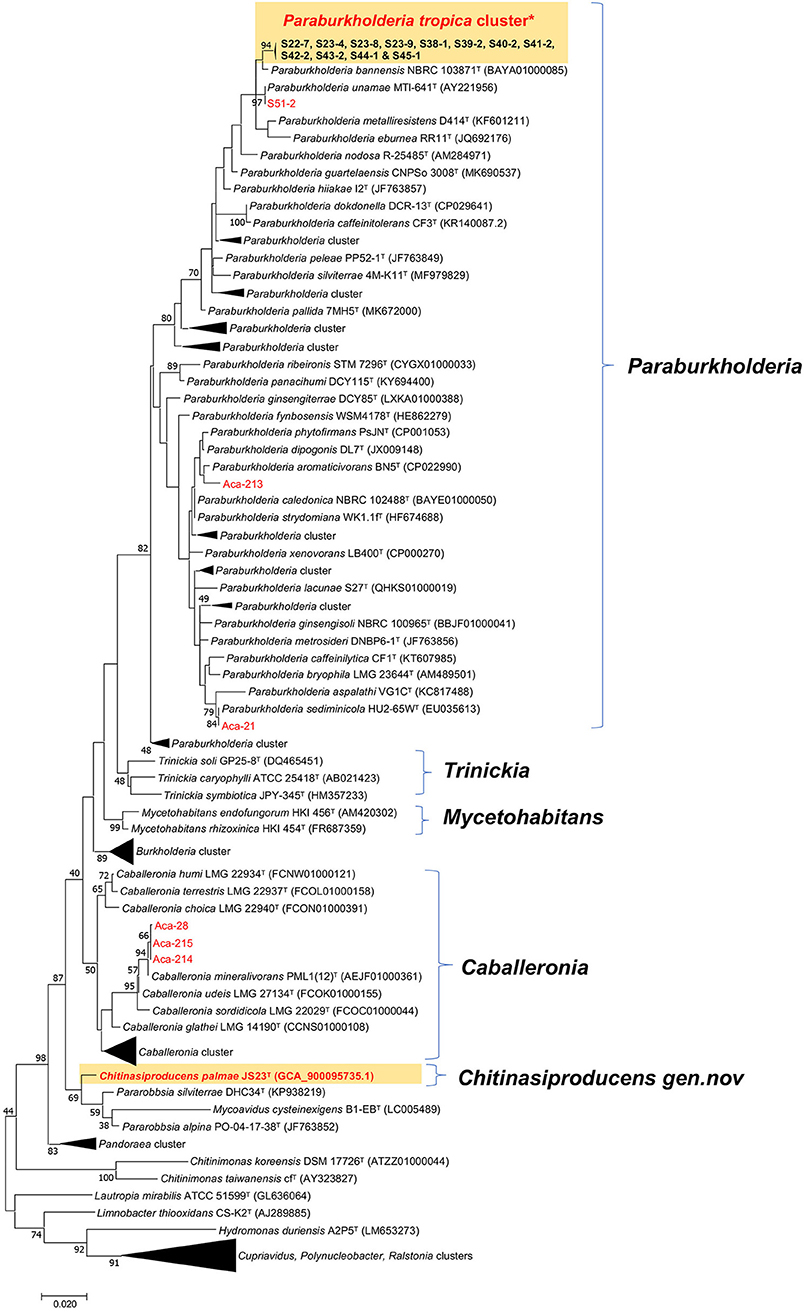
Figure 1. Maximum-likelihood tree based on 16S rRNA gene sequences. Positions of isolates from tree crops and close relatives of the genera Caballeronia, Chitinasiproducens, and Paraburkholderia (family Burkholderiaceae) are shown. GenBank accession numbers are given in parentheses. Bootstrap values (expressed as percentages of 500 replications) greater than 40% are shown at the branch points. Bar, 0.005 substitutions per nucleotide position. *Twelve strains (S22-7, S23-4, S23-9, S23-8, S38-1, S39-2, S40-2, S41-2, S42-2, S43-2, S44-1, and S45-1) closely related to P. tropica cluster and all isolated strains in the present study are indicated in red.
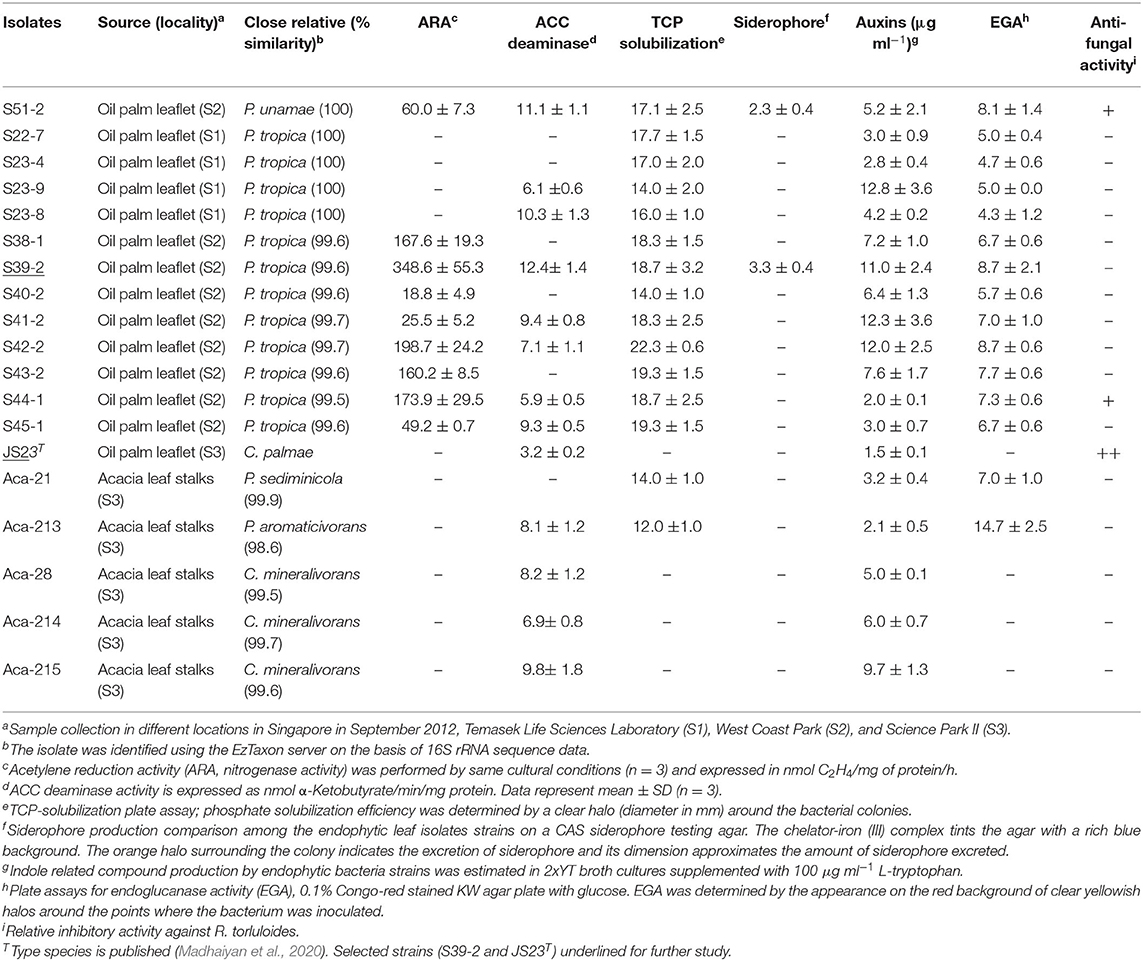
Table 1. Screening for growth-promoting traits of endophytic isolates from oil palm and acacia leaves.
