- Liaoning Key Laboratory of Genetics and Breeding for Cruciferous Vegetable Crops, College of Horticulture, Shenyang Agricultural University, Shenyang, Liaoning, China
Introduction: Chinese cabbage is one of the most important vegetable crops in China. However, the clubroot disease caused by the infection of Plasmodiophora brassicae (P. brassicae) has seriously affected the yield and quality of Chinese cabbage. In our previous study, BrUFO gene was found to be significantly up-regulated in diseased roots of Chinese cabbage after inoculation with P. brassicae. UFO (UNUSUAL FLORAL ORGANS) have the properties of substrate recognition during ubiquitin-mediated proteolysis. A variety of plant can activate immunity response through the ubiquitination pathway. Therefore, it is very important to study the function of UFO in response to P. brassicae.
Methods: In this study, The expression pattern of BrUFO Gene was measured by qRT-PCR and In situ Hybridization (ISH). The expression location of BrUFO in cells was determined by subcellular localization. The function of BrUFO was verified by Virus-induced Gene Silencing (VIGS). proteins interacting with BrUFO protein were screened by yeast two-hybrid.
Results: Quantitative real-time polymerase chain reactions (qRT-PCR) and in situ hybridization analysis showed that expression of BrUFO gene in the resistant plants was lower than that in susceptible plants. Subcellular localization analysis showed that BrUFO gene was expressed in the nucleus. Virus-induced gene silencing (VIGS) analysis showed that silencing of BrUFO gene reduced the incidence of clubroot disease. Six proteins interacting with BrUFO protein were screened by Y2H assay. Two of them (Bra038955, a B-cell receptor-associated 31-like protein and Bra021273, a GDSL-motif esterase/acyltransferase/lipase Enzyme) were confirmed to strongly interact with BrUFO protein.
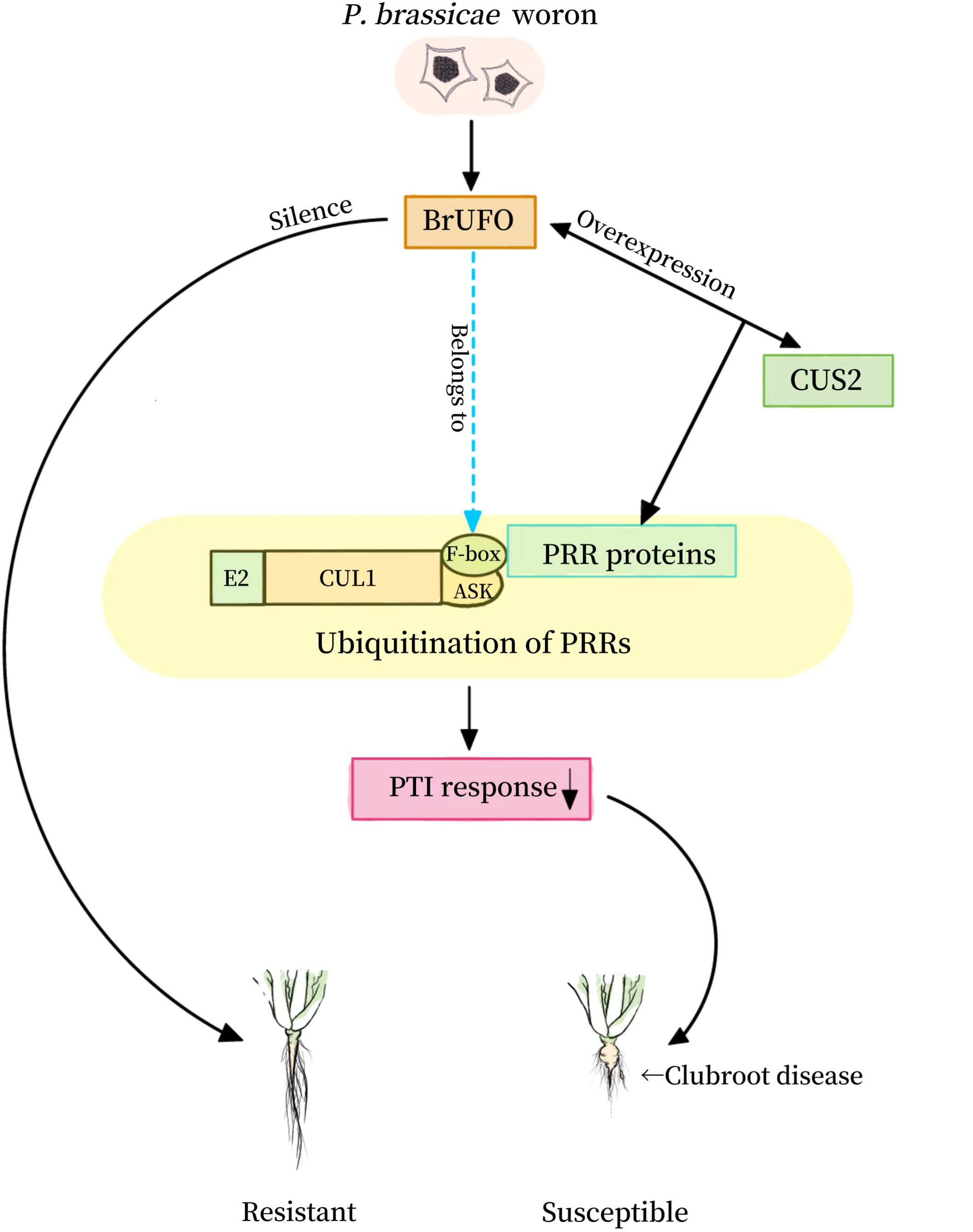
Discussion: BrUFO gene should be a key gene of chinese cabbage against the infection of P. brassicae. BrUFO gene silencing improves the resistance of plants to clubroot disease. BrUFO protein may interact with CUS2 to induce ubiquitination in PRR-mediated PTI reaction through GDSL lipases, so as to achieve the effect of Chinese cabbage against the infection of P. brassicae.
1 Introduction
Clubroot disease is a worldwide soil-borne disease caused by Plasmodiophora brassicae Woron, which mainly infects Chinese cabbage, pakchoi, cabbage, radish, broccoli, mustard, rape and other cruciferous plants (Dixon, 2009; Wei et al., 2016; Yang et al., 2020). In recent years, clubroot disease has rapidly expanding around the world, leading to a significant decline in the yield and quality of Chinese cabbage, and has become one of a main diseases (Long et al., 2019; Zhang et al., 2020). It is safe, efficient and economical to cultivate resistant varieties by using resistance genes, and it is also one of the important ways to fundamentally solve the problems of clubroot disease (Sun et al., 2005).
In our previous study, a gene was found significantly up-regulated expressed on infected Chinese cabbage roots by P. brassicae, with a more than 5 times different expression between inoculated and uninoculated roots. Bioinformatics analysis showed that it has 91.26% sequence homology with Arabidopsis UFO (Unusual Floral Organs), so we named it BrUFO. UFO, as a part of the ubiquitin E3 complex, specifically degrades proteins negatively regulated by AP3 (APETALA 3) and those proteins that control normal growth in flower primordia (Lee et al., 1997). UFO genes are involved in regulating floral meristem and floral organ characteristics during flower development (Lee et al., 1997). In addition, UFOs may also play a role in early petal formation (Durfee et al., 2003). Therefore, UFO regulates multiple stages of flower development in the SCF complex.
UFO is an F-box protein containing F-box motifs and have the properties of substrate recognition during ubiquitin-mediated proteolysis. These proteins play an important role in many physiological processes such as cell time-phase conversion, signal transduction and development (Hershko, 1996). F-box proteins are SCF (SKP-Cullin-F-box) E3 ubiquitin ligase complexes that mediate ubiquitination and subsequent proteasome degradation of the target protein. Ubiquitination is one of the biochemical reactions that regulate the degradation of egg white matter in plants, and its functions are related to plant growth, development, biological stress and abiotic stress. Plants defend themselves against pathogen infections through a variety of mechanisms (Wielkopolan and Obrepalska-Steplowska, 2016). In recent years, scientists have found that several key ubiquitination enzymes (E1, E2, E3) are involved in plant disease resistance (Kravtsova-Ivantsiv and Ciechanover, 2011). A variety of plant can activate PTI (pamp to trigger immunity) response through ubiquitination pathway to inhibit pathogen infection (Sandra and Birgit, 2009). Many studies have shown that ubiquitination plays an important role in the PRR (People Recombinant Protein)-mediated PTI response. Ubiquitin-dependent protein degradation pathway is an effective regulatory mechanism for protein degradation in all organisms. It has been found that the infection of some twin viruses can interfere with the activity of E3 ubiquitin ligase through JA and GA pathways, thus providing favorable conditions for infection of the pathogen (Lozano-Durán et al., 2011; Ji et al., 2017; Jia et al., 2016; Kushwaha et al., 2017; Li et al., 2019). In addition, GDSL protein family plays an important role in jasmonic acid (JA) antagonistic SA signaling pathway. Many studies have proved that some GDSL proteins can enhance plant resistance to some pathogens through the SA resistance pathway (Hong et al., 2008; Kwon et al., 2009; Kim et al., 2014; Gao et al., 2017). But there are no reports about the direct relationship between UFO and disease resistance. Therefore, it is very important to study the function of UFO in response to P. brassicae.
