- 1Department of Information Technology, Faculty of Computing and Information Technology, King Abdulaziz University, Jeddah, Saudi Arabia
- 2Department of Information Systems, Faculty of Computing and Information Technology, King Abdulaziz University, Jeddah, Saudi Arabia
- 3Department of Computer Science and Engineering, SNS College of Engineering, Coimbatore, India
- 4Research and Development, Information Communication Technology Academy, Chennai, India
- 5Department of Computer Science and Engineering, Institute of Chartered Financial Analysts of India Foundation of Higher Education, Hyderabad, India
- 6Higher Colleges of Technology, Women's College, Abu Dhabi, United Arab Emirates
In this paper, a data mining model on a hybrid deep learning framework is designed to diagnose the medical conditions of patients infected with the coronavirus disease 2019 (COVID-19) virus. The hybrid deep learning model is designed as a combination of convolutional neural network (CNN) and recurrent neural network (RNN) and named as DeepSense method. It is designed as a series of layers to extract and classify the related features of COVID-19 infections from the lungs. The computerized tomography image is used as an input data, and hence, the classifier is designed to ease the process of classification on learning the multidimensional input data using the Expert Hidden layers. The validation of the model is conducted against the medical image datasets to predict the infections using deep learning classifiers. The results show that the DeepSense classifier offers accuracy in an improved manner than the conventional deep and machine learning classifiers. The proposed method is validated against three different datasets, where the training data are compared with 70%, 80%, and 90% training data. It specifically provides the quality of the diagnostic method adopted for the prediction of COVID-19 infections in a patient.
Introduction
The novel coronavirus disease 2019 (COVID-19) is a pandemic outbreak (1). COVID-19 patients are classified essentially based on computerized tomography (CT) lung images, and it is used widely for testing. The healthcare institutions fitted with CT scans help in the process of image acquisition and classification of CT images at a faster rate. However, the need for an expert medical practitioner is hence required for the verification of the final results, which increases the time of computation (2). On the other hand, the supervised learning models (3–10) can be utilized for classifying the patients from the CT images.
Infections based on CT images are not classified using very little unattended methods (11–24). We have developed a model that mainly includes supervised and unsupervised learning models in order to improve the classification process. The aim is to classify the infected patients automatically based on their CT images.
In this paper, a DeepSense algorithm is utilized to diagnose COVID-19 infections among the medical community. The deep learning method is designed as a combination of convolutional neural network (CNN) and recurrent neural network (RNN) that reduces the classifier burden on optimal classification of the multidimensional data features.
The main contribution of the work includes the following:
(a) The authors develop a combined CNN and RNN to classify the medical image datasets.
(b) The experimental results are conducted to measure the correctness in terms of its accuracy, precision, and recall values against artificial neural network (ANN), feedforward neural network (FFNN), back propagation neural network (BPNN), deep neural network (DNN), and RNN.
The outline of the paper is presented as follows: The Methods section provides the details of the ensemble classifier. The DeepSense Model section evaluates the entire work. The Results and Discussions section concludes the work with future enhancement.
Methods
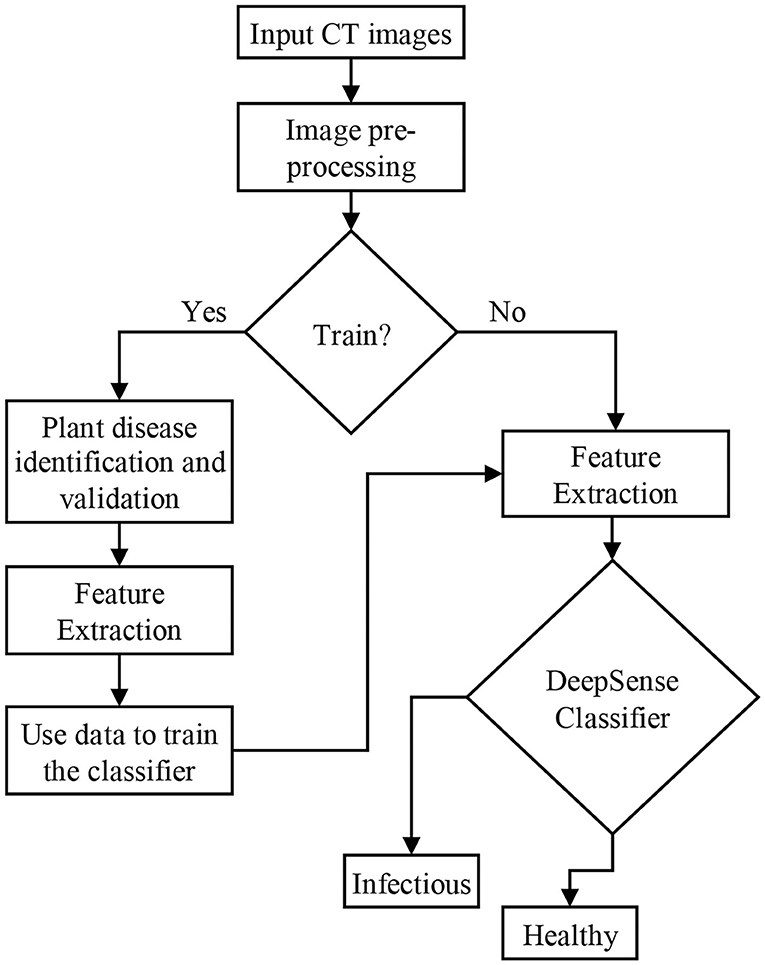
The deep learning model namely DeepSense algorithm is a combination of CNN and RNN designed to improve the performance of the classification accuracy. DeepSense learning is regarded as a module for accurate predictions of lung infections caused by the COVID-19 virus. Figure 1 shows the architecture of the proposed classification model using the DeepSense algorithm.