Nitrogenase Activity
The 12 strains that belong to P. tropica showed relatively high nitrogenase activity, ranging from 18.8 to 348.6 nmol C2H4 (mg protein)−1 h−1, while the activity was completely absent in four strains even after 96 h of incubation (Table 1). Of the ARA-positive strains, 11 of the 12 strains (91.7%) were derived from oil palm and eight were identified as P. tropica. The remaining strains were identified as to B. anthina, B. vietnamiensis and P. unamae. None of four acacias isolates showed ARA activity. The nifH genes from the Burkholderia strains that had ARA formed a tight cluster with those from other plant-associated Burkholderia species (Supplementary Figure 2). Comparisons of the nifH sequences generated in this study with those available in GenBank revealed that they shared 96–99% sequence identity with P. tropica Ppe8T (Reis et al., 2004). The nifH gene was not present in the genome of strain JS23T as determined by PCR and this has been confirmed by genome sequencing (Madhaiyan et al., 2020).
Nitrogenases are usually highly sensitive to oxygen and ammonium (). Both molybdenum-dependent and molybdenum-independent nitrogenases are stringently regulated at the transcriptional and post-translational levels (Dixon and Kahn, 2004). Acetylene reduction assay in a medium supplemented with 1 mM or 10% oxygen showed that, with the exception of five strains (S39-2, S43-2, S38-1, S44-1, and S53-1), none had significant nitrogenase activity when reached over 1 mM (Figure 2). Surprisingly, nitrogenase activity significantly increased with the supplementation of 10% oxygen in all the strains with the exception of strains S40-2 and S41-2 (Figure 2).
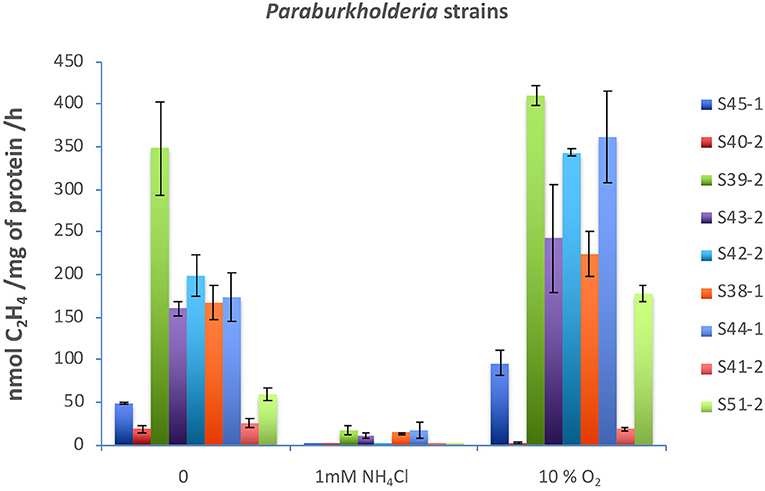
Figure 2. Effects of (2 mM) and O2 (10%) on nitrogenase activity. All strains were incubated in 40 ml of N-free medium in a 120 ml vial at 30°C. was added to 2 mM, and O2 gas was injected to 10% v/v. Acetylene reduction assay was performed by injecting purified acetylene into the bottles sealed with gas-tight serum stoppers to yield 15% acetylene (v/v); this was followed by incubation for up to 96 h at 30°C. Gas samples (0.5 ml) were removed after incubation and analyzed by gas chromatography with a flame ionization detector and GS-alumina column. Paraburkholderia strains tested: S45-1, S40-2, S39-2, S43-2, S42-2, S38-1, S44-1, S41-2, and S51-2.
Indole-Related Compounds and ACC Deaminase Production
Apart from N-fixation, the production of phytohormones, such as indole compounds and ethylene catabolizing ACC deaminase, contribute to the PGP function. Previous studies have demonstrated auxin production in P. kururiensis, P. phytofirmans, and P. unamae (Caballero-Mellado et al., 2007; Sun et al., 2009). We found that all the isolates produced indole compounds from the precursor L-tryptophan (Table 1). The activity was higher in the extracts of P. unamae S51-2, P. tropica S39-2, and P. tropica S23-8, producing 11.1, 12.4, and 10.3 nmol α-ketobutyrate mg−1 h−1 proteins, respectively. In addition, most strains showed ACC deaminase activity as evidenced by their ability to grow on media containing 3 mM ACC with the exception of six strains (Table 1). Sequence analysis of PCR products using the specific primer pair F1936/F1938 (Blaha et al., 2006) confirmed the presence of acdS genes in P. unamae S51-2, P. aromaticivorans Aca-213, and P. tropica S39-2.
Phosphorus Solubilization and Siderophore Production
Phosphorus solubilization is an important PGP trait and it is very common among Paraburkholderia species (Ghosh et al., 2016; Kaur et al., 2016). In the presence of inorganic phosphate in NBRIP medium (Nautiyal, 1999), all N-fixing strains produced a clear zone on the plates, indicating solubilization of inorganic phosphate (Table 1 and Supplementary Figure 3A). These strains were taxonomically placed in B. anthina, B. vietnamiensis, P. unamae, and P. tropica. Two of the N-fixing strains also produced siderophores, which are responsible for chelating Fe3+ ion (Table 1 and Supplementary Figure 3B).
Endoglucanase and Anti-microbial Activity
Subsequent to their colonization of the root surface, endophytic bacteria usually enter the inner tissues. Such endophytic bacteria secrete cell wall-degrading endo-1,4-glucanase which is believed to be important for plant colonization (Compant et al., 2005; Reinhold-Hurek et al., 2006; Fan et al., 2016). In this study, four strains showed clear endoglucanase activity when cultured in KW medium supplemented with CMC and glucose (Table 1). Three strains, C. palmae JS23T, P. tropica S44-1, and P. unamae S51-2, weakly inhibited the growth of the red yeast, Rhodosporidium toruloides. None of the isolates was able to inhibit the growth of endophytic bacterium K. sacchari R4-368 (Table 1).
Colonization of Lemon Eucalyptus Seedlings
Considering their PGP characteristics, strains S39-2 and JS23T originally isolated from oil palm were selected to gain further insights on their interactions with their host. Strain S39-2 had high nitrogenase activity and endoglucanase activity, while strain JS23T lacked both traits. Both strains produced ACC deaminase and IAA (Table 1). To facilitate the tracking of plant colonization by the bacterial strains, they were labeled with GFP by integrating a T5::egfp gene cassette into the chromosome via Tn5 transposition. The selected GFP-tagged strains were confirmed to have indistinguishable cell morphology and growth rate from the respective parent (Figures 3a,b) and showed high fluorescence (Figure 3c).