In this study, the function and possible regulatory mechanism of Chinese cabbage BrUFO in response to the infection of P. brassicae was analyzed. These results provide a theoretical basis for using BrUFO gene to improve the resistance of Chinese cabbage to P. brassicae.
2 Materials and methods
2.1 Material
Plant materials used in this project were susceptible variety ‘SN742’ and resistant variety ‘SN205’of Chinese cabbage. The pathogen used in this study was physiological race NO.4 of P. brassicae isolated in previous work (Lv et al., 2021). The plants were cultured at 23°C with illumination for 12 h per day and 60% humidity. Roots samples were collected on the 14th day after inoculation with P. brassicae (when zoospores of P. brassicae can be observe d in the root hairs under the ordinary light microscope), and on the 40th day after inoculation with P. brassicae (when obvious club symptoms are appear on roots), and then stored at -80°C after flash-freezing in liquid nitrogen.
2.2 Method
2.2.1 qRT-PCR analysis of BrUFO
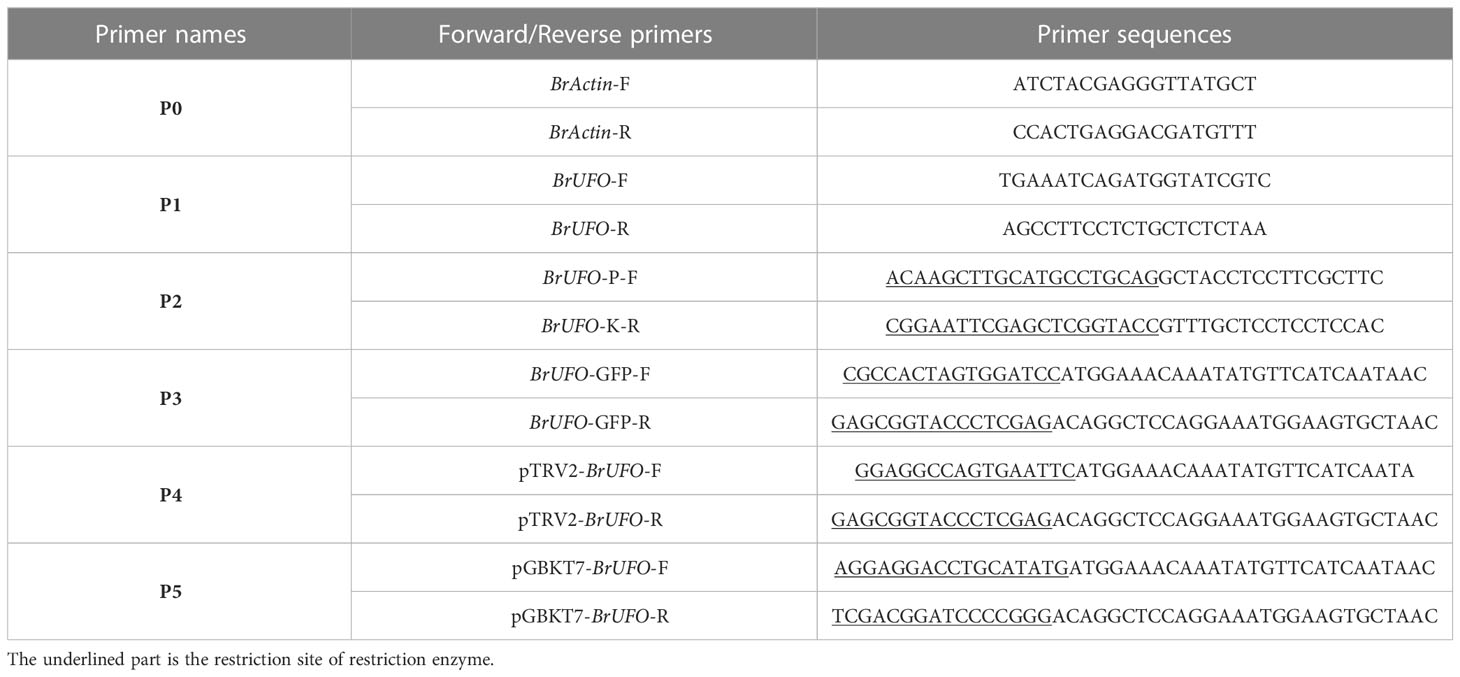
In this study, the expression level of BrUFO in the roots of susceptible and resistant materials at different stages after inoculation with P. brassicae was quantitatively measured by qRT-PCR. In detail, total RNA was extracted using a Steady Pure Plant RNA Extraction Kit (Ecri Bio, Hunan, China), and RNA was reverse-transcribed into cDNA using HiScript 1st Strand cDNA Synthesis Kit (Vazyme, Nanjing, China). Specific primer P1 was designed according to the coding sequence (CDS) of Chinese cabbage BrUFO, and the BrActin gene was used as the internal control (the primers of P0 are shown in Table 1). qRT-PCR analysis was performed on the QuantStudio™ 6 Flex Real-Time PCR System (Life Technologies™, China) with ChamQ Universal SYBR qPCR Master Mix (Vazyme, Nanjing, China).
The study was arranged with three replicates for each different treatment. Duncan multi-range test (α = 0.05) was used to evaluate the expression levels of BrUFO. Data were analyzed by SPSS 11.5 (IBM, Armonk, NY, USA), and OriginPro 7.5 (OriginLab Corp., Northampton, MA, USA) was used to make graphs.
2.2.2 In situ Hybridization analysis of BrUFO
In situ Hybridization (ISH) was performed to verify the results of qRT-PCR. For probes construction: specific primer P2 containing PstI and KpnI restriction sites were designed according to the CDS sequence of BrUFO (Table 1), and a 263 bp sequence was amplified and purified, then digested by PstI and KpnI. The digested product was ligated to the pSPT18 vector, and the resulting construct transformed, amplified, digested by PstI or KpnI. Probes were constructed using a DIG RNA Labeling Kit (SP6/T7; Roche, Basel, Switzerland) for in vitro transcription. For preparation of paraffin sections of root tissues: the roots of susceptible and resistant materials at different stages after inoculation with P.brassicae were fixed with 4% Paraformaldehyde solution (PFA) (Rnase-free) solution. Uninoculated roots were used as control. Evacuate the samples with a vacuum pump until sanking into the bottom of the fixing solution, and then submerged in paraffin overnight at 4°C (Ji et al., 2017). Fixed the embedded samples on the slicer for slicing (cut at least three slices each treatment), and paraffin sections containing samples were placed in DEPC-H2O at 42°C to attach to slides, and dried overnight. Paraffin sections were dewaxed in a series of ethanol:xylene mixtures (100% xylene, 1:3 ethanol:xylene, 1:1 ethanol:xylene, 3:1 ethanol:xylene, and 100% ethanol) for 5 minutes in each mixture.
The dewaxed plant sections hybridized with BrUFO probe, and then were stained and observed under optical microscope (ECLIPSE Ci-L, Nikon) following Ji et al. (2017).
2.2.3 Subcellular localization analysis of BrUFO
The expression location of BrUFO in cells was determined by subcellular localization. The full-length cDNA of BrUFO was amplified using primer P3 with BamHI and XhoI restriction sites (Table 1) from the roots of susceptible materials, and then ligated to pBWA(V)BS-GFP to obtained pBWA(V)BS-BrUFO-GFP. pBWA(V)BS-BrUFO-GFP was transformed into agrobacterium GV3101, and then cultured overnight, centrifuged. The cell pellet was resuspended in infection solution (containing 10 mM MgCl2, 10 mM MES, 15 μL AS, pH = 5.6). The obtained suspension (OD600 = 1.0) was injected into 4-week-old tobacco leaves. After 2 days of dark culture, the leaves were observed under a laser confocal microscope (TCS SP8, Leica, Germany).
