- Department of Life Sciences, Yeungnam University, Gyeongsan, South Korea
Hepatica is a small genus of Ranunculaceae with medicinal and horticultural value. We characterized nine complete chloroplast (cp) genomes of Hepatica, which ranged from 159,549 to 161,081 bp in length and had a typical quadripartite structure with a large single-copy region (LSC; 80,270–81,249 bp), a small single-copy region (SSC; 17,029–17,838 bp), and two copies of inverted repeat (IR; 31,008–31,100 bp). The cp genomes of Hepatica possess 76 protein-coding genes (PCGs), 29 tRNAs, and four rRNA genes. Comparative analyses revealed a conserved ca. 5-kb IR expansion in Hepatica and other Anemoneae; moreover, multiple inversion events occurred in Hepatica and its relatives. Analyses of selection pressure (dN/dS) showed that most of the PCGs are highly conserved except for rpl20 and rpl22 in Hepatica falconeri, Hepatica americana, and Hepatica acutiloba. Two genes (rps16 and infA) were identified as pseudogenes in Hepatica. In contrast, rpl32 gene was completely lost. The phylogenetic analyses based on 76 PCGs resolved the phylogeny of Hepatica and its related genera. Non-monophyly of Anemone s.l. indicates that Hepatica should be reclassified as an independent genus. In addition, Hepatica nobilis var. japonica is not closely related to H. nobilis.
Introduction
Chloroplast (cp) is associated with photosynthesis and is considered to have originated as endosymbiotic cyanobacteria (Timmis et al., 2004). The cp is usually uniparentally inherited, with multiple copies per cell, and has a slower evolutionary rate than the nuclear and mitochondrial genomes (Drouin et al., 2008). The cp genome is a circular molecule with a quadripartite structure with two inverted repeats (IR) separated by large single-copy (LSC) and small single-copy (SSC) regions (Palmer, 1985; Jansen and Ruhlman, 2012). The land plant cp genomes are highly conserved in terms of gene content, order, and organization (Palmer, 1991; Jansen and Ruhlman, 2012).
Previous phylogenetic analyses have supported three subtribes in Anemoneae (Ehrendorfer and Samuel, 2001; Mikeda et al., 2006; Wang et al., 2009; Xie et al., 2011; Hoot et al., 2012; Lehtonen et al., 2016; Jiang et al., 2017; Liu et al., 2018b). In Clematidinae, almost all satellite genera of Clematis (e.g., Naravelia and Archiclematis) were nested within Clematis in a previous study (Wang et al., 2009; Lehtonen et al., 2016; Jiang et al., 2017; Liu et al., 2018a). In subtribe Anemoninae, there is a discrepancy regarding the classification of Anemone. Hoot and Palmer (1994), Hoot et al. (2012), and Hoot (1995) suggested a broad concept for the genus and merged Hepatica, Pulsatilla, Oreithales, Knowltonia, and Barneoudia into Anemone based on their molecular phylogenetic results inferred from nrITS and cpDNA data. Ehrendorfer (1995) preferred a narrow concept and suggested the subdivision of the genus into several genera. Furthermore, Jiang et al. (2017) suggested that Hepatica is regarded as an independent genus and Anemone sections Anemonidium, Keiska, and Omalocarpus should be subsumed into Hepatica. Liu et al. (2018b) suggested that subtribe Anemoninae be separated into at least three genera: Anemoclema, Anemone s.l. (including Pulsatilla and Pulsatilloides), and Hepatica (including sect. Omalocarpus, sect. Anemonidium, and sect. Keiskea).
Structural rearrangements and inversions within the cp genome of tribe Anemoneae have been reported based on genetic mapping by restriction enzyme sites (Hoot and Palmer, 1994; Johansson, 1999). Recently, several cp genomes of Ranunculaceae have been published (Park et al., 2015, 2020; Szczecińska and Sawicki, 2015; Li et al., 2016; Park and Park, 2016, 2020; Jiang et al., 2017; Liu et al., 2018a, b; Zhang et al., 2015). Liu et al. (2018a, b) and Zhai et al. (2019) reported the complete cp genome sequences of several members of tribe Anemoneae: Anemoclema, Clematis, Hepatica, Naravelia, and Pulsatilla; they discovered a 4.4-kb expansion of the IR and multiple inversions across Anemoneae.