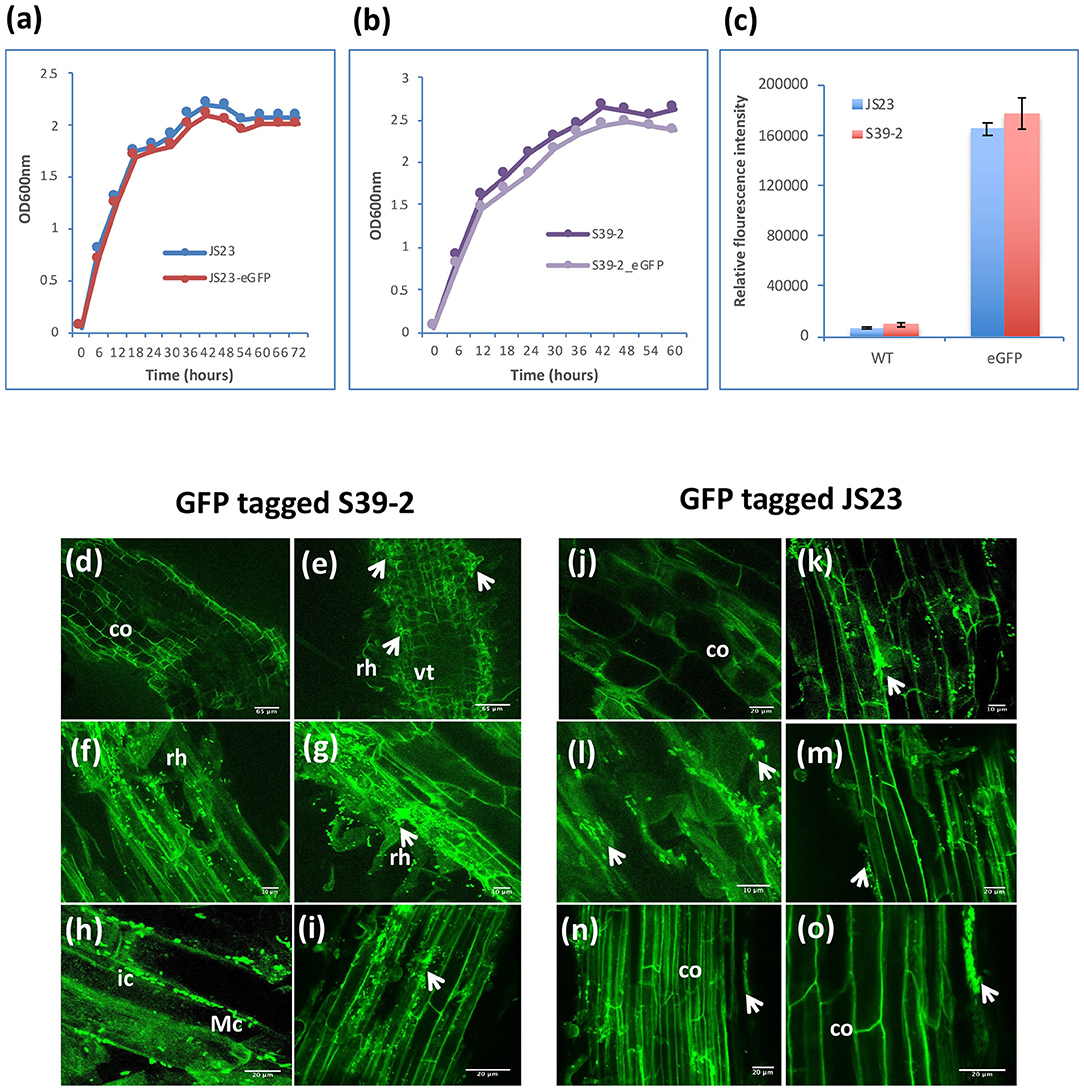
Figure 3. Colonization of GFP-tagged S39-2 and JS23T. (a, b) Cell density of wild-type (WT) and GFP-tagged JS23T and S39-2 in liquid culture (2xYT medium). (c) Relative fluorescence intensity of WT and GFP-tagged cells in culture medium was quantified in Tecan plate reader (Tecan i-Control, infinite 200; 476 nm excitation wavelength and 506 nm emission wavelength) using 96 flat bottom white polystyrol plate (Corning). Error bars are standard deviations (n = 3). (d, j) uninoculated roots. (e–i) Root tissues inoculated with S39-2-gfp on 15 DAI (d–f) and 45 DAI (h, i). (k–o) JS23T-gfp inoculated root tissues on 15 DAI (k, l) and 45 DAI (m–o). DAI, days after inoculation; rh, root hairs; co, cortical cells; arrowhead indicate micro-colonies (Mc); vt, vascular tissue; ic, intercellular colonization. Scale bars = 10 μm (f, g, k, and l), 20 μm (h, i, j, m, n, and o), and 65 μm (d, e).
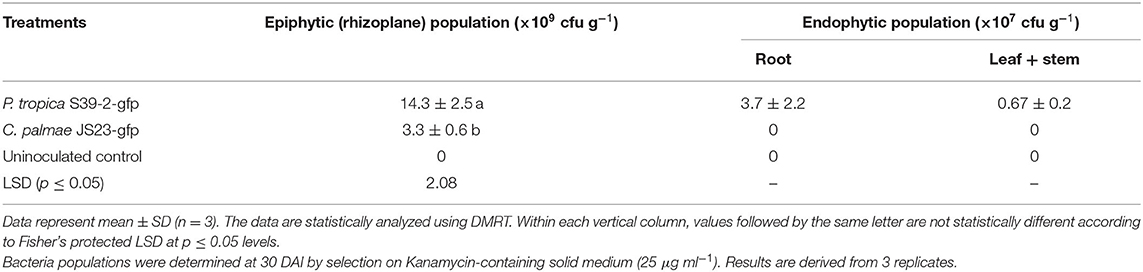
Inoculation of GFP-tagged strains on eucalyptus seedlings under sterile conditions showed that both C. palmae JS23-gfp and P. tropica S39-2-gfp were able to efficiently colonize the rhizosphere of the seedlings. Micro-colonies of JS23-gfp and S39-2-gfp could be found on the surface of roots or root hairs (Figures 3d–g, j–m). The rhizoplane colonization pattern of strains JS23-gfp and S39-2-gfp was similar to previous reports where rhizosphere bacteria colonize the nutrient rich zones of root hairs, emerging lateral roots or root tips that support bacterial proliferation (Walker et al., 2003; Bais et al., 2006). However, persistent intercellular colonization was observed only with S39-2-gfp (Figures 3h,i), but not with JS23-gfp (Figures 3n,o). It was observed that JS23-gfp strain had a weaker GFP signal on the root surface and the signal in internal tissue was only obvious at 15 DAI. Surprisingly, GFP signal in the internal tissues disappeared at 45 DAI in JS23-gfp inoculated plants. To further confirm the GFP imaging data, the titers of JS23-gfp and S39-2-gfp in different parts of the plants were determined. Strain JS23T had significantly lower cell density on the rhizoplane, and the endophytic population was not observed at 30 DAI (Table 2). This is in stark contrast to strain S39-2, which had 3.7 ± 2.2 and 0.67 ± 0.2 × 107 cfu g−1 tissues in roots and shoots (stem + leaf), respectively, at 30 DAI.