2.2.4 Function verification of BrUFO in Chinese cabbage by virus-induced gene silencing
The function of BrUFO was verified by Virus-induced Gene Silencing (VIGS). In detail, a 300bp cDNA fragment of BrUFO was connected to pTRV2 to constructed pTRV2-BrUFO silenced vector. pTRV2-00 (empty vector) and pTRV2- BrUFO were transferred into A. tumefaciens strain GV3101 separately using the freeze-thaw method (Zhou et al., 2021). The infection solution containing vectors pTRV2-00, pTRV2- BrUFO were mixed respectively with the infection solution containing pTRV1 vector (1:1, vol/vol) to constructed pTRV1+pTRV2-00 or pTRV1+pTRV2- BrUFO mixed infection solution. Germinated seeds were treated with above two kinds of mixed infection solution respectively by the agrobacterium vacuum infiltration method (Yue et al., 2021), wild-type (untreated) plants served as controls. 60 seeds were infected in each group (the experiment was repeated three times). The seeds treated with the mix infection solution were cultured in a 25 °C incubator with a 16 h light/8 h dark cycle. When the gene silencing was confirmed, the roots were inoculated with P. brassicae suspension. When the typical swelling symptom of clubroot disease was obvious, the expression level of BrUFO was measured by qRT-PCR with a specific primer pair P4 (Table 1). All treatments were performed in triplicate with five randomly selected plants per treatment for each replicate. Duncan’s multiple range tests (α=0.05) were conducted to evaluate significant differences. Data were analyzed by SPSS 11.5 (IBM, Armonk, NY, USA), and OriginPro 7.5 (OriginLab Corp., Northampton, MA, USA) was used to produce graphs. At the same time, when clubroot symptom was showed, the incidence and level of clubroot disease were analyzed using a completely random design with three replicates and 30 seedlings per experimental unit. The infection progress of inoculated seedlings was analyzed by Fisher’s least significant difference test at a significance threshold of p <0.05.
2.2.5 Yeast two-hybrid (Y2H) analysis
In order to screen the interaction proteins of BrUFO, the full-length sequence of BrUFO with NdeI and XmaI restriction sites were amplificated with primer P5 (Table 1) and connected to pGBKT7 to obtained recombinant vector pGBKT7-BrUFO. Self-activation and toxicity tests of the PGBKT7-BrUFO were performed to exclude the self-expression influence.
Y2H screening was conducted using yeast mating according to the Matchmaker Gold Y2H System user manual (Clontech, USA). Strains carrying PGBKT7-BrUFO were mixed with the library strain and screened on SD/-LEU/- TRP/X-α-Gal/AbA (DDO/X/A) medium. All single blue colonies were selected and streaked on SD/-ADE/-HIS/- LEU/-TRP/X-α-Gal/AbA (QDO/X/A) medium. Positive blue colonies were assessed by PCR, and sequenced. The sequencing results were compared with the BLAST database using BRAD (http://brassicadb.cn/#/) to obtain potential interaction proteins of BrUFO.
2.2.6 Point-to-point interaction verification between candidate proteins and BrUFO
The full-length cDNA sequences of candidate proteins were cloned and connected to PGADT7 to construct PGADT7 recombinant vectors respectively. Each PGADT7 recombinant vector and PGBKT7-BrUFO recombinant vector were co-transfected into Y2H Gold competent yeast cells respectively, and then cultivated in DDO medium at 30°C for 72-96 h. Selected the well-grown monocolones, and resuspensed them with 30 μL of sterile ultra-pure water. Incubated them on the QDO/A/X medium upside down at 30°C for 72-96 h, and observed the growth of colones.
3 Results
3.1 The expression pattern analysis of BrUFO gene
qRT-PCR results of BrUFO in the roots of Chinese cabbage showed that the expression level of BrUFO in the inoculation-treated group (T) was higher than that in the uninoculated control group (CK) (Figure 1A). In addition, the expression level of BrUFO was higher in susceptible materials than that in resistant materials. Thus, it was concluded that low expression of BrUFO likely enhances the resistance of plants to P. brassicae.
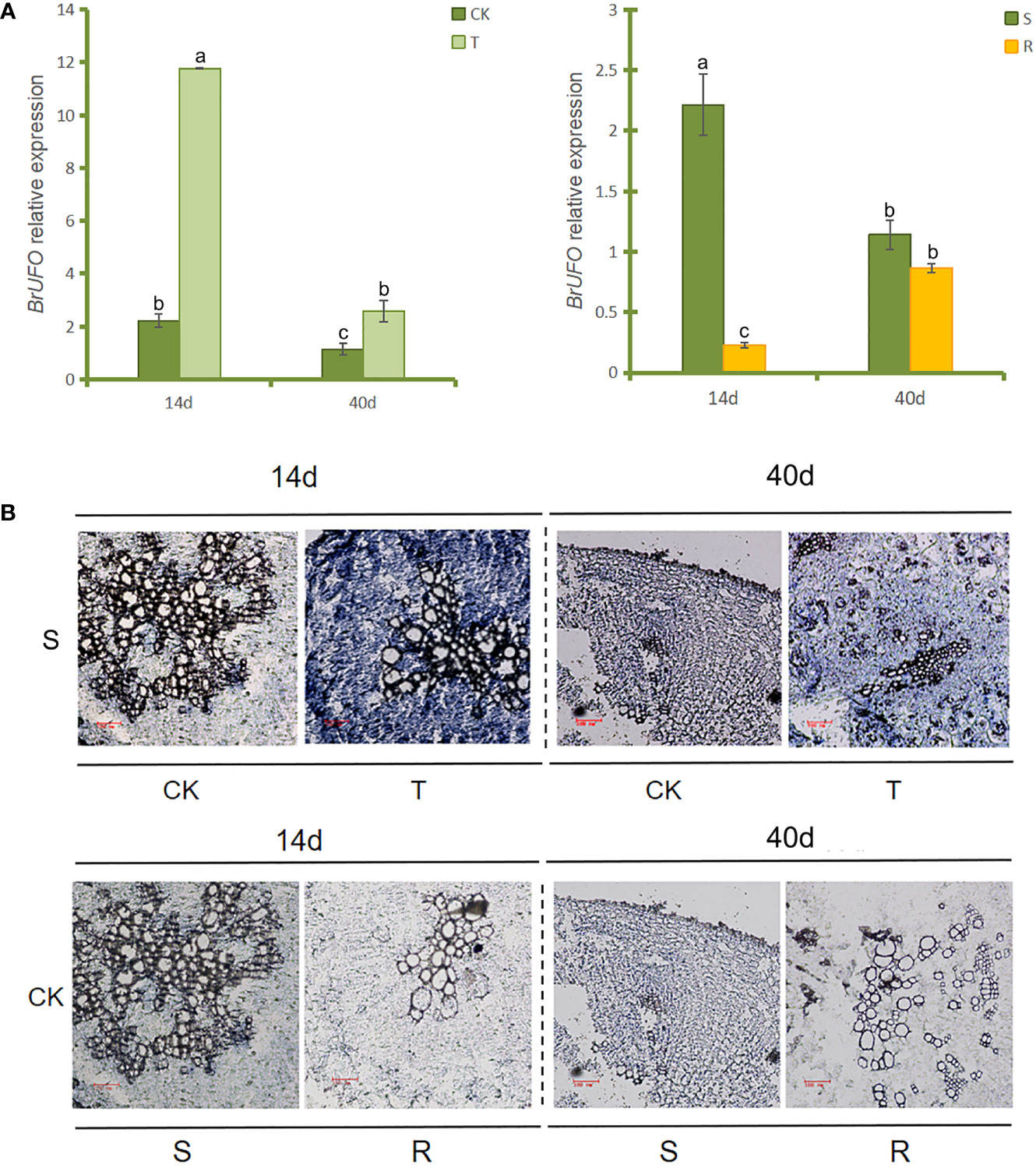
Figure 1 BrUFO expression patterns. (A) qRT-PCR analysis of BrUFO. Results are mean ± standard deviation (n = 3). Each set of data represents the mean of its three replicates, and the error line represents the standard deviation of its three replicates. Letters indicate significant differences at p ≤0.05 according to Duncan’s multiple range test. (B) In situ hybridization analysis of BrUFO. Roots samples were collected on the 14th day after inoculation with P. brassicae (when zoospores of P. brassicae can be observe d in the root hairs under the ordinary light microscope), and on the 40th day after inoculation with P. brassicae (when obvious club symptoms are appear on roots); S, susceptible material of Chinese cabbage susceptible material’SN742’; R, Chinese cabbage resistant material ‘SN205’; CK, control roots not inoculated with Plasmodiophora brassicae; T, treated roots inoculated with Plasmodiophora brassicae.