Deepsense Model
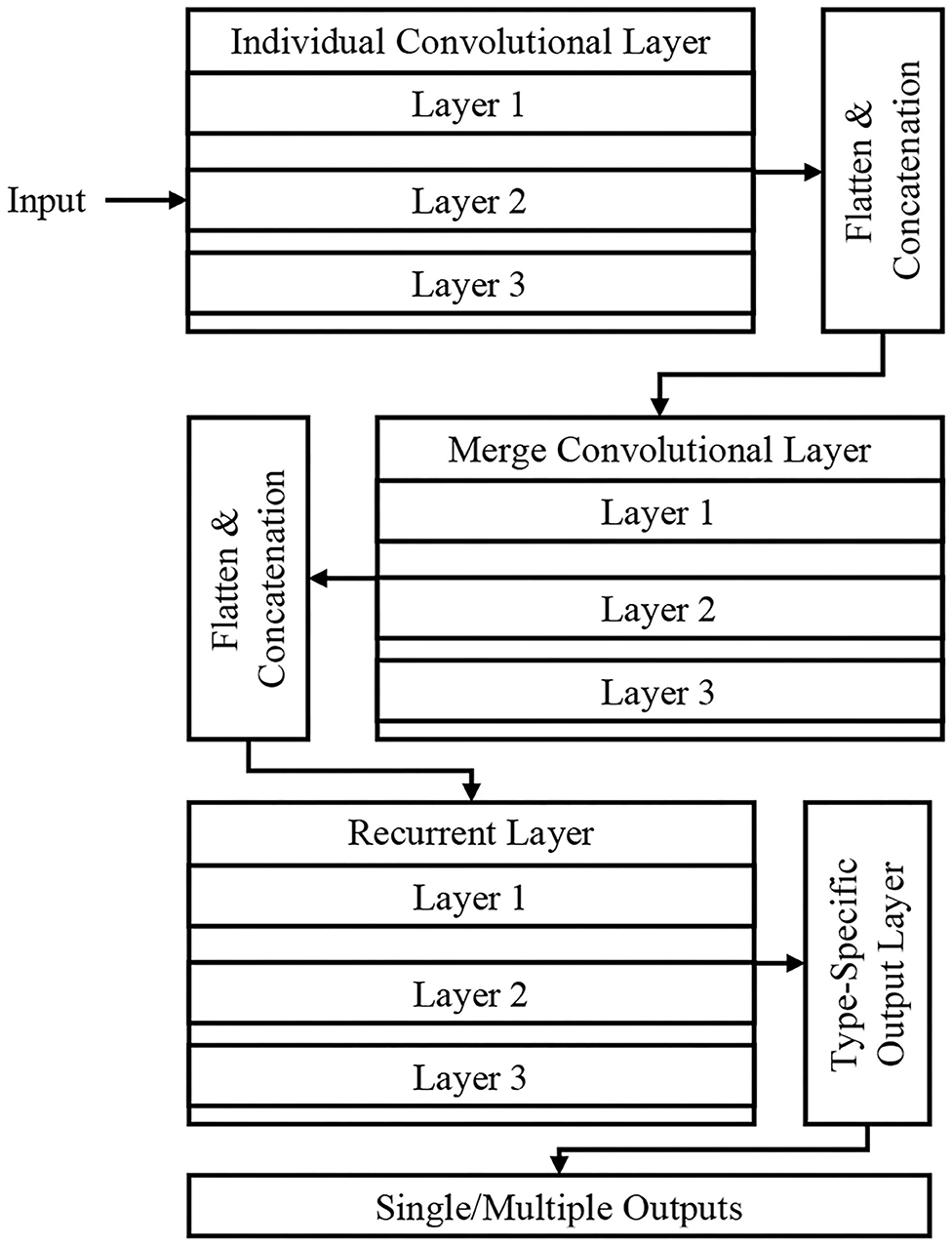
Figure 2 shows the DeepSense DNN (25) model that has three components, including convolutional, recurrent, and output layer that are stacked upon one another. The convolutional and recurrent layers are regarded as the significant building blocks (Figure 1), and the output layer is considered as a specific layer that classifies the images. The DeepSense DNN model is designed for the classification of input CT images for COVID-19-related infections.