Plant Growth Promoting Effect on Tree Species
Strains S39-2 and JS23T were inoculated on eucalyptus seedlings. After 60 days, the inoculated plants displayed a ~30.4% increase in plant height over the control plants. The number of leaves and branches, leaf area, and chlorophyll contents were also significantly higher in the inoculated plants (Figures 4A–E). Compared to the mock-inoculated plants, strain S39-2-inoculated plants showed a significantly higher biomass yield, with a 60.4% increase in root biomass and a 44.2% increase in shoot biomass (Figure 4F). Strain S39-2 displayed a clear in planta nitrogenase activity, which was associated with a 53.5% increase in leaf nitrogen content over control plants. In contrast, clear in planta AR-activity was not observed in control plants. Consistent with this finding, the leaf nitrogen content was not altered (Figures 4G,H). The trace level of AR-activity detected in JS23T inoculated plants was not significantly different from the control and is likely to have resulted from aerial cross-contamination during the cultivation. Surprisingly, this non-diazotroph with relatively low auxin production displayed significant PGP activity in eucalyptus.
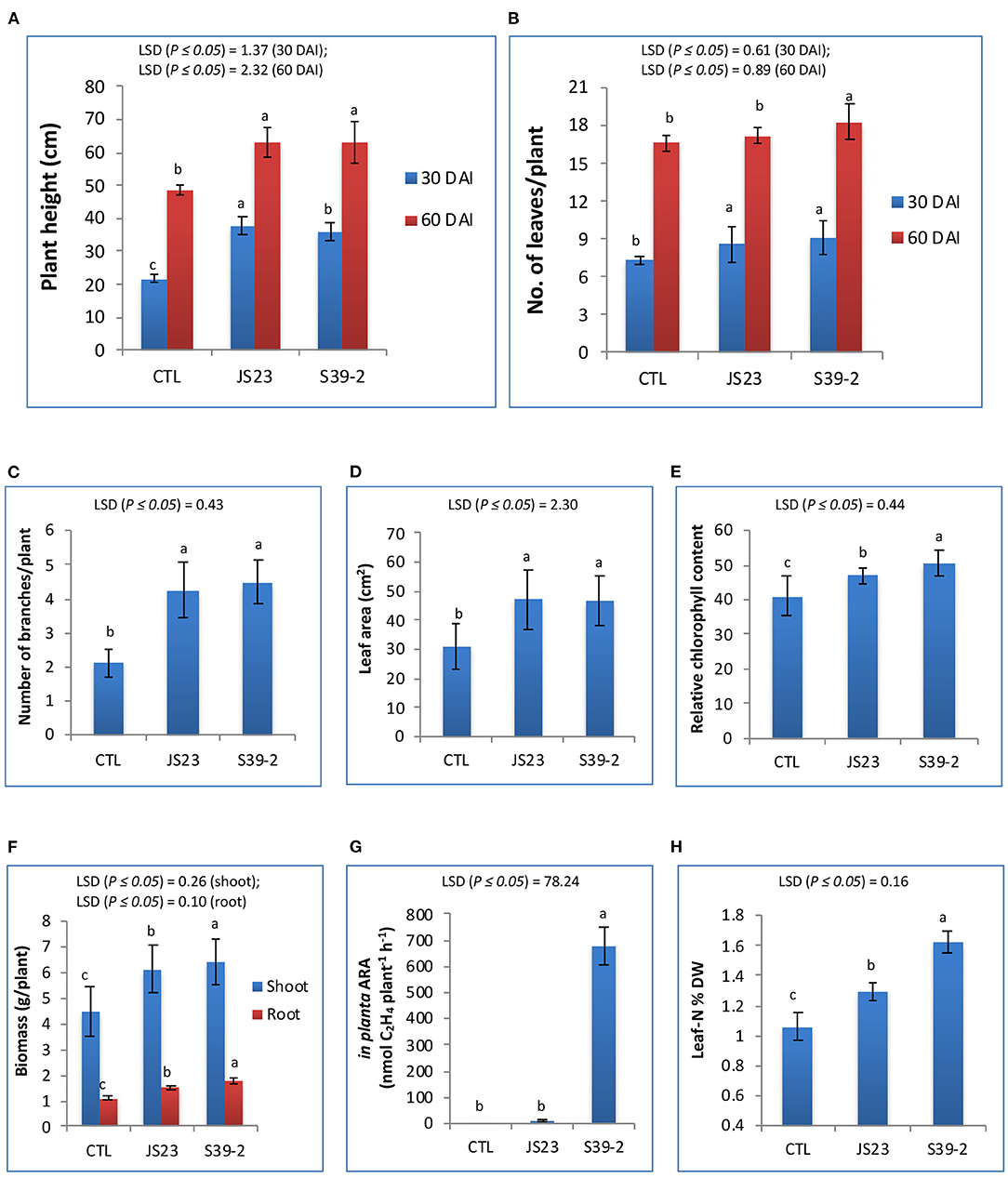
Figure 4. PGP effects of non-diazotrophic and diazotrophic isolates on eucalyptus seedlings. The effect on plant height (A), number of leaves (B), number of branches (C), leaf area (D), relative chlorophyll content (E), total biomass of leaves, stems and roots (F), in planta AR activity (G) and leaf N content (H) was measured at 30 and 60 DAI with strain JS23T in lemon eucalyptus grown in small pot containing 750 g of red soil. Mean values ± SD are presented (n = 18). The data were statistically analyzed using DMRT. Different letters indicate significant differences of the treatments according to least significant difference (LSD) at p ≤ 0.05 levels. DAI, days after inoculation; CTL, uninoculated control.
Similarly, both strains were able to promote the growth of oil palm seedlings (Table 3). The growth promotion results were statistically significant at 30, 60, and 90 DAI. Strain S39-2 treated seedlings had a higher bacterial population in roots (21.8 ± 2.7 × 105 cfu g−1 tissues) than in leaflets (6.3 ± 0.54 × 105 cfu g−1 tissues) while no bacteria were recovered at 30 DAI from strain JS23T treated seedlings (Table 3). Furthermore, we tested the PGP activity of strains JS23T and S39-2 on Jatropha curcas, a tree plant that holds promise for biofuel production. Strain JS23T showed very little PGP activity while S39-2 showed strong PGP activity (Table 4). Strain S39-2 was able to colonize the root, stem, and leaf tissues of Jatropha, yielding population densities of 63.8 ± 4.8 × 106, 9.8 ± 1.8 × 106, and 0.57 ± 0.02 × 106 cfu g−1 tissues, respectively, at 60 DAI, while JS23T cells were not recovered from the surface-sterilized root, stem, or leaf tissues (Table 4).
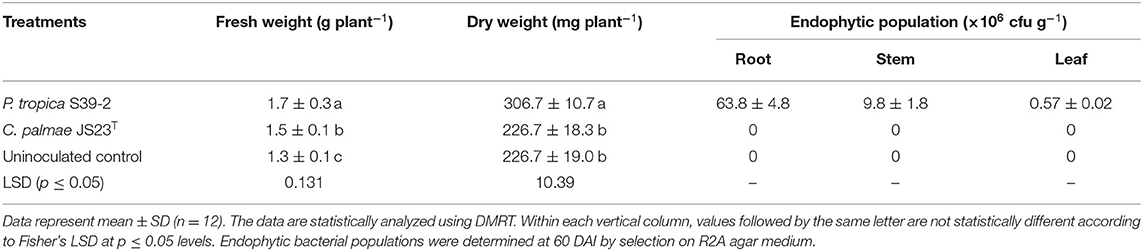
Table 4. Endophytic population of Burkholderia related strains in various tissues of Jatropha at 60 DAI under sterile N-limiting conditions.