Hepatica Mill. is a small genus of Ranunculaceae comprising plants that are valuable to medicine and horticulture. Hepatica is distinguished from Anemone L. by the length of the peduncle and simple and entire leaves (Hoot et al., 2012). Hepatica comprises 11 taxa and is distributed in temperate regions of the northern hemisphere (Ulbrich, 1906; Nakai, 1937a, b; Meusel et al., 1965; Tamura, 1995). Although Hepatica is widely distributed, most species are local endemics (Jalas and Suominen, 1976). The genus is most diverse in East Asia, with four species and two varieties (Pfosser et al., 2011). Nakai (1937a), Nakai (1952) reported three taxa, including two Korean endemics—Hepatica insularis Nakai and Hepatica maxima (Nakai) Nakai—divided into two groups: Hepatica asiatica Nakai and H. insularis with annual leaves and H. maxima with biennial leaves. H. asiatica is widespread from the Korean peninsula to Manchuria, H. insularis is restricted to southern Korea and Jeju Island, and H. maxima is endemic to Ulleung Island. Two taxa—H. nobilis var. japonica Nakai and H. nobilis var. pubescens (Hiroe) Hiroe—are considered varieties of Hepatica nobilis Mill. and are distributed in Japan (Nakai, 1937a, b; Hiroe, 1957). Hepatica henryi (Oliv.) Steward is restricted to central west China (Oliver, 1887; Wang, 1980). Hepatica falconeri (Thomson) Yuz. is found in the Kashmir and Pamir regions (Shishkin, 1937; Tamura, 1995; Ogisu et al., 2002). Hepatica americana (DC.) Ker Gawl. and Hepatica acutiloba DC. occur in central to northeastern North America (Steyermark and Steyermark, 1960). Two Hepatica are distributed in Europe: H. nobilis var. nobilis—the type species of Hepatica—is widespread in Europe, and Hepatica transsilvanica Fuss is a local endemic in the alpine regions of Transylvania, Romania.
Previous phylogenetic investigations of Hepatica have used morphological, cytological, and molecular approaches (Kurita, 1955; Hoot and Palmer, 1994; Ogisu et al., 2002; Woo et al., 2002; Jiang et al., 2018; Zhai et al., 2019). However, only a few species of Hepatica have been included (Hoot and Palmer, 1994; Meyer et al., 2010), and relationships within Hepatica remain ambiguous (Pfosser et al., 2011; Jiang et al., 2018).
The cp genome has been reported for two Hepatica species, H. henryi and H. maxima. The Hepatica cp genome has undergone several inversions, and intracellular gene transfer events were detected (Liu et al., 2018b; Zhai et al., 2019; Park and Park, 2020). Therefore, it is necessary to uncover the cp genome characteristics of Hepatica to resolve their phylogenetic relationships. For this purpose, we sequenced, assembled, and analyzed the cp genomes of nine taxa of Hepatica. This study aims to (1) identify the genomic characteristics of these taxa, (2) discover their cp genome structures and determine structural variation by comparing them with the cp genomes of nine Anemoneae and one outgroup [Oxygraphis glacialis (Fischer ex de Candolle) Bunge], and (3) clarify the phylogenetic relationship of Hepatica using 76 protein-coding genes (PCGs).
Materials and Methods
Plant Sampling, DNA Isolation, and Sequencing
Nine Hepatica taxa were collected from the field, herbaria, or flower companies (Supplementary Table 1). The living material was replanted in the greenhouse of the Yeungnam University Herbarium (YNUH), Gyeongsan, South Korea. We generated chloroplast genome sequences by isolating total genomic DNA from fresh tissue with a DNeasy Plant Mini Kit (Qiagen Inc., Valencia, CA, United States). From the herbarium materials, DNA was extracted using a modified CTAB method (Allen et al., 2006). The sequencing was outsourced to Phyzen1 (Seongnam, South Korea), generating 150-bp paired-end reads from a library of 350- and 550-bp inserts on an Illumina Hiseq 2500 platform (Illumina, San Diego, CA, United States).