The DeepSense network avoids gradient exploding and improves the rate of convergence using residual learning, adjustable learning rate, and gradient clipping that helps in optimizing the process of training.
The features are extracted through the DeepSense model that increases the reconstruction accuracy and reduces the time of training. Such optimization helps in obtaining the rich text information, and it has better ability for classification.
Convolutional Layers
The convolutional layers have three different parts that include individual convolutional subnets for input from CT device X(k), where k is the number of CT device. The other subnets include a merged convolutional subnet for K convolutional subnets' outputs.
For a time interval t, the matrix X(k) is used as an input to the DNN architecture that extracts the relationship of X(k,t), which includes the relationships lying inside the frequency domain. The sensor measurement interactions include entire dimension, where the frequency domain usually has several local patterns. These interactions are studied using 2D filters and produces the output X(k,1,t) based on the local patterns and dimensions in frequency domain. The high-level relationships are learned hierarchically using the application of a 1D filter. The matrix is then flattened into a vector, and they are concatenated to produce the input for RNN layers. The activation function in the convolutional layer is a rectified linear unit (ReLU) function, and batch normalization eliminates the internal covariate shift.
Recurrent Layers
The RNN architecture learns the needed features having long-term dependencies (long paths). The study uses Gated Recurrent Unit (GRU) on long and short path selection to reduce well the network complexity. A set of three layers stacked in GRU is used in this paper that uses time flow that runs the stacked GRU incrementally for faster input data processing. The recurrent layer outputs vector series {x(r,t)} where t = 1,2,···, T for the process of classification at the output layer.
Output Layer
For the purpose of classification, {x(r,t)} is selected as the feature vector, and this layer converts the vector of variable length into fixed length. The final feature is generated by averaging the features over a specific time interval based on long or short paths, x(r) . Finally the probability of predicted category is generated by feeding the averaging features into the softmax layer.