Discussion
Endophytic Bacteria From Acacias and Oil Palm Phyllosphere and Their PGP Characteristics
Plant productivity is closely related to soil nutrient and climatic conditions. Since fertilizer application is not normally practiced in forestry, improvements in genotypes and the rhizospheric microbiome are critical for boosting tree productivity. The phyllosphere harbors diverse bacteria and the dominant bacterial taxa usually include diazotrophs and methylotrophs (methanol- and other one-carbon compound-consuming). This study focused on the diversity of Burkholderia-related genera within leaf tissues of acacias and oil palm. It is evident that members of the genus Paraburkholderia were dominant, followed by Caballeronia. Notably, Chitinasiproducens palmae is a new addition to the existing list of Burkholderia-related and tree plants associated bacteria (Madhaiyan et al., 2020). Members of N2-fixing Paraburkholderia species have often been isolated as rhizospheric and/or in root endophytic bacteria from various crops (Viallard et al., 1998; Compant et al., 2005; Theocharis et al., 2012). Our studies added a new insight into the diversity of Burkholderia-related bacteria in plant leaves, and our data suggested that they play an important role in tree crop health and growth.
Consistent with many reports on endophytes, the colonies formed were yellowish, round, smooth, and convex, 1–2 mm in diameter (Estrada-De Los Santos et al., 2001). The observed congruence between phylogenetic trees of 16S rRNA and nifH genes suggest that the common ancestor of Paraburkholderia was a diazotroph, and this function has been inherited in most species with a few exceptions (Suárez-Moreno et al., 2012). The ability to fix nitrogen is a major feature of plant-growth promoting Paraburkholderia species (Martínez-Aguilar et al., 2008). Although Paraburkholderia species were isolated from enriched N-free media, some strains were negative for the AR assay since the ability to grow on N-free medium or the presence of the nifH gene does not necessarily warrant nitrogenase activity (Madhaiyan et al., 2015). It is likely that these bacteria are not true diazotrophs despite their ability to grow in a culture medium without added nitrogen (Castanheira et al., 2016). The strains might have lost the ability during the purification procedure since it has been speculated that dinitrogen fixers grow best in the presence of other heterotrophic bacteria which support nitrogen fixers by physical or biochemical activities (Wiegel and Schlegel, 1976). Paraburkholderia isolates from oil palm belonging to P. tropica had higher ARA (Table 1), but none of the isolates from acacias reduced acetylene. It is surprising that the N-fixing activity was not significantly inhibited with 10% oxygen supplementation in many of the leaf isolates, suggesting the possibility of oxygen resistance mechanism in the leaf endophytes (Figure 3).
Phytohormones regulate many aspects of plant growth, development, and responses to stress. Previous studies have reported IAA production in different Paraburkholderia species, such as P. kururiensis, P. phytofirmans, and P. unamae (Caballero-Mellado et al., 2007; Sun et al., 2009). Our data with P. tropica are consistent with these reports as the majority of strains strongly produce auxins. ACC deaminase activity was found in all three genera of strains reported here although some strains appeared to have lost the gene (Table 1), suggesting their common role in inhibiting ethylene-mediated leaf senescence. This is consistent with previous reports on Burkholderia and Paraburkholderia that were of non-leaf origin (Suárez-Moreno et al., 2012). The member of the genus Paraburkholderia usually has a strong capacity to solubilize inorganic phosphates. On the other hand, this is rarely found in Caballeronia and Chitinasiproducens (Table 1). Siderophore production was also rarely encountered in the leaf isolates, suggesting that it play a minor role in the phyllosphere.
Plant Colonization and Growth Promotion by Strains S39-2 and JS23
The strains selected for plant inoculation studies, S39-2 and JS23T, significantly promoted the growth of seedlings of oil palm and eucalyptus despite the fact that JS23T had only weak auxin production, moderate ACC deaminase activity, and total lack of nitrogenase activity (Table 1). Further, JS23T failed to persistently colonize the internal tissues of lemon eucalyptus, palm, and Jatropha curcas. This suggests that JS23T promoted plant growth mainly with the ACC deaminase activity in the rhizosphere. It remains possible that these bacteria promoted plant growth via other mechanisms in the oil palm rhizosphere, where its inoculation led to a significant increase in leaf number and plant height throughout the 90-day observation period (Table 3). Unlike S39-2, JS23T displayed only weak PGP activity in Jatropha curcas. This suggests that there was a close interaction between the plants and the bacteria with regard to its colonization and growth promotion.
Although the mechanisms for these plant-bacteria interactions await further investigations, our GFP-tagged strains provided some interesting hints. JS23-gfp and S39-2-gfp differed significantly in their pattern of colonization in lemon eucalyptus (Figure 3). The most obvious difference was the long-term colonization pattern: while S39-2-gfp was able to sustain long-term colonization as an endophyte, JS23-gfp appeared to trigger some defense response that led to its gradual removal from the host tissues. Coincidently, this strain was also able to inhibit the growth of red yeast. In addition, JS23-gfp appeared to trigger a similar defense response in oil palm and Jatropha curcas as no bacteria was observed in the leaves of both plants.
Conclusion
Paraburkholderia tropica S39-2 isolated as a leaf endophyte from oil palm possesses multiple plant growth promotion traits and is able to colonize and promote the growth of oil palm and Jatropha. It may be developed as an efficient bioinoculant for tree species to reduce fertilizer usage; improve crop productivity, and reduce greenhouse gas emission. The strains S39-2 and JS23T can also serve as valuable tools for further studies of bacteria–host interactions of the Burkholderia related taxa, particularly with regard to understanding endophyte-plant interactions and their utilization for enhancing the sustainability of trees and conventional crop production.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, given in the article.
Author Contributions
MM and TA performed all the experiments, isolation, characterization, and bioassays for bacteria and plants performed data analysis and prepared the manuscript. GS edited the manuscript. LC participated in plant inoculation experiments and prepared plant materials. LJ designed the research plans and supervised the whole study and revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the Temasek Foundation and the Singapore Economy Development Board (EDB).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We also thank Wang Jing Fang, and Microscopy and Imaging Facility, TLL, Singapore, for their assistance with confocal laser scanning microscopy.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fsufs.2021.618305/full#supplementary-material
Supplementary Figure 1. pUT-tac-aph-sGFP plasmid (8776 bp). aph: kanamycin resistance gene; bla: ampicilin resistance gene; sfGFP: super-folder GFP.
Supplementary Figure 2. Phylogenetic tree based on nifH gene sequences showing the position of isolates from tree crops. GenBank accession numbers are given in parentheses. Bootstrap values (expressed as percentages of 1000 replications) greater than 50% are shown at the branch points. Bar, 0.05 substitutions per nucleotide position. The gold box represents nifH positive strains, eight strains closely related to P. tropica, one strain each from P. unamae and B. vietnamiensis. The blue box represents symbiotic Paraburkholderia strains.
Supplementary Figure 3. (A) Phosphate solubilizing endophytic leaf isolates on NBRIP agar medium at 7 days. (B) CAS agar plate: siderophore production comparison among the endophytic leaf isolates on a CAS siderophore testing agar. The chelator–iron (III) complex tints the agar with a rich blue background. The orange halo surrounding the colony indicates the excretion of siderophore, and its dimension approximates the amount of siderophore excreted. Strains: P. tropica strains S43-2, S42-2, S45-1, S44-1, S23-8, S22-7, S23-9, S23-4, S39-2, S38-1, S40-2, and S41-2, C. palmae strain JS23T, P. unamae strain S51-2, P. sediminicola strain Aca-21, P. aromaticivorans strain Aca-213, B. anthina strains S25-8 and S21-2, B. vietnamiensis strain S53-1, and C. mineralivorans strains Aca-28, Aca-214 and Aca-215.