In situ hybridization was used to verify the expression patterns of BrUFO in the roots of Chinese cabbage. The results revealed blue hybridization signals were showed in inoculated roots in both sampling periods (Figure 1B). Furthermore, the signal of BrUFO was stronger in susceptible materials than that in resistant materials when the plants was not been infected by P. brassicae, but low expression of BrUFO makes the difference indistinguishable by naked eye. Thus, the results of in situ hybridization were consistent with that of qRT-PCR.
UFO is an F-box protein (Samach et al., 1999; Wang et al., 2003), and some F-box proteins are located in the nucleus, such as VVF-Box5 of the grape F-Box family is mainly located in the nucleus of onion epidermal cells (Yu et al., 2018). The F-box protein of P. euphratica (Ren et al., 2017) and F-box family protein SIF-Box18 in millet (Huo et al., 2014) are also located in the nucleus. In order to clarify the action site of BrUFO in Chinese cabbage, subcellular localization was performed. The results showed that the leaves containing pBWA(V)BS-GFP (empty vector) showed fluorescence signals in the nucleus, cell membrane and cytoplasm. However, the leaves containing pBWA(V)BS-BrUFO-GFP recombinant vector only showed fluorescence signals in the nucleus. After staining with DAPI, the leaves containing pBWA(V)BS-GFP or pBWA(V)BS-BrUFO-GFP emitted a red signal on nucleus (Figure 2), which was the location of the GFP channel. Therefore, it was inferred that BrUFO was located in the nucleus. This result provided a basis for screening proteins potentially interacting with BrUFO from the Y2H nuclear library.
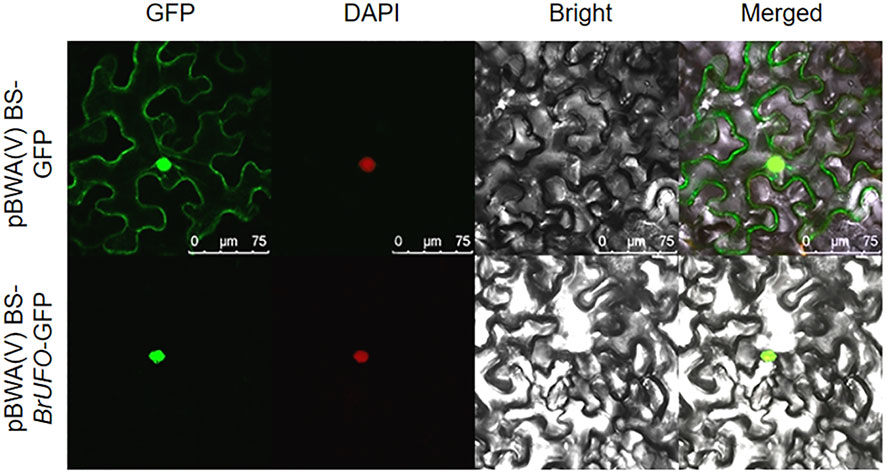
Figure 2 Subcellular localization of BrUFO. The green fluorescence signal is from GFP; the red fluorescence signal is from DAPI; the yellow fluorescence signal is the result of superposition of green and red fluorescence signals.
3.2 The functional validation of BrUFO
The VIGS technique was used to verify the function of BrUFO in Chinese cabbage. The results showed that BrUFO began to be silenced on the 15th day after pTRV1+pTRV2-BrUFO VIGS treatment, which showed as the expression levels of BrUFO in plants harbouring pTRV1+pTRV2-BrUFO were lower than in pTRV1+pTRV2-00 (empty vector) and untreated wild-type plants. On the 25th day after VIGS treatment, the expression of BrUFO was significantly down-regulated in plants harbouring pTRV1+pTRV2-BrUFO. When the expression level of BrUFO in pTRV1+pTRV2-00 plants was set as 1, the expression levels in pTRV1+pTRV2-BrUFO plants was only 0.4, and the silencing efficiency was 60% (Figure 3A). On the 40th day after VIGS treatment (i.e. the 25th day after inoculation with P. brassicae), obvious swelling symptoms were showed on the taproots and fibrous roots of two control plants. To pTRV1+pTRV2-BrUFO plants, only mild swelling symptoms were evident on the fibrous roots, but not on taproots (Figure 3B). The average clubroot disease incidence (20 plants per treatment) of untreated, pTRV1+pTRV2-00 and pTRV1+pTRV2-BrUFO treated plants were 11, 42.25 and 7.25, respectively. And their average incidence rates were 35%, 85% and 15%, respectively (Table 2). pTRV1+pTRV2-00 (empty vector) treatment significantly influenced the resistance of plants, while the silence of BrUFO restored the resistance of plants. These results suggest that the decrease of BrUFO expression can enhance the resistance of Chinese cabbage to P. brassicae.
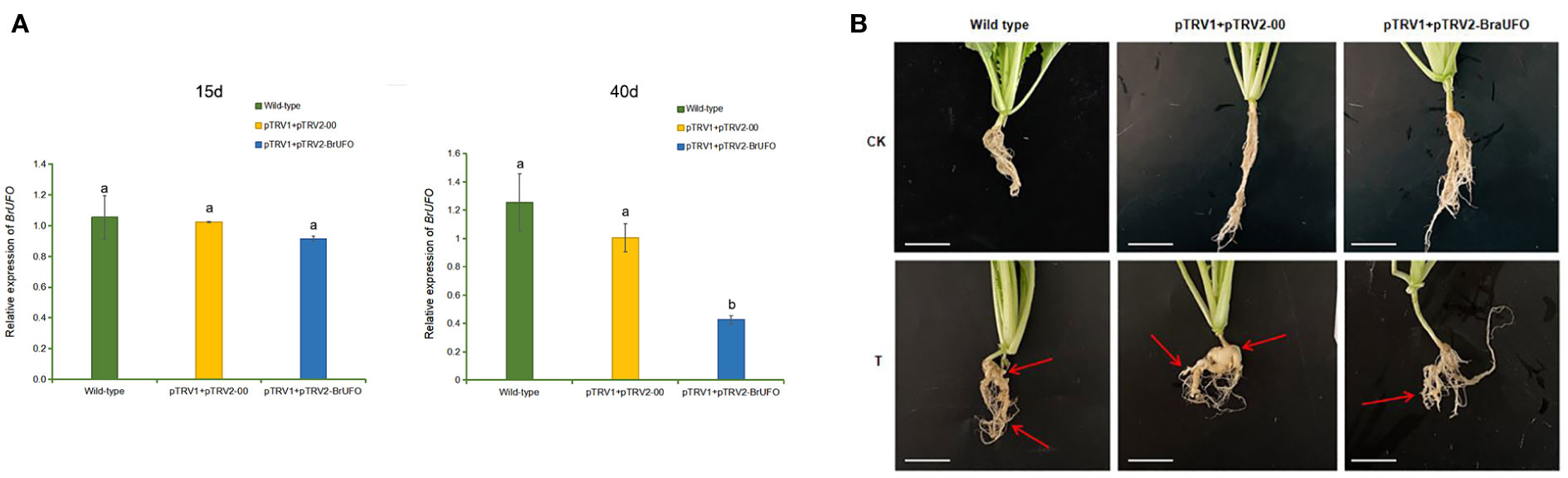
Figure 3 Functional validation of BrUFO in Chinese cabbage responses to infection by Plasmodiophora brassicae. (A) Expression of BrUFO in the roots of Chinese cabbage after VIGS treatment. Results are mean ± standard deviation (n = 3). Each set of data represents the mean of its three replicates, and the error line represents the standard deviation of its three replicates. Letters indicate significant differences at p ≤0.05 according to Duncan’s multiple range test. (B) Symptoms of clubroot disease in VIGS-treated roots on day 40 after VIGS treatment. CK, control plants not inoculated with P. brassicae; T, plants treated with P. brassicae. On the 40th day after VIGS treatment (i.e. the 25th day after inoculation with P. brassicae), obvious clubroot symptoms were showed on the taproots and fibrous roots of both control plants. To pTRV1+pTRV2-BrUFO plants, only mild swelling symptoms were evident on the fibrous roots, but not on taproots. Scale bar=2 cm.