Type-Specific Layer
For the customization of the DeepSense layer to operate the process of classification, we specifically use the following process:
Step 1: Identify the input image
Step 2: Preprocessing input image for temporal and spectral noise
Step 3: Extract the features related to COVID-19 infections
Step 4: Apply DeepSense classifier for optimal classifier.
Results and Discussions
This section provides the results of comparison between the machine/deep learning classifiers for predicting COVID-19 infections using IEEE8023 (26), COVID-CT-Dataset (27), and COVID-19 Open Research Dataset Challenge (CORD-19) (28) datasets.
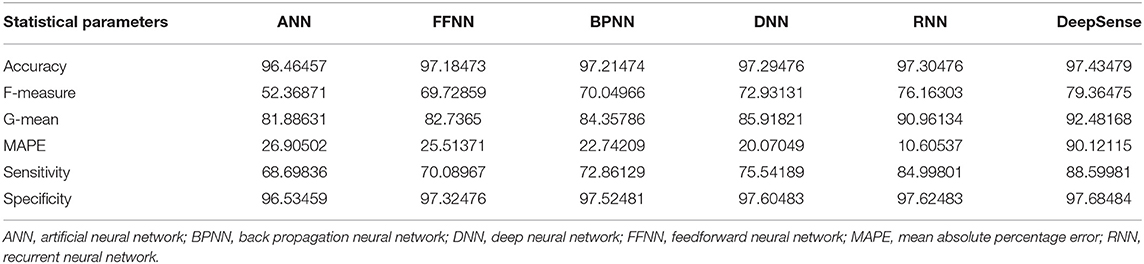
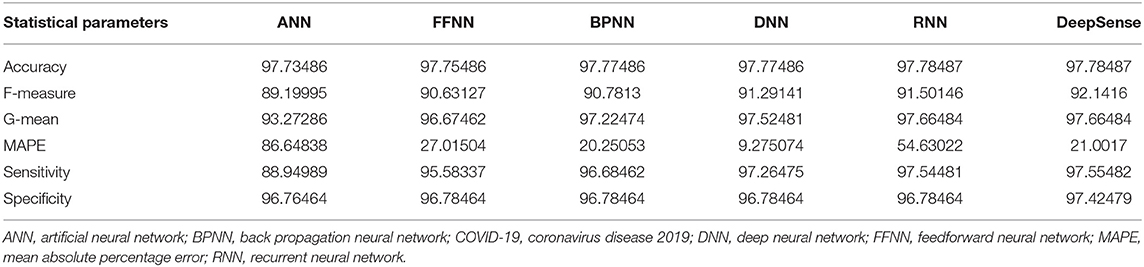
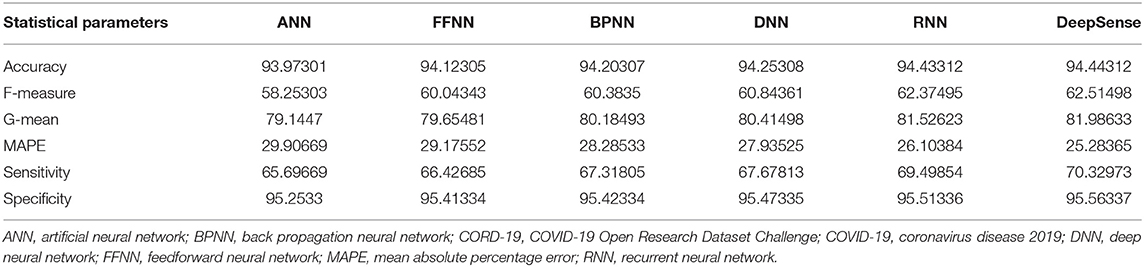
IEEE8023 has the image collection from various sources including COVID-19 or viral and bacterial pneumonias in the form of CT images. COVID-CT has 349 COVID-19 CT images from 216 patients and 463 non-COVID-19 CTs. CORD-19 has collected the CT image resources from 52,000 scholarly articles.
The study is experimented using a 10-fold cross validation, which is tested with all these three base classifiers.
Experiment
The performance measures for evaluating the DeepSense classifier is estimated against various metrics: accuracy, geometric mean (G-mean), F-measure, precision, percentage error, specificity, and sensitivity.
Accuracy for optimal classification is given below:
where:
TP is defined as the true positive
TN is defined as the true negative
FP is defined as the false positive
FN is defined as the false negative
F-measure of the DeepSense classifier is defined as follows:
G-mean of the DeepSense classifier is defined as follows:
Mean absolute percentage error (MAPE) of the DeepSense classifier is defined as follows:
Where
At is defined as the actual class
Ft is defined as the predicted class, and
n is defined as the fitted points
Sensitivity of the DeepSense is defined as:
Specificity of the DeepSense is defined as:
Analysis
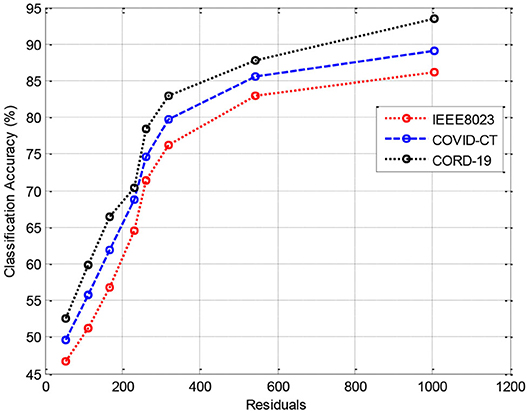
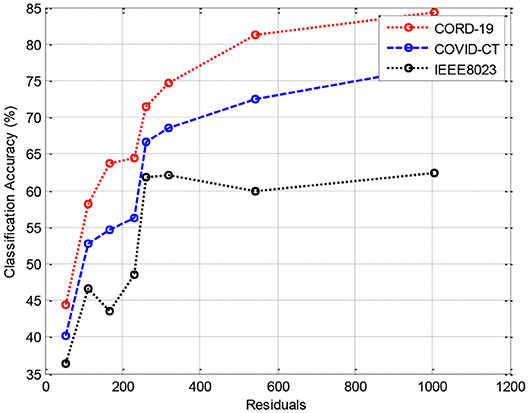
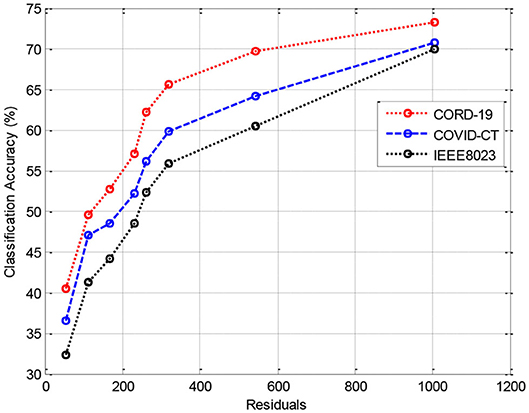
In this section, we provide the results of various meta-ensemble classifiers that include FFNN (29), ANN (25), DNN (30), BPNN (31), and RNN (32). The proposed method is validated against three different datasets, where the training data are compared with 70% (Figure 3), 80% (Figure 4), and 90% (Figure 5) training data.