References
Aizawa, T., Ve, N. B., Nakajima, M., and Sunairi, M. (2010). Burkholderia heleia sp. nov., a nitrogen-fixing bacterium isolated from an aquatic plant, Eleocharis dulcis, that grows in highly acidic swamps in actual acid sulfate soil areas of Vietnam. Int. J. Syst. Evol. Microbiol. 60, 1152–1157. doi: 10.1099/ijs.0.015198-0
Bais, H. P., Weir, T. L., Perry, L. G., Gilroy, S., and Vivanco, J. M. (2006). The role of root exudates in rhizosphere interactions with plants and other organisms. Annu. Rev. Plant Biol. 57, 233–266. doi: 10.1146/annurev.arplant.57.032905.105159
Barka, E. A., Belarbi, A., Hachet, C., Nowak, J., and Audran, J.-C. (2000). Enhancement of in vitro growth and resistance to gray mould of Vitis vinifera co-cultured with plant growth-promoting rhizobacteria. FEMS Microbiol. Lett. 186, 91–95. doi: 10.1111/j.1574-6968.2000.tb09087.x
Blaha, D., Prigent-Combaret, C., Mirza, M. S., and Moënne-Loccoz, Y. (2006). Phylogeny of the 1-aminocyclopropane-1-carboxylic acid deaminase-encoding gene acdS in phytobeneficial and pathogenic Proteobacteria and relation with strain biogeography. FEMS Microbiol. Ecol. 56, 455–470. doi: 10.1111/j.1574-6941.2006.00082.x
Bringel, F., and Couée, I. (2015). Pivotal roles of phyllosphere microorganisms at the interface between plant functioning and atmospheric trace gas dynamics. Front. Microbiol. 6:486. doi: 10.3389/fmicb.2015.00486
Caballero-Mellado, J., Martínez-Aguilar, L., Paredes-Valdez, G., and Estrada-De Los Santos, P. (2004). Burkholderia unamae sp. nov., an N2-fixing rhizospheric and endophytic species. Int. J. Syst. Evol. Microbiol. 54, 1165–1172. doi: 10.1099/ijs.0.02951-0
Caballero-Mellado, J., Onofre-Lemus, J., Estrada-De Los Santos, P., and Martínez-Aguilar, L. (2007). The tomato rhizosphere, an environment rich in nitrogen-fixing Burkholderia species with capabilities of interest for agriculture and bioremediation. Appl. Environ. Microbiol. 73, 5308–5319. doi: 10.1128/AEM.00324-07
Castanheira, N., Dourado, A. C., Kruz, S., Alves, P. I. L., Delgado-Rodríguez, A. I., Pais, I., et al. (2016). Plant growth-promoting Burkholderia species isolated from annual ryegrass in Portuguese soils. J. Appl. Microbiol. 120, 724–739. doi: 10.1111/jam.13025
Cheng, A. C., and Currie, B. J. (2005). Melioidosis: epidemiology, pathophysiology, and management. Clin. Microbiol. Rev. 18, 383–416. doi: 10.1128/CMR.18.2.383-416.2005
Coenye, T., and Vandamme, P. (2003). Diversity and significance of Burkholderia species occupying diverse ecological niches. Environ. Microbiol. 5, 719–729. doi: 10.1046/j.1462-2920.2003.00471.x
Compant, S., Reiter, B., Sessitsch, A., Nowak, J., Clément, C., and Barka, E. A. (2005). Endophytic colonization of Vitis vinifera L. by plant growth-promoting bacterium Burkholderia sp. strain PsJN. Appl. Environ. Microbiol. 71, 1685–1693. doi: 10.1128/AEM.71.4.1685-1693.2005
Conn, K. L., Lazarovits, G., and Nowak, J. (1997). A gnotobiotic bioassay for studying interactions between potatoes and plant growth-promoting rhizobacteria. Can. J. Microbiol. 43, 801–808. doi: 10.1139/m97-117
De Meyer, S. E., Cnockaert, M., Ardley, J. K., Maker, G., Yates, R., Howieson, J. G., et al. (2013). Burkholderia sprentiae sp. nov., isolated from Lebeckia ambigua root nodules. Int. J. Syst. Evol. Microbiol. 63, 3950–3957. doi: 10.1099/ijs.0.048777-0
De Meyer, S. E., Cnockaert, M., Ardley, J. K., Van Wyk, B.-E., Vandamme, P. A., and Howieson, J. G. (2014). Burkholderia dilworthii sp. nov., isolated from Lebeckia ambigua root nodules. Int. J. Syst. Evol. Microbiol. 64, 1090–1095. doi: 10.1099/ijs.0.058602-0
DeLong, E. F. (1992). Archaea in coastal marine environments. Proc. Natl. Acad. Sci. USA. 89, 5685–5689. doi: 10.1073/pnas.89.12.5685
Dixon, R., and Kahn, D. (2004). Genetic regulation of biological nitrogen fixation. Nat. Rev. Microbiol. 2, 621–631. doi: 10.1038/nrmicro954
Döbereiner, J., Baldani, V. L., and Reis, V. M. (1995). “Endophytic occurrence of diazotrophic bacteria in non-leguminous crops,” in Azospirillum VI and related Microorganisms. Berlin: Springer, 3–14.
Dobritsa, A. P., Kutumbaka, K. K., and Samadpour, M. (2016). Reclassification of Paraburkholderia panaciterrae (Farh et al. 2015). Dobritsa and Samadpour 2016 as a later synonym of Paraburkholderia ginsengiterrae (Farh et al. 2015). Dobritsa and Samadpour 2016. Int. J. Syst. Evol. Microbiol. 66, 4085–4087. doi: 10.1099/ijsem.0.001314
Dobritsa, A. P., and Samadpour, M. (2016). Transfer of eleven Burkholderia species to the genus Paraburkholderia and proposal of Caballeronia gen. nov., a new genus to accommodate twelve species of Burkholderia and Paraburkholderia. Int. J. Syst. Evol. Microbiol. 66, 2836–2846. doi: 10.1099/ijsem.0.001065
Dworkin, M., and Foster, J. W. (1958). Experiments with some microorganisms which utilize ethane and hydrogen. J. Bacteriol. 75:592.