Chloroplast Genome Assembly and Gene Annotation
The obtained raw data were filtered using an NGS QC Tool Kit (Patel and Jain, 2012) by trimming the adaptors and filtering low-quality reads using default options. After filtering the raw data, clean reads were assembled using SOAPdenovo2 (Lou et al., 2012). The complete chloroplast genome sequences were annotated using GeSeq with chloroplast genomes of nine Anemoneae species (Supplementary Table 1; Tillich et al., 2017). tRNA genes were verified with the tRNAscan--SE search server2 (Lowe and Chan, 2016). PCGs were defined as putatively functional if they followed two criteria: (1) presence of an open reading frame with the complete conserved domain, verified by the NCBI Conserved Domains Database (CDD3), and (2) absence of internal stop codons. The circular maps of Hepatica chloroplast genomes were drawn using OGDRAW4 (Lohse et al., 2013).
Comparative Analyses of Chloroplast Genomes
The cp genomes of Hepatica were compared to nine Anemoneae cp genomes, with one Ranunculeae cp genome as an outgroup (Supplementary Table 1). In order to evaluate similarity, mVISTA was used to compare the cp genome of Hepatica species to the other Anemoneae cp genomes with the LAGAN mode, which produces true multiple alignments regardless of whether they contain inversions or not (Frazer et al., 2004). The IR boundaries were illustrated and compared to those of Ranunculeae species. We aligned cp genome sequences using MAFFT (Katoh and Standley, 2013) and examined the sequence divergence among the Hepatica species through a sliding window analysis computing nucleotide variability (pi) in DnaSP v.5.0 (Librado and Rozas, 2009). For the sequence divergence analysis, we applied a window size of 600 bp with a 200-bp step size. Genes with similar functions were grouped following a previous study to infer the non-synonymous to synonymous substitution rate ratio (dN/dS; Chang et al., 2006) using PAML v4.9, with Anemone flaccida set as the outgroup. Analyses were performed using genes with the same functions (atp, ndh, pet, psa, psb, rpl, rpo, and rps) and singular genes (ccsA, clpP, cemA, and matK). To identify cp genome rearrangements in Hepatica, the complete cp genome alignments for 10 Hepatica and the references—nine Anemoneae and one Oxygraphis—were performed using progressiveMauve v.2.3.1 (Darling et al., 2004) in Geneious Prime 2019. Inverted repeat B was removed from all cp genomes before the alignments. Locally collinear blocks (LCBs) generated by the Mauve alignment were numbered to estimate genome rearrangements.
Phylogenetic Analyses
Phylogenetic analysis was performed using all the 76 PCGs in the cp genome. The genes were extracted from cp genomes and aligned using MAFFT (Katoh and Standley, 2013); the alignments were then concatenated in Geneious Prime 2019.2.1. We conducted phylogenetic analyses using RAxML, v. 8.2.4, with 1,000 bootstrap replicates for evaluating the node support. These analyses used the GTR model with GAMMA+I, selected by jModelTest, v. 2.1.9. We also used Bayesian inference (BI) implemented in MrBayes, v.3.2 (Ronquist et al., 2012). To determine the best-fitting substitution model, the Akaike information criterion implemented in jModelTest, v. 2.1.9, was used. The GTR GAMMA+I model was selected. Markov chain Monte Carlo analysis was run for 1,000,000 generations. The first 25% of the trees were discarded as burn-in, and the remaining trees were used to generate a majority-rule consensus tree. The maximum likelihood (ML) and BI analyses were visualized using FigTree, v. 1.4.35.