Figure 3 shows the results of classification accuracy of CORD-19 datasets for all residuals are higher, and with increasing residuals, the accuracy increases. Same is the case for other training sets; however, with 80% datasets, the accuracy is fluctuating due to the extraction of on-optimal features from IEEE8023 datasets.
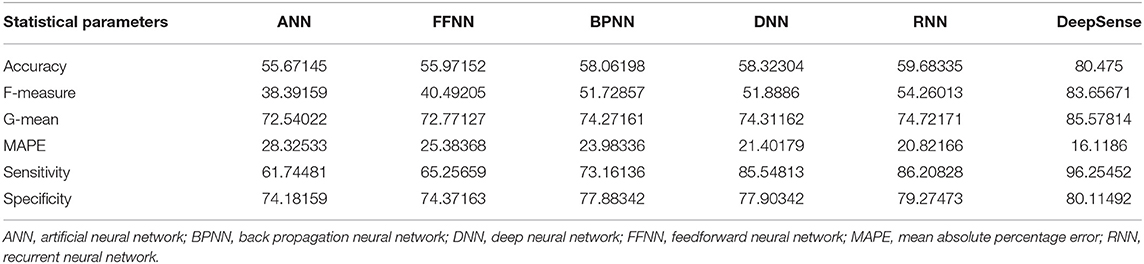
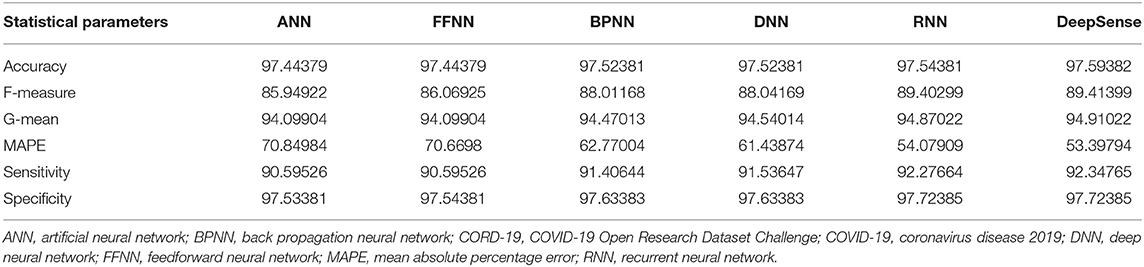
Tables 1, 4, 7 provide the results of statistical parameters on predicting COVID-19 infections over 70, 80, and 90% training data over IEEE8023 datasets.
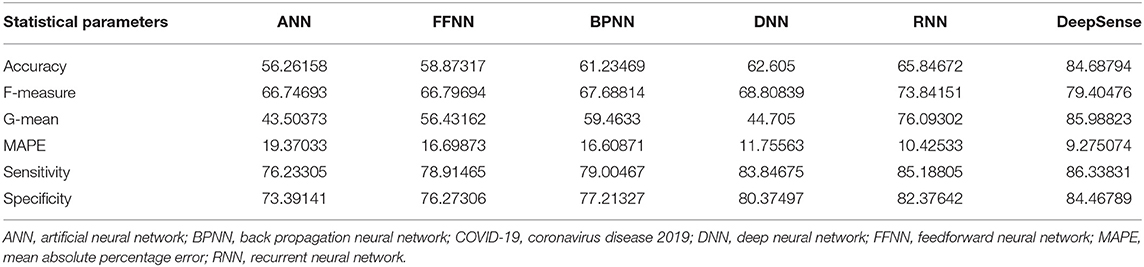
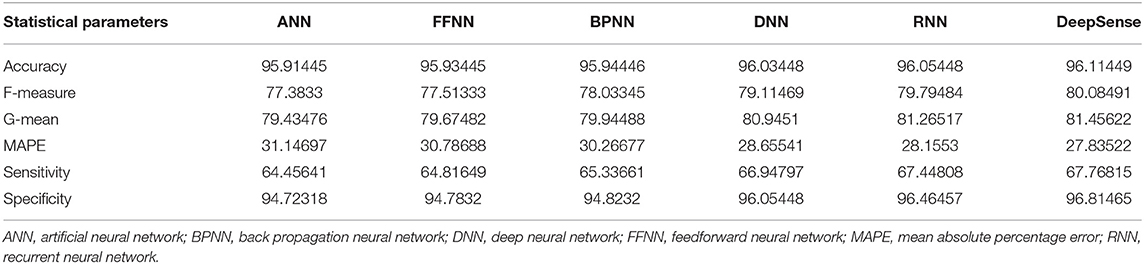
Tables 2, 5, 8 provide the results of statistical parameters on predicting COVID-19 infections over 70, 80, and 90% training data over COVID-CT datasets.