Estrada-De Los Santos, P., Bustillos-Cristales, R. O., and Caballero-Mellado, J. (2001). Burkholderia, a genus rich in plant-associated nitrogen fixers with wide environmental and geographic distribution. Appl. Environ. Microbiol. 67, 2790–2798. doi: 10.1128/AEM.67.6.2790-2798.2001
Fan, X., Yang, R., Qiu, S., Cai, X., Zou, H., and Hu, F. (2016). The endo-β-1, 4-glucanase of Bacillus amyloliquefaciens is required for optimum endophytic colonization of plants. J. Microbiol. Biotechnol. 26, 946–952. doi: 10.4014/jmb.1512.12055
Frommel, M. I., Nowak, J., and Lazarovits, G. (1991). Growth enhancement and developmental modifications of in vitro grown potato (Solanum tuberosum spp. tuberosum) as affected by a nonfluorescent Pseudomonas sp. Plant Physiol. 96, 928–936. doi: 10.1104/pp.96.3.928
Gao, W., Liu, Y., Giometti, C. S., Tollaksen, S. L., Khare, T., Wu, L., et al. (2006). Knock-out of SO1377 gene, which encodes the member of a conserved hypothetical bacterial protein family COG2268, results in alteration of iron metabolism, increased spontaneous mutation and hydrogen peroxide sensitivity in Shewanella oneidensis MR-1. BMC Genomics 7:76. doi: 10.1186/1471-2164-7-76
Ghosh, R., Barman, S., Mukherjee, R., and Mandal, N. C. (2016). Role of phosphate solubilizing Burkholderia spp. for successful colonization and growth promotion of Lycopodium cernuum L. (Lycopodiaceae) in lateritic belt of Birbhum district of West Bengal, India. Microbiol. Res. 183, 80–91. doi: 10.1016/j.micres.2015.11.011
Gobert, A., and Plassard, C. (2008). “The beneficial effect of mycorrhizae on N utilization by the host-plant: Myth or reality?,” in Mycorrhiza. Berlin: Springer, 209–240. doi: 10.1007/978-3-540-78826-3_11
Hallmann, J., Quadt-Hallmann, A., Mahaffee, W., and Kloepper, J. (1997). Bacterial endophytes in agricultural crops. Can. J. Microbiol. 43, 895–914. doi: 10.1139/m97-131
Honma, M., and Shimomura, T. (1978). Metabolism of 1-aminocyclopropane-1-carboxylic acid. Agric. Biol. Chem. 42, 1825–1831.
Hu, J., and Ji, L. (2016). Draft genome sequences of Rhodosporidium toruloides strains ATCC 10788 and ATCC 10657 with compatible mating types. Genome Announc. 4, e00098–e00016. doi: 10.1128/genomeA.00098-16
Iniguez, A. L., Dong, Y., and Triplett, E. W. (2004). Nitrogen fixation in wheat provided by Klebsiella pneumoniae 342. Mol. Plant-Microbe Int. 17, 1078–1085. doi: 10.1094/MPMI.2004.17.10.1078
Kaur, C., Selvakumar, G., and Ganeshamurthy, A. N. (2016). Draft genome sequence of phosphate-solubilizing bacterium Paraburkholderia tropica strain P-31 isolated from pomegranate (Punica granatum) rhizosphere. Genome Announc. 4, e00844–e00816. doi: 10.1128/genomeA.00844-16
Kim, B., and Wimpenny, J. (1981). Growth and cellulolytic activity of Cellulomonas flavigena. Can. J. Microbiol. 27, 1260–1266. doi: 10.1139/m81-193
Kim, O.-S., Cho, Y.-J., Lee, K., Yoon, S.-H., Kim, M., Na, H., et al. (2012a). Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int. J. Syst. Evol. Microbiol. 62, 716–721. doi: 10.1099/ijs.0.038075-0
Kim, S., Lowman, S., Hou, G., Nowak, J., Flinn, B., and Mei, C. (2012b). Growth promotion and colonization of switchgrass (Panicum virgatum) cv. Alamo by bacterial endophyte Burkholderia phytofirmans strain PsJN. Biotechnol. Biofuels 5, 1–10. doi: 10.1186/1754-6834-5-37
Koh, L. P., and Wilcove, D. S. (2007). Cashing in palm oil for conservation. Nature 448, 993–994. doi: 10.1038/448993a
Kost, T., Stopnisek, N., Agnoli, K., Eberl, L., and Weisskopf, L. (2013). Oxalotrophy, a widespread trait of plant-associated Burkholderia species, is involved in successful root colonization of lupin and maize by Burkholderia phytofirmans. Front. Microbiol. 4:421. doi: 10.3389/fmicb.2013.00421
Lindow, S. E., and Brandl, M. T. (2003). Microbiology of the phyllosphere. Appl. Environ. Microbiol. 69, 1875–1883. doi: 10.1128/AEM.69.4.1875-1883.2003
Madhaiyan, M., Alex, T. H. H., Te Ngoh, S., Prithiviraj, B., and Ji, L. (2015). Leaf-residing Methylobacterium species fix nitrogen and promote biomass and seed production in Jatropha curcas. Biotechnol. Biofuels 8, 1–14. doi: 10.1186/s13068-015-0404-y
Madhaiyan, M., Peng, N., and Ji, L. (2013a). Complete genome sequence of Enterobacter sp. strain R4-368, an endophytic N-fixing gammaproteobacterium isolated from surface-sterilized roots of Jatropha curcas L. Genome Announc. 1, e00544–e00513. doi: 10.1128/genomeA.00544-13
Madhaiyan, M., Peng, N., Te Ngoh, S., Hsin, C., Lin, C., Lin, F., et al. (2013b). Improvement of plant growth and seed yield in Jatropha curcas by a novel nitrogen-fixing root associated Enterobacter species. Biotechnol. Biofuels 6, 1–13. doi: 10.1186/1754-6834-6-140
Madhaiyan, M., See-Too, W. S., Ee, R., Saravanan, V. S., Wirth, J. S., Alex, T. H. H., et al. (2020). Chitinasiproducens palmae gen. nov., sp. nov., a new member of the family Burkholderiaceae isolated from leaf tissues of oil palm (Elaeis guineensis Jacq.). Int. J. Syst. Evol. Microbiol. 70, 2640–2647. doi: 10.1099/ijsem.0.004084
Martínez-Aguilar, L., Díaz, R., Peña-Cabriales, J. J., Estrada-De Los Santos, P., Dunn, M. F., and Caballero-Mellado, J. (2008). Multichromosomal genome structure and confirmation of diazotrophy in novel plant-associated Burkholderia species. Appl. Environ. Microbiol. 74, 4574–4579. doi: 10.1128/AEM.00201-08
Martínez-Aguilar, L., Salazar-Salazar, C., Méndez, R. D., Caballero-Mellado, J., Hirsch, A. M., Vásquez-Murrieta, M. S., et al. (2013). Burkholderia caballeronis sp. nov., a nitrogen fixing species isolated from tomato (Lycopersicon esculentum) with the ability to effectively nodulate Phaseolus vulgaris. Anton Leeuw. Int. J. G. 104, 1063–1071. doi: 10.1007/s10482-013-0028-9
Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plantarum 15, 473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x
Nautiyal, C. S. (1999). An efficient microbiological growth medium for screening phosphate solubilizing microorganisms. FEMS Microbiol. Lett. 170, 265–270. doi: 10.1111/j.1574-6968.1999.tb13383.x
Nierman, W. C., DeShazer, D., Kim, H. S., Tettelin, H., Nelson, K. E., Feldblyum, T., et al. (2004). Structural flexibility in the Burkholderia mallei genome. Proc. Natl. Acad. Sci. USA. 101, 14246–14251. doi: 10.1073/pnas.0403306101