Results
Chloroplast Genome Organization
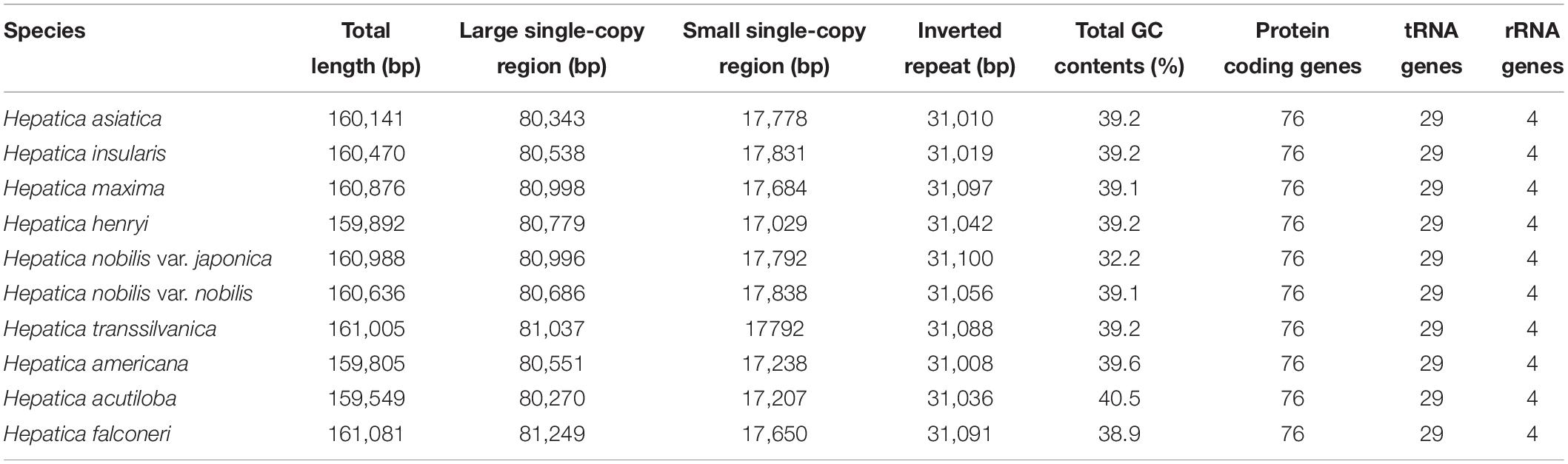
The complete cp genomes of the nine Hepatica taxa ranged from 159,549 bp (H. acutiloba) to 161,081 bp (H. falconeri; Table 1 and Figure 1). The cp genomes had a typical quadripartite structure consisting of LSC 80,270 bp (H. acutiloba) to 81,249 bp (H. falconeri) in length, SSC 17,029 bp (H. henryi) to 17,838 bp (H. nobilis) in length, and two copies of IR 31,008 bp (H. americana) to 31,100 bp (H. nobilis var. japonica) in length, respectively (Table 1). The gene content of Hepatica cp genome was identical in all species: 76 PCGs, 29 tRNAs, and four rRNAs. Of these 109 genes, 56 were related to self-replication (four in rRNAs and 29 in tRNAs), including eight genes related to large subunits and 11 related to small subunits. Forty-three genes were involved in photosynthesis, including six associated with ATP synthase, 11 with NADH dehydrogenase, six with the cytochrome b/f complex, five with the PSI system, 15 with the PSII system, and one with Rubisco. In addition, nine genes were annotated as having other (clpP, ccsA, accD, cemA, and matK) or unknown functions (ycf1, ycf2, ycf3, and ycf4). Fifteen genes had one intron (atpF, ndhA, ndhB, petB, petd, rpl16, rpl2, rpoC1, rps12, trnA-UGC, trnG-GCC, trnI-GAU, trnK-UUU, trnL-UAA, and trnV-UAC), and two had two introns (clpP and ycf3; Supplementary Table 2). The GC contents of Hepatica cp genomes were 32.2–40.5%.
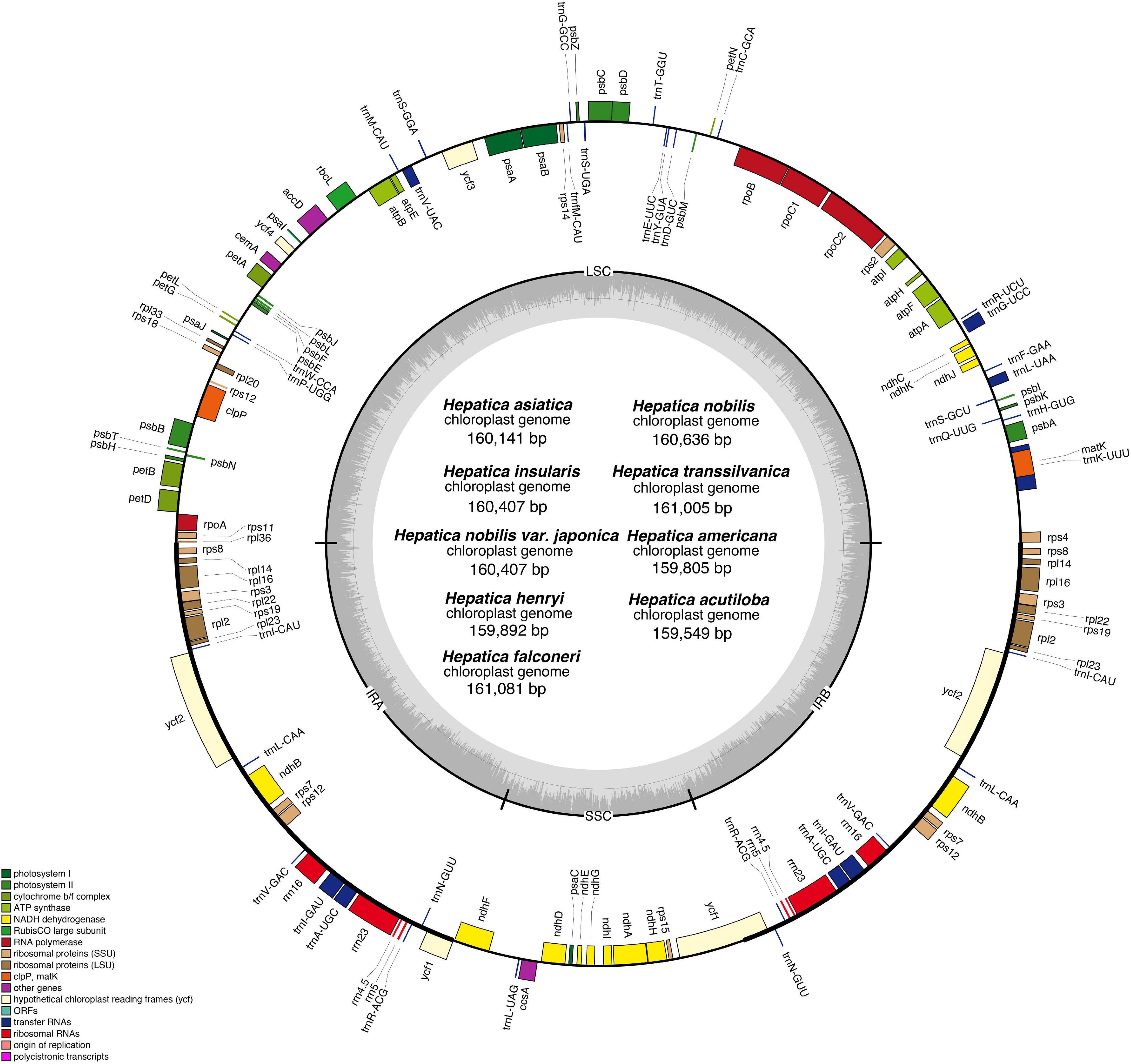
Figure 1. Complete chloroplast genome of genus Hepatica. Genes drawn inside the circle are transcribed clockwise, while the genes drawn outside are counterclockwise. The gray plot in the inner circle corresponds to the GC content.