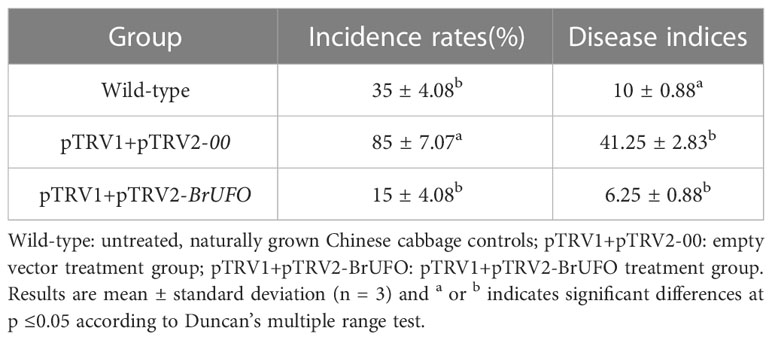
Table 2 Disease indices and incidence rates of BrUFO Silenced Chinese cabbage infected with P. brassicae.
3.3 Y2H screening for interaction proteins of BrUFO
To assess the self-activation ability of bait vector PGBKT7-BrUFO, PGBKT7-BrUFO and pGADT7-T were co-transfected into Y2H competent cells. The results showed that white colonies could grow on DDO solid plates, but no colonies grew after single colonies were diluted and plated on QDO/A/X solid plates as well as negative controls (pGBKT7-Lam + pGADT7-T). However, positive controls (pGBKT7-53 + pGADT7-T) yielded blue colonies on QDO/A/X solid plates (Figure 4A). These results indicate that the expressed protein of PGBKT7-BrUFO did not possess self-activation ability. The toxicity test results for bait vector PGBKT7-BrUFO showed that the growth of Y187 colonies containing PGBKT7-BrUFO were similar to those of colones containing PGBKT7-T on SDO solid plates (Figure 4A). These results proved that the pGBKT7-BrUFO protein was not toxic, and it could therefore be used for subsequent library screening.
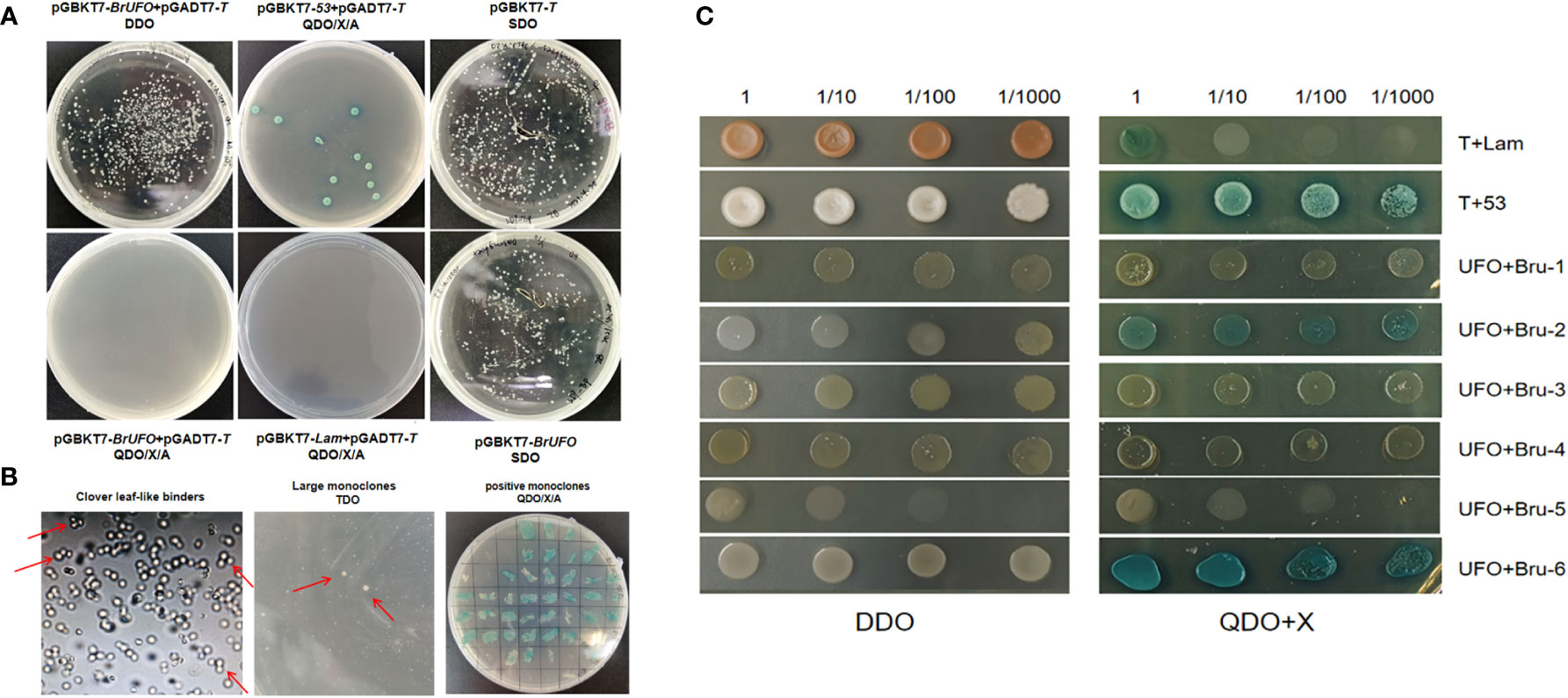
Figure 4 Yeast two hybrid test for BrUFO. (A) Self-activation analysis and toxicity testing of pGBKT7-BrUFO. (B) Screening of potentially interacting proteins with BrUFO from the yeast library. Left, Cloverleaf-like binders on TDO medium. Middle, Large monoclonal colonies in TDO transferred to QDO medium. Right, Positive interaction of proteins with BrUFO on DDO/X/A medium. (C) Validation of proteins interacting with BrUFO. Co-transformation results for proteins potentially interacting with BrUFO on DDO medium and QDO+X-α-Gal medium.