Nunn, D. N., and Lidstrom, M. E. (1986). Phenotypic characterization of 10 methanol oxidation mutant classes in Methylobacterium sp. strain AM1. J. Bacteriol. 166, 591–597. doi: 10.1128/jb.166.2.591-597.1986
Pillay, V., and Nowak, J. (1997). Inoculum density, temperature, and genotype effects on in vitro growth promotion and epiphytic and endophytic colonization of tomato (Lycopersicon esculentum L.) seedlings inoculated with a pseudomonad bacterium. Can. J. Microbiol. 43, 354–361. doi: 10.1139/m97-049
Pinto-Tomás, A. A., Anderson, M. A., Suen, G., Stevenson, D. M., Chu, F. S., Cleland, W. W., et al. (2009). Symbiotic nitrogen fixation in the fungus gardens of leaf-cutter ants. Science 326, 1120–1123. doi: 10.1126/science.1173036
Reinhold-Hurek, B., Hurek, T., Claeyssens, M., and Van Montagu, M. (1993). Cloning, expression in Escherichia coli, and characterization of cellulolytic enzymes of Azoarcus sp., a root-invading diazotroph. J. Bacteriol. 175, 7056–7065. doi: 10.1128/jb.175.21.7056-7065.1993
Reinhold-Hurek, B., Maes, T., Gemmer, S., Van Montagu, M., and Hurek, T. (2006). An endoglucanase is involved in infection of rice roots by the not-cellulose-metabolizing endophyte Azoarcus sp. strain BH72. Mol. Plant-Microbe Interact. 19, 181–188. doi: 10.1094/MPMI-19-0181
Reis, V., Estrada-De Los Santos, P., Tenorio-Salgado, S., Vogel, J., Stoffels, M., Guyon, S., et al. (2004). Burkholderia tropica sp. nov., a novel nitrogen-fixing, plant-associated bacterium. Int. J. Syst. Evol. Microbiol. 54, 2155–2162. doi: 10.1099/ijs.0.02879-0
Reiter, B., and Sessitsch, A. (2006). Bacterial endophytes of the wildflower Crocus albiflorus analyzed by characterization of isolates and by a cultivation-independent approach. Can. J. Microbiol. 52, 140–149. doi: 10.1139/w05-109
Sawana, A., Adeolu, M., and Gupta, R. S. (2014). Molecular signatures and phylogenomic analysis of the genus Burkholderia: proposal for division of this genus into the emended genus Burkholderia containing pathogenic organisms and a new genus Paraburkholderia gen. nov. harboring environmental species. Front. Genet. 5:429. doi: 10.3389/fgene.2014.00429
Schwyn, B., and Neilands, J. (1987). Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 160, 47–56. doi: 10.1016/0003-2697(87)90612-9
Shah, S., Li, J., Moffatt, B. A., and Glick, B. R. (1998). Isolation and characterization of ACC deaminase genes from two different plant growth-promoting rhizobacteria. Can. J. Microbiol. 44, 833–843. doi: 10.1139/w98-074
Sheu, S.-Y., Chen, M.-H., Liu, W. Y., Andrews, M., James, E. K., Ardley, J. K., et al. (2015). Burkholderia dipogonis sp. nov., isolated from root nodules of Dipogon lignosus in New Zealand and Western Australia. Int. J. Syst. Evol. Microbiol. 65, 4716–4723. doi: 10.1099/ijsem.0.000639
Sheu, S.-Y., Chou, J.-H., Bontemps, C., Elliott, G. N., Gross, E., Dos Reis Junior, F. B., et al. (2013). Burkholderia diazotrophica sp. nov., isolated from root nodules of Mimosa spp. Int. J. Syst. Evol. Microbiol. 63, 435–441. doi: 10.1099/ijs.0.039859-0
Smethurst, P., Holz, G., Moroni, M., and Baillie, C. (2004). Nitrogen management in Eucalyptus nitens plantations. For. Ecol. Manag. 193, 63–80. doi: 10.1016/j.foreco.2004.01.023
Steenkamp, E. T., Van Zyl, E., Beukes, C. W., Avontuur, J. R., Chan, W. Y., Palmer, M., et al. (2015). Burkholderia kirstenboschensis sp. nov. nodulates papilionoid legumes indigenous to South Africa. Syst. Appl. Microbiol. 38, 545–554. doi: 10.1016/j.syapm.2015.09.003
Suárez-Moreno, Z. R., Caballero-Mellado, J., Coutinho, B. G., Mendonça-Previato, L., James, E. K., and Venturi, V. (2012). Common features of environmental and potentially beneficial plant-associated Burkholderia. Microb. Ecol. 63, 249–266. doi: 10.1007/s00248-011-9929-1
Sun, Y., Cheng, Z., and Glick, B. R. (2009). The presence of a 1-aminocyclopropane-1-carboxylate (ACC) deaminase deletion mutation alters the physiology of the endophytic plant growth-promoting bacterium Burkholderia phytofirmans PsJN. FEMS Microbiol. Lett. 296, 131–136. doi: 10.1111/j.1574-6968.2009.01625.x
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Theocharis, A., Bordiec, S., Fernandez, O., Paquis, S., Dhondt-Cordelier, S., Baillieul, F., et al. (2012). Burkholderia phytofirmans PsJN primes Vitis vinifera L. and confers a better tolerance to low nonfreezing temperatures. Mol Plant-Microbe Int. 25, 241–249. doi: 10.1094/MPMI-05-11-0124
Viallard, V., Poirier, I., Cournoyer, B., Haurat, J., Wiebkin, S., Ophel-Keller, K., et al. (1998). Burkholderia graminis sp. nov., a rhizospheric Burkholderia species, and reassessment of [Pseudomonas] phenazinium,[Pseudomonas] pyrrocinia and [Pseudomonas] glathei as Burkholderia. Int. J. Syst. Evol. Microbiol. 48, 549–563. doi: 10.1099/00207713-48-2-549
Vincent, J. (1970). The cultivation, isolation and maintenance of rhizobia. A manual for the practical study of the root-nodule bacteria. Hoboken: Blackwell Scientific Publications, 1–13.
Walker, T. S., Bais, H. P., Grotewold, E., and Vivanco, J. M. (2003). Root exudation and rhizosphere biology. Plant Physiol. 132, 44–51. doi: 10.1104/pp.102.019661
Wiegel, J., and Schlegel, H. (1976). Enrichment and isolation of nitrogen fixing hydrogen bacteria. Arch. Microbiol. 107, 139–142. doi: 10.1007/BF00446833
Keywords: Paraburkholderia tropica, Chitinasiproducens palmae, N-fixing leaf endophytes, oil palm, acacia
Citation: Madhaiyan M, Selvakumar G, Alex TH, Cai L and Ji L (2021) Plant Growth Promoting Abilities of Novel Burkholderia-Related Genera and Their Interactions With Some Economically Important Tree Species. Front. Sustain. Food Syst. 5:618305. doi: 10.3389/fsufs.2021.618305
Received: 16 October 2020; Accepted: 03 August 2021;
Published: 07 September 2021.
Edited by:
Everlon Cid Rigobelo, São Paulo State University, BrazilReviewed by:
Asma Imran, National Institute for Biotechnology and Genetic Engineering, PakistanMaura Santos Reis De Andrade Da Silva, Universidade Estadual de São Paulo, Brazil
Copyright © 2021 Madhaiyan, Selvakumar, Alex, Cai and Ji. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lianghui Ji, jilh@tll.org.sg; Munusamy Madhaiyan, mmadhaiyan@hotmail.com
 Munusamy Madhaiyan
Munusamy Madhaiyan Govindan Selvakumar
Govindan Selvakumar Tan HianHwee Alex1
Tan HianHwee Alex1  Lianghui Ji
Lianghui Ji