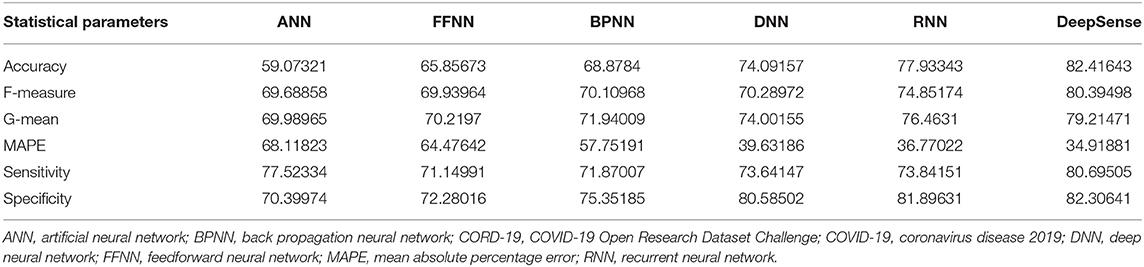
Tables 3, 6, 9 provide the results of statistical parameters on predicting COVID-19 infections over 70, 80, and 90% training data over CORD-19 datasets.
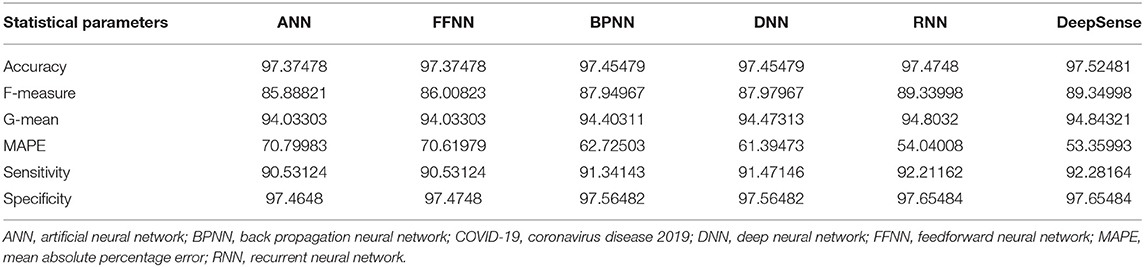
Table 8. Results of statistical parameters for COVID-CT with 90% training data on 1,000 images classifier.
Evaluation Criteria
The simulation results show that the DeepSense classifier has higher classification accuracy than the existing meta-ensemble classifiers. In addition, the CORD-19 datasets offer optimum selection of features to increase classification accuracy by 90% training data over 80 or 90%. The other measurements are optimal for CORD-19 than the other selection tools. Furthermore, MAPE is less than the other methods in the deep learning model.
The result shows that the CORD-19 datasets are more accurate than RNN and DNN. The results also show that the classification accuracy with IEEE8023 as a functional selection tool decreases at some point as the number of residues increases compared to COVID-CT and CORD-19. The class of infections is therefore accurately determined with the proposed classification.
Conclusions and Future Work
In this paper, a DeepSense algorithm is designed for the classification of COVID-19 infections. The DeepSense algorithm helps in optimal classification of multidimensional features from CT images. The classifier combined with hybrid deep learning classifier, namely, CNN and RNN, helps in improving the prediction of events from a medical image. The extraction of optimal features from the feature extraction model helps the classifier to optimally detect whether the patient is infected or not. The experimental results show that the proposed method has higher accuracy than the other methods. In the future, the model can be designed with an ensemble data model to classify the highly rated multidimensional dataset.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary materials, further inquiries can be directed to the corresponding author/s.
Author Contributions
AK: visualization and investigation. AOK: data curation, software, and validation. SK: methodology, data curation, review and editing, and supervision. YN: conceptualization, methodology, writing original draft, software, and data curation. SM: software and validation. GT: writing—review and editing and supervision.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Pathak Y, Shukla PK, Tiwari A, Stalin S, Singh S, Shukla PK. Deep transfer learning based classification model for COVID-19 disease. Ing Rech BioMed. (2020). doi: 10.1016/j.irbm.2020.05.003. [Epub ahead of print].