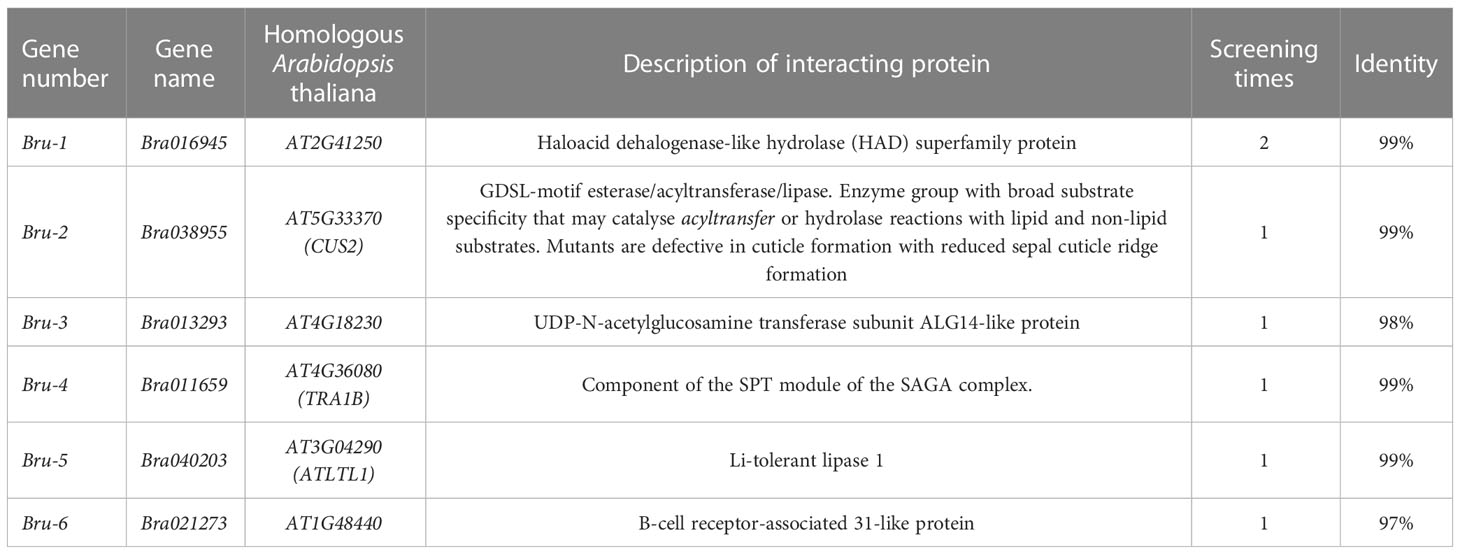
When the Y2H yeast monoclone containing PGBKT7-BrUFO was hybridized with the Chinese cabbage yeast library induced by clubroot, typical cloverleaf-like binders were observed under the light microscope, and 366 large monoclonal colonies on TDO medium were picked and cultivated on QDO/X/A medium (Figure 4B). Positive monoclones on QDO medium were sequenced and subjected to BLAST to analyze the function using the NCBI database, and six proteins were eventually identified (Table 3); These proteins include: Bru-1, Haloacid dehalogenase-like hydrolase (HAD) superfamily protein (detected twice); Bru-2 (CUS2), GDSL-motif esterase/acyltransferase/lipase (detected once); Bru-3, UDP-N-acetylglucosamine transferase subunit ALG14-like protein (detected once); Bru-4, Component of the SPT module of the SAGA complex (detected once); Bru-5, Li-tolerant lipase 1 protein (detected once) and Bru-6, B-cell receptor-associated 31-like protein (detected once).
3.4 The validation result of proteins interacting with BrUFO
To validate the interactions between candidate proteins and BrUFO, co-transfection results showed these colonies could grow on DDO medium (Figure 4C). However, on QDO/X medium, only PGADT7-Bru-6+PGBKT7-BrUFO, and PGADT7-Bru-2+PGBKT7-BrUFO yielded blue clones as well as pGADT7-T + PGBKT7-53 (positive controls). Contrast with positive controls, stains harbouring other candidate proteins as well as pGAD7-T + PGBKT7-T (negative controls) did not grow colonies (Figure 4C). So it is determined that Bru-2 and Bru-6 interact with BrUFO.
4 Discussion
4.1 BrUFO should be a key gene of Chinese cabbage against the infection of P. brassicae
A series of changes will occur when plants are infected by pathogens, such as the changes of gene expression (Coll et al., 2011). For example, the infection of magnaporthe oryzae resulted in the increase of expression levels of Bd4g02060, Bd1g21177, Bd5g06390, Bd3g41480, and Bd3g28280 in rice within 48 hours (Peng et al., 2021). Both CsRSF1 and CsRSF1 could respond to the S. fuliginea infection. In the early stage of S. fuliginea infection, the expression levels of CsRSF1 and CsRSF1 were higher in the resistant varieties, which had a positive regulatory effect on the S. fuliginea infection (Wang X et al., 2021). In this study, qRT-PCR and in situ hybridization results showed that BrUFO was significantly up-regulated expressed on the roots of Chinese cabbage infected by P. brassicae, and BrUFO expression levels were higher in susceptible materials than that in resistant materials (Figure 1).
Plant innate immune response system is mainly composed of PTI and ETI. Many studies have shown that ubiquitination plays an important role in the PRR-mediated PTI response (Sandra and Birgit, 2009). Some F-Box proteins function in plant defence responses dependent on ubiquitin. UFO is an F-box protein (Samach et al., 1999; Wang et al., 2003), and some F-Box proteins, such as Coronatine insensitive 1 (COI1) and Suppressor of nim1-1 (SON1) have the function in plant defence responses such as disease resistance and insect resistance (Xie et al., 1998; Kim and Delaney, 2002). Arabidopsis SON1 is an F-box protein that negatively regulates the resistance of pathogenic downy fungi and leaf spot bacteria through a selective protein degradation pathway dependent on ubiquitin (Kim and Delaney, 2002). These results suggest that BrUFO as an F-box protein played role in different types of disease resistance in plants.
4.2 BrUFO silencing improves the resistance of plants to clubroot disease
VIGS is a rapid and efficient method for studying gene function. It has the advantages of a short cycle, simple operation, and the capacity for high-throughput, and it is independent of a mature transgenic system (Burch-Smith et al., 2004). It is widely used in plant growth and development, such as pistil development, flower development, fruit cracking, and ripening (Zhao et al., 2015; Qi et al., 2018), as well as gene function identification related to flower colour, organ formation (Wang P. et al., 2021), plant disease resistance and abiotic stress (Zhao et al., 2015; Liu et al., 2019), signal transduction, and metabolite synthesis and regulation. Currently, more than 30 vectors have been developed using VIGS technology. Among them, tobacco rattle virus (TRV) is the most widely used VIGS vector in many herbs and some woody plants (Ye et al., 2009; Annette and Matthias, 2010; Huang et al., 2012; Sahu et al., 2012; Jiang et al., 2014). In this study, pTRV1+pTRV2-BrUFO was firstly used to silence BrUFO. The results showed that BrUFO was effectively silenced, and the incidence of clubroot disease was reduced in silenced plants, suggesting that BrUFO silencing could improve the resistance of plants to P. brassicae.
4.3 Regulation mechanism of BrUFO in Chinese cabbage against Plasmodiophora brassicae
A useful way to explore intracellular signal transduction is to study interactions between proteins (Waadt et al., 2010). The Y2H system is widely used to identify and verify interactions between proteins (Zhang et al., 2021). In this study, Bru-6 (Bra021273) and Bru-2 (Bra038955) were screened and confirmed to interact with BrUFO. Bru-6 is a receptor-associated 31-like protein but little is known about it. Bru-2 is CUS2, a GDSL-like esterase/acyltransferase/lipase protein.
GDSL lipases play important roles in many physiological and biochemical processes such as plant growth and development (Takahashi et al., 2009; Dong et al., 2016; Ji et al., 2017), organ morphogenesis, lipid metabolism (Clauß et al., 2011; Ding et al., 2019a; Ding et al., 2019b) and stress reaction (Hong et al., 2008; Kwon et al., 2009; Lee et al., 2009; Kim et al., 2014). In addition, expression of GDSL lipase genes in some plants can be induced by pathogens, hormones such as salicylic acid (SA), ethylene and jasmonic acid (JA), and abiotic stress factors, suggesting that they may be involved in plant disease resistance and stress responses (Jakab et al., 2003; Lee and Cho, 2003; Hong et al., 2008; Kwon et al., 2009; Kim et al., 2014; Gao et al., 2017). GDSL protein family plays an important role in jasmonic acid (JA) antagonistic SA signaling pathway. In addition, many studies have proved that some GDSL proteins can enhance plant resistance to some pathogens through the SA resistance pathway (Hong et al., 2008; Kwon et al., 2009; Kim et al., 2014; Gao et al., 2017). The overexpression of AtGDSL1 decreased the content of reactive oxygen species (ROS) and salicylic acid (SA), which enhanced the resistance of Brassica napus to S. sclerotiorum (Ding et al., 2020). The expression of two GDSL lipases, OsGLIP1 and OsGLIP2, in rice immune response were inhibited by pathogen infection and salicylic acid (SA) treatment. The simultaneous downregulation of OsGLIP1 and OsGLIP2 increased the resistance of rice to Xoo and M. oryzae (Gao et al., 2017). Therefore, it is speculated that BrUFO may interact with CUS2 to induce ubiquitination in PRR-mediated PTI reaction through GDSL lipases, so as to achieve the effect of Chinese cabbage against the infection of P. brassicae (Figure 5). However, this should be verified in follow-up studies.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.
Author contributions
BZ: Conceptualization, Methodology, Data curation, Writing-original draft, Formal analysis, Investigation, Writing-review & editing. HF: Formal analysis, Methodology, Data curation, Investigation. WG and XW: Formal analysis, Data curation. JZ: Formal analysis. RJ: Funding acquisition, Conceptualization, Formal analysis, Supervision, Writing-review & editing. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China under grant 31972412 and 32272717.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Annette, B., Matthias, L. (2010). VIGS-genomics goes functional. Trends Plant Sci. 15 (1), 1–4. doi: 10.1016/j.tplants.2009.09.002
Burch-Smith, T. M., Anderson, J. C., Martin, G. B., Dinesh-Kumar, S. P. (2004). Applications and advantages of virus-induced gene silencing for gene function studies in plants. Plant J. 39 (5), 734–746. doi: 10.1111/j.1365-313X.2004.02158.x
Clauß, K., Roepenack-Lahaye, E., Böttcher, C., Roth, M., Welti, R., Erban, A., et al. (2011). Overexpression of sinapine esterase BnSCE3 in oilseed rape seeds triggers global changes in seed metabolism. Plant Physiol. 155 (3), 1127–1145. doi: 10.1104/pp.110.169821
Coll, N. S., Epple, P., Dangl, J. L. (2011). Programmed cell death in the plant immune system. Cell Death dif. 18 (8), 1247–1256. doi: 10.1038/cdd.2011.37
Ding, L., Guo, X., Li, M., Fu, Z., Yan, S., Zhu, K., et al. (2019a). Improving seed germination and oil contents by regulating the GDSL transcriptional level in Brassica napus. Plant Cell Rep. 38 (2), 243–253. doi: 10.1007/s00299-018-2365-7
Ding, L., Li, M., Guo, X., Tang, M., Cao, J., Wang, Z., et al. (2020). Arabidopsis GDSL1 overexpression enhances rapeseed sclerotinia sclerotiorum resistance and the functional identification of its homolog in Brassica napus. Plant Biot. J. 18 (5), 1255–1270. doi: 10.1111/pbi.13289
Ding, L., Li, M., Wang, W., Cao, J., Wang, Z., Zhu, K., et al. (2019b). Advances in plant GDSL lipases: from sequences to functional mechanisms. Acta Physiol. Plant 41 (9), 1–11. doi: 10.1007/s11738-019-2944-4
Dixon, G. R. (2009). The occurrence and economic impact of plasmodiophora brassicae and clubroot disease. J. Plant Growth Reg. 28, 194–202. doi: 10.1007/s00344-009-9090-y
Dong, X., Yi, H., Han, C., NouIll, S., Hur, Y. (2016). GDSL esterase/lipase genes in Brassica rapa l.: genome-wide identification and expression analysis. Mol. Gen. Gen. 291 (2), 531–542. doi: 10.1007/s00438-015-1123-6
Durfee, T., Roe, J. L., Sessions, R. A., Inouye, C., Serikawa, K., Feldmann, K. A., et al. (2003). The f-box-containing protein UFO and agamous participate in antagonistic pathways governing early petal development in Arabidopsis. Proc. Natl. Acad. Sci.USA. 100, 8571–8576. doi: 10.1073/pnas.1033043100
Gao, M., Yin, X., Yang, W., Lam, S., Tong, X., Liu, J. (2017). GDSL lipases modulate immunity through lipid homeostasis in rice. PloS Pathog. 13 (11), e1006724. doi: 10.1371/journal.ppat.1006724
Hershko, A. (1996). The ubiquitin system for protein degradation. Bioch. Society. 24 (4), 627S–627S. doi: 10.1042/bst024627sc
Hong, J., Choi, H., Hwang, I., Kim, D., Kim, N., Choi, D., et al. (2008). Function of a novel GDSL-type pepper lipase gene, CaGLIP1, in disease susceptibility and abiotic stress tolerance. Planta 227 (3), 539–558. doi: 10.1007/S00425-007-0637-5
Huang, C., Qian, Y., Li, Z., Zhou, X. (2012). Virus-induced gene silencing and its application in plant functional genomics. Sci. Chin. Life Sci. 55 (2), 99–108. doi: 10.1007/s11427-012-4280-4
Huo, D., Zheng, W., Li, P., Xu, Z., Zhou, Y., Chen, M., et al. (2014). Identification, classification and drought response of f-box gene family in foxtail millet. J. crops. 40 (09), 1585–1594. doi: 10.3724/SP.J.1006.2014.01585
Jakab, G., Manrique, A., Zimmerli, L., Métraux, J. P., Mauchi-Mani, B. (2003). Molecular characterization of a novel lipase-like pathogen-inducible gene family of Arabidopsis. Plant Physiol. 132, 2230–2239. doi: 10.1104/pp.103.025312
Ji, R., Wang, H., Xin, X., Peng, S., Hur, Y., Li, Z., et al. (2017). BrEXL6, a GDSL lipase gene of Brassica rapa, functions in pollen development. Biol. Plant 61 (4), 682–694. doi: 10.1007/s10535-017-0735-6
Jia, Q., Liu, N., Xie, K., Dai, Y., Han, S., Zhao, X., et al. (2016). CLCuMuB βC1 subverts ubiquitination by interacting with NbSKP1s to enhance geminivirus infection in Nicotiana benthamiana. PloS Pathog. 12 (6), e1005668. doi: 10.1371/journal.ppat.1005668
Jiang, Y., Ye, S., Wang, L., Duan, Y., Lu, W., Liu, H., et al. (2014). Heterologous gene silencing induced by tobacco rattle virus (TRV) is efcient for pursuing functional genomics studies in woody plants. Plant Cell Tissue Organ Cult. 116 (2), 163–174. doi: 10.1007/s11240-013-0393-0
Kim, H. S., Delaney, T. P. (2002). Arabidopsis SON1 is an f-box protein that regulates a novel induced defense response independent of both salicylic acid and systemic acquired resistance. Plant Cell. 14, 1469–1482. doi: 10.1105/tpc.001867
Kim, G. K., Kwon, S. J., Jang, Y. J., Chung, J. H., Myung, H. N., Park, O. K. (2014). GDSL lipase 1 regulates ethylene signaling and ethylene-associated systemic immunity in Arabidopsis. FEBS Letters. 588 (9), 1652–1658. doi: 10.1016/j.febslet.2014.02.062
Kravtsova-Ivantsiv, Y., Ciechanover, A. (2011). “Ubiquitination and degradation of proteins,” in Gel-free proteomics. methods molec. biol, vol. 753 . Eds. Gevaert, K., Vandekerckhove, J. (Humana Press), 335–357. doi: 10.1007/978-1-61779-148-2_23
Kushwaha, N. K., Bhardwaj, M., Chakraborty, S. (2017). The replication initiator protein of a geminivirus interacts with host monoubiquitination machinery and stimulates transcription of the viral genome. PloS Pathog. 13 (8), e1006587. doi: 10.1371/journal.ppat.1006587
Kwon, S. J., Jin, H. C., Lee, S., Nam, M. H., Chung, J. H., Kwon, S. I., et al. (2009). GDSL lipase-like 1 regulates systemic resistance associated with ethylene signaling in Arabidopsis. Plant J. 58 (2), 235–245. doi: 10.1111/j.1365-313X.2008.03772.x
Lee, K., Cho, T. (2003). Characterization of a salicylic acid and pathogen induced lipase like gene in Chinese cabbage. J. Bioc. Mol. Biol. 36 (5), 433–441. doi: 10.5483/bmbrep.2003.36.5.433
Lee, D., Kim, B., Kwon, S., Jin, H., Park, O. (2009). Arabidopsis GDSL lipase 2 plays a role in pathogen defense via negative regulation of auxin signaling. Biochem. Biophys. Res. communic. 379 (4), 1038–1042. doi: 10.1016/j.bbrc.2009.01.006
Lee, I., Wolfe, D. S., Nilsson, O., Weigel, D. (1997). A leafy co-regulator encoded by UNUSUAL FLORAL ORGANS. Curr. Biol. 7 (2), 95–104. doi: 10.1016/s0960-9822(06)00053-4
Li, P., Liu, C., Deng, W., Yao, D., Pan, L., Li, Y., et al. (2019). Plant begomoviruses subvert ubiquitination to suppress plant defenses against insect vectors. PloS pathogens. 15 (2), e1007607. doi: 10.1371/journal.ppat.1007607
Liu, J., Lei, Y., Zhang, Z., Hu, G., Tang, Y., Zhang, N., et al. (2019). Resistance of GhWRKY48 negatively regulated cotton against verticillium dahliae. Acta agricult. Bor. 34 (5), 99–105.