2. Singh D, Kumar V, Kaur M. Classification of COVID-19 patients from chest CT images using multi-objective differential evolution–based convolutional neural networks. Eur J Clin Microbiol Infect Dis. (2020) 39:1379–89. doi: 10.1007/s10096-020-03901-z
3. Gao XW, James-Reynolds C, Currie E. Analysis of tuberculosis severity levels from ct pulmonary images based on enhanced residual deep learning architecture. Neurocomputing. (2020) 392:233–44. doi: 10.1016/j.neucom.2018.12.086
4. Gerard SE, Patton TJ, Christensen GE, Bayouth JE, Reinhardt JM. FissureNet: a deep learning approach for pulmonary fissure detection in CT images. IEEE Trans Med Imaging. (2018) 38:156–66. doi: 10.1109/TMI.2018.2858202
5. Hagerty JR, Stanley RJ, Almubarak HA, Lama N, Kasmi R, Guo P, et al. Deep learning and handcrafted method fusion: higher diagnostic accuracy for melanoma dermoscopy images. IEEE J Biomed Health Inform. (2019) 23:1385–91. doi: 10.1109/JBHI.2019.2891049
6. Nardelli P, Jimenez-Carretero D, Bermejo-Pelaez D, Washko GR, Rahaghi FN, Ledesma-Carbayo MJ, et al. Pulmonary artery–vein classification in CT images using deep learning. IEEE Trans Med Imaging. (2018) 37:2428–40. doi: 10.1109/TMI.2018.2833385
7. Pannu HS, Singh D, Malhi AK. Improved particle swarm optimization based adaptive neuro-fuzzy inference system for benzene detection. CLEAN Soil Air Water. (2018) 46:1700162. doi: 10.1002/clen.201700162
8. Xia K, Yin H, Qian P, Jiang Y, Wang S. Liver semantic segmentation algorithm based on improved deep adversarial networks in combination of weighted loss function on abdominal CT images. IEEE Access. (2019) 7:96349–58. doi: 10.1109/ACCESS.2019.2929270
9. Xie Y, Xia Y, Zhang J, Song Y, Feng D, Fulham M, et al. Knowledge-based collaborative deep learning for benign-malignant lung nodule classification on chest CT. IEEE Trans Med Imaging. (2018) 38:991–1004. doi: 10.1109/TMI.2018.2876510
10. Zreik M, Van Hamersvelt RW, Wolterink JM, Leiner T, Viergever MA, Išgum I. A recurrent CNN for automatic detection and classification of coronary artery plaque and stenosis in coronary CT angiography. IEEE Trans Med Imaging. (2018) 38:1588–98. doi: 10.1109/TMI.2018.2883807
11. Carrillo-Larco RM, Castillo-Cara M. Using country-level variables to classify countries according to the number of confirmed COVID-19 cases: an unsupervised machine learning approach. Wellcome Open Res. (2020) 5:56. doi: 10.12688/wellcomeopenres.15819.2
12. Hemdan EED, Shouman MA, Karar ME. Covidx-net: a framework of deep learning classifiers to diagnose covid-19 in x-ray images. arXiv. (2020). p. 1–14.
13. Khmaissia F, Haghighi PS, Jayaprakash A, Wu Z, Papadopoulos S, Lai Y, et al. An Unsupervised machine learning approach to assess the ZIP code level impact of COVID-19 in NYC. arXiv. (2020). p. 1–16.