Long, T., Zhang, Y., Yang, N., Cao, C., Zhang, G., Zhang, S., et al. (2019). Progress in biological control of Chinese cabbage clubroot disease. Hubei Agricult. Sci. 58 (5), 5–8. doi: 10.14088/j.cnki.issn0439-8114.2019.05.001
Lozano-Durán, R., Rosas-Díaz, T., Gusmaroli, G., Bejarano, E. R. (2011). Geminiviruses subvert ubiquitination by altering CSN-mediated derubylation of SCF E3 ligase complexes and inhibit jasmonate signaling in Arabidopsis thaliana. Plant Cell. 3), 1014–1032. doi: 10.1105/tpc.110.080267
Lv, M., Liu, Y., Wu, Y., Zhang, J., Liu, X., Ji, R., et al. (2021). An improved technique for isolation and characterization of single-spore isolates of Plasmodiophora brassicae. Plant Disease. 105, 3932–3938. doi: 10.1094/PDIS-03-21-0480-RE
Peng, W., Song, N., Li, W., Yan, M., Huang, C., Yang, Y., et al. (2021). Integrated analysis of microrna and target genes in brachypodium distachyon infected by magnaporthe oryzae by small rna and degradome sequencing. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.742347
Qi, D., Zhang, Y., Zhao, Z., Zhang, W., Chen, Y., Lian, Y., et al. (2018). Optimization of vitus-induced gene silencing system and function identification of smIAA19 gene in eggplant. Acta horticult. Sinica. 45 (4), 691–701. doi: 10.16420/j.issn.0513-353x.2017-0643
Ren, Y., Jia, H., Xu, X., Li, J., Lu, M., Fan, J., et al. (2017). Identification and functional analysis of f - box gene from populus euphratica. J. Mol. Plant Breed. 15 (5), 1655–1662. doi: 10.13271/j.mpb.015.001655
Sahu, P. P., Puranik, S., Khan, M., Prasad, M. (2012). Recent advances in tomato functionalgenomics: utilization of VIGS. Protoplasma 249 (4), 1017–1027. doi: 10.1007/s00709-012-0421-7
Samach, A., Klenz, J. E., Kohalmi, S. E., Risseeuw, E., Haughn, G. W., Crosby, W. L. (1999). The UNUSUAL FLORAL ORGANS gene of Arabidopsis thaliana is an f-box protein required for normal patterning and growth in the floral meristem. Plant J. 20, 433–445. doi: 10.1016/J.IJBIOMAC.2020.12.102
Sandra, P., Birgit, K. (2009). Plant systems for recognition of pathogen-associated molecular patterns. Semin. Cell Dev. Biol. 20 (9), 1025–1031. doi: 10.1016/j.semcdb.2009.06.002
Sun, B., Shen, X., Guo, H., Zhou, Y. (2005). Research progress in clubroot of crucifers and resistance breeding. Chin. Veg. 4, 34–37.
Takahashi, K., Shimada, T., Kondo, M., Tamai, A., Mori, M., Nishimura, M., et al. (2009). Ectopic expression of an esterase, which is a candidate for the unidentified plant cutinase, causes cuticular defects in Arabidopsis thaliana. Plant Cell Physiol. 51 (1), 123–131. doi: 10.1093/pcp/pcp173
Waadt, R., Schmidt, L. K., Lohse, M., Hashimoto, K. J., Bock, R., Kudla, J. (2010). Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta. Plant J. 56 (3), 505–516. doi: 10.1111/j.1365-313X.2008.03612.x
Wang, X., Chen, Q., Huang, J., Meng, X., Cui, N., Yu, Y., et al. (2021). Nucleotide-binding leucine-rich repeat genes CsRSF1 and CsRSF2 are positive modulators in the cucumis sativus defense response to Sphaerotheca fuliginea. Int. J. Mol. Sci. 22 (8), 3986. doi: 10.3390/ijms22083986
Wang, X., Feng, S., Nakayama, N., Crosby, W. L., Irish, V., Deng, X., et al. (2003). The COP9 signalosome interacts with SCFUFO and participates in Arabidopsis flower development. Plant Cell. 15, 1071–1082. doi: 10.1105/tpc.009936
Wang, P., Liu, P., Geng, D., Wang, K., Hao, Q. (2021). Establishing virus induced gene silencing(VIGS)system of polination and fertilization gene PoMYBs in tree peony. J. Qingdao Agricult. Univ. (Nat. Sci. edit.). 38 (03), 199–204+217.
Wei, X., Xu, W., Yuan, Y., Yao, Q., Zhao, Y., Wang, Z., et al. (2016). Genome-wide investigation of microRNAs and their targets in brassica rapa ssp. pekinensis root with plasmodiophora brassicae infection. Horticult. Plant J. 2 (4), 209–216. doi: 10.1016/j.hpj.2016.11.004
Wielkopolan, B., Obrepalska-Steplowska, A. (2016). Three-way inter-action among plants, bacteria, and coleopteran insects. Planta 244, 313–332. doi: 10.1007/s00425-016-2543-1
Xie, D. X., Feys, B. F., James, S., Nieto-Rostro, M., Turner, J. G. (1998). COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280, 1091–1094. doi: 10.1126/science.280.5366.1091
Yang, L., Fang, Z., Zhang, Y., Zhuang, M., Lü, H., Wang, Y., et al. (2020). Recent advances of disease and stress resistance breeding of cabbage in china. Acta Horticult. Sinica. 47 (9), 1678–1688. doi: 10.16420/j.issn.0513-353x.2020-0494
Ye, J., Qu, J., Bui, H., Chua, N. (2009). Rapid analysis of jatropha curcas gene functions by virus-induced gene silencing. Plant Biotechnol. J. 7 (9), 964–976. doi: 10.1111/j.1467-7652.2009.00457.x
Yu, L., Zhou, S., Zhang, S., Ji, Y. J., Liu, Y. (2018). Gene structure and expression analysis of f-box gene VvF-BOX5 in grapevine (Vitis vinefera l.). J. Plant Genet. Res. 12 (02), 361–369. doi: 10.13430/j.carol.carroll.nki.JPGR.2018.02.019
Yue, N., Li, Y., Sun, Y., Pan, P., Pan, Y., Zheng, Y., et al. (2021). VIGS silencing effects of VIGS silencing SlDCL2 and SlDCL4 destroy tomato TY-1/TY-3 resistance to tomato yellow leaf curl virus (TYLCV). J. Nuc. Agricult. Sci. 35 (11), 2493–2500.
Zhang, J., Ge, W., Chang, H., Xin, X., Ji, R. (2021). Discovery of BrATG6 and its potential role in Brassica rapa l. resistance to infection by Plasmodiophora brassicae. Gene 791, 145711. doi: 10.1016/J.GENE.2021.145711
Zhang, H., Zhang, S., Li, F., Zhang, S., Li, G., Ma, X., et al. (2020). Research progress on clubroot disease resistance breeding of Brassica rapa. Acta Hortic. Sinica. 47 (9), 1648–1662. doi: 10.16420/j.issn.0513-353x.2020-0681
Zhao, Z., Liu, F., Zhang, Y., Qi, D., Chen, Y., Lian, Y. (2015). VIGS expression vector construction and expression analyses of SmMsrA gene in eggplant. Acta Hortic. sinica. 42 (8), 1495–1504. doi: 10.16420/j.issn.0513-353x.2015-0177
Keywords: Chinese cabbage, BrUFO, Plasmodiophora brassicae, VIGS, Yeast two-hybrid
Citation: Zhang B, Feng H, Ge W, Wang X, Zhang J and Ji R (2023) BrUFO positively regulates the infection of Chinese cabbage by Plasmodiophora brassicae. Front. Plant Sci. 14:1128515. doi: 10.3389/fpls.2023.1128515
Received: 27 December 2022; Accepted: 23 February 2023;
Published: 09 March 2023.
Edited by:
Jia-He Wu, Institute of Microbiology, Chinese Academy of Sciences (CAS), ChinaReviewed by:
Yuxiang Yuan, Henan Academy of Agricultural Sciences (HNAAS), ChinaXiaochun Wei, Henan Academy of Agricultural Sciences (HNAAS), China
Copyright © 2023 Zhang, Feng, Ge, Wang, Zhang and Ji. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ruiqin Ji, ruiqinji@syau.edu.cn
 Bo Zhang
Bo Zhang Hui Feng
Hui Feng Wenjie Ge
Wenjie Ge Ruiqin Ji
Ruiqin Ji