14. Pereira RM, Bertolini D, Teixeira LO, Silla CN Jr, Costa YM. COVID-19 identification in chest X-ray images on flat and hierarchical classification scenarios. Comput Methods Programs Biomed. (2020) 194:105532. doi: 10.1016/j.cmpb.2020.105532
15. Wang X, Deng X, Fu Q, Zhou Q. A weakly-supervised framework for COVID-19 classification and lesion localization from chest CT. IEEE Trans. Med. Imag. 39:26150–25. doi: 10.1109/TMI.2020.2995965
16. Gupta D, Rodrigues JJ, Sundaram S, Khanna A, Korotaev V, de Albuquerque VHC. Usability feature extraction using modified crow search algorithm: a novel approach. Neural Comput Appl. (2018) 32:10915–925. doi: 10.1007/s00521-018-3688-6
17. Gupta D, Ahlawat AK. Usability determination using multistage fuzzy system. Procedia Comput Sci. (2016) 78:263–70. doi: 10.1016/j.procs.2016.02.042
18. Khamparia A, Saini G, Gupta D, Khanna A, Tiwari S, de Albuquerque VHC. Seasonal crops disease prediction and classification using deep convolutional encoder network. Circuits Syst Signal Process. (2020) 39:818–36. doi: 10.1007/s00034-019-01041-0
19. Khamparia A, Singh A, Anand D, Gupta D, Khanna A, Kumar NA, et al. A novel deep learning-based multi-model ensemble method for the prediction of neuromuscular disorders. Neural Comp Appl. (2018) 32:11083–95. doi: 10.1007/s00521-018-3896-0
20. Gochhayat SP, Kaliyar P, Conti M, Tiwari P, Prasath VBS, Gupta D, et al. LISA: Lightweight context-aware IoT service architecture. J Clean Prod. (2019) 212:1345–56. doi: 10.1016/j.jclepro.2018.12.096
21. Khan FQ, Musa S, Tsaramirsis G, Buhari SM. SPL features quantification and selection based on multiple multi-level objectives. Appl Sci. (2019) 9:2212. doi: 10.3390/app9112212
22. Raj RJS, Shobana SJ, Pustokhina IV, Pustokhin DA, Gupta D, Shankar K. Optimal feature selection-based medical image classification using deep learning model in internet of medical things. IEEE Access. (2020) 8:58006–17. doi: 10.1109/ACCESS.2020.2981337
23. Tsaramirsis K, Tsaramirsis G, Khan FQ, Ahmad A, Khadidos AO, Khadidos A. More agility to semantic similarities algorithm implementations. Int J Environ Res Public Health. (2020) 17:267. doi: 10.3390/ijerph17010267
24. Pustokhina IV, Pustokhin DA, Gupta D, Khanna A, Shankar K, Nguyen GN. An effective training scheme for deep neural network in edge computing enabled Internet of medical things (IoMT) systems. IEEE Access. (2020) 8:107112–23. doi: 10.1109/ACCESS.2020.3000322
25. Saritas MM, Yasar A. Performance analysis of ANN and naive bayes classification algorithm for data classification. Int J Intell Syst Appl Eng. (2019) 7:88–91. doi: 10.18201/ijisae.2019252786
26. IEEE8023. Available online at: https://github.com/ieee8023/covid-chestxray-dataset (accessed March 3, 2020).
27. Zhao J, Zhang Y, He X, Xie P. COVID-CT-dataset: a CT scan dataset about COVID-19. arXiv. (2020). p. 1–15.
28. COVID-19, Open Research Dataset Challenge (CORD-19): Available online at: https://www.kaggle.com/allen-institute-for-ai/CORD-19-research-challenge (accessed August 17, 2020).
29. Alrikabi HA, Annajjar W, Alnasrallah AM, Mustafa ST, Rahim MSM. Using FFNN classifier with HOS-WPD method for epileptic seizure detection. in 2019 IEEE 9th International Conference on System Engineering and Technology (ICSET). Shah Alam: IEEE (2019). p. 360–3. doi: 10.1109/ICSEngT.2019.8906408
30. Nurmaini S, UmiPartan R, Caesarendra W, Dewi T, NaufalRahmatullah M, Darmawahyuni A, et al. An automated ECG beat classification system using deep neural networks with an unsupervised feature extraction technique. Appl Sci. (2019) 9:2921. doi: 10.3390/app9142921
31. Dandu JR, Thiyagarajan AP, Murugan PR, Govindaraj V. Brain and pancreatic tumor segmentation using SRM and BPNN classification. Health Technol. (2020) 10:187–95. doi: 10.1007/s12553-018-00284-2
Keywords: DeepSense, artificial intelligence, convolutional neural network, CT images, prediction, COVID-19
Citation: Khadidos A, Khadidos AO, Kannan S, Natarajan Y, Mohanty SN and Tsaramirsis G (2020) Analysis of COVID-19 Infections on a CT Image Using DeepSense Model. Front. Public Health 8:599550. doi: 10.3389/fpubh.2020.599550
Received: 27 August 2020; Accepted: 16 October 2020;
Published: 20 November 2020.
Edited by:
Deepak Gupta, Maharaja Agrasen Institute of Technology, IndiaReviewed by:
Abdulsattar Abdullah Hamad, University of Tikrit, IraqGeorge Halikias, City University of London, United Kingdom
Mohammad Yamin, King Abdulaziz University, Saudi Arabia
Copyright © 2020 Khadidos, Khadidos, Kannan, Natarajan, Mohanty and Tsaramirsis. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Srihari Kannan, harionto@gmail.com
 Adil Khadidos1
Adil Khadidos1 Srihari Kannan
Srihari Kannan