Comparative Analyses and Nucleotide Substitution Rates
The mVISTA analysis revealed that the cp genomes of Hepatica species were conserved generally across the 10 taxa with a few variable regions, mostly restricted to non-coding regions (Supplementary Figure 1).
The average pi-values were estimated to be 0.00262, with a range from 0 to 0.02074 (Supplementary Figure 2). The most variable region was found in the SSC region with an average pi = 0.0619. The LSC and IR regions were less variable with pi = 0.00323 and 0.00083, respectively. The most variable regions (pi > 0.01) included eight intergenic regions (trnY-trnD, trnG-grnS, trnR-trnN, Ψycf1-ndhF, ndhF-trnL, trnL-ccsA, and rps15-ycf1) and one coding region (ycf1).
The length of the IR region ranged from 31,010 to 31,100 bp, and the gene contents of the IR region were conserved in all Hepatica species (Figure 2). In Hepatica, the LSC/IRa boundary (JLA) was located between rpl36 and ΨinfA, and the LSC/IRb boundary (JLB) was located on rps4. The IRa/SSC and IRb/SSC boundaries (JSA and JSB) were located on ycf1 or between the 5′ ends of truncated ycf1 and ndhF. The IR junction regions of Hepatica species are similar to the Anemoneae species. In Oxygraphis, the IR junctions (JLA and JLB) were located on rpl2, whereas in Anemoneae species, IR regions had been expanded to LSC regions ∼5 kb including ΨinfA. Moreover, the IR/SSC boundaries of all Ranunculaceae were located on ycf1 or between the 5′ ends of truncated ycf1 and ndhF. In this study, the IR expansion event was found to be common to all Anemoneae including Hepatica, and the IR expansion has resulted in the duplication of six genes (rps8, rpl14, rpl16, rps3, rpl22, and rps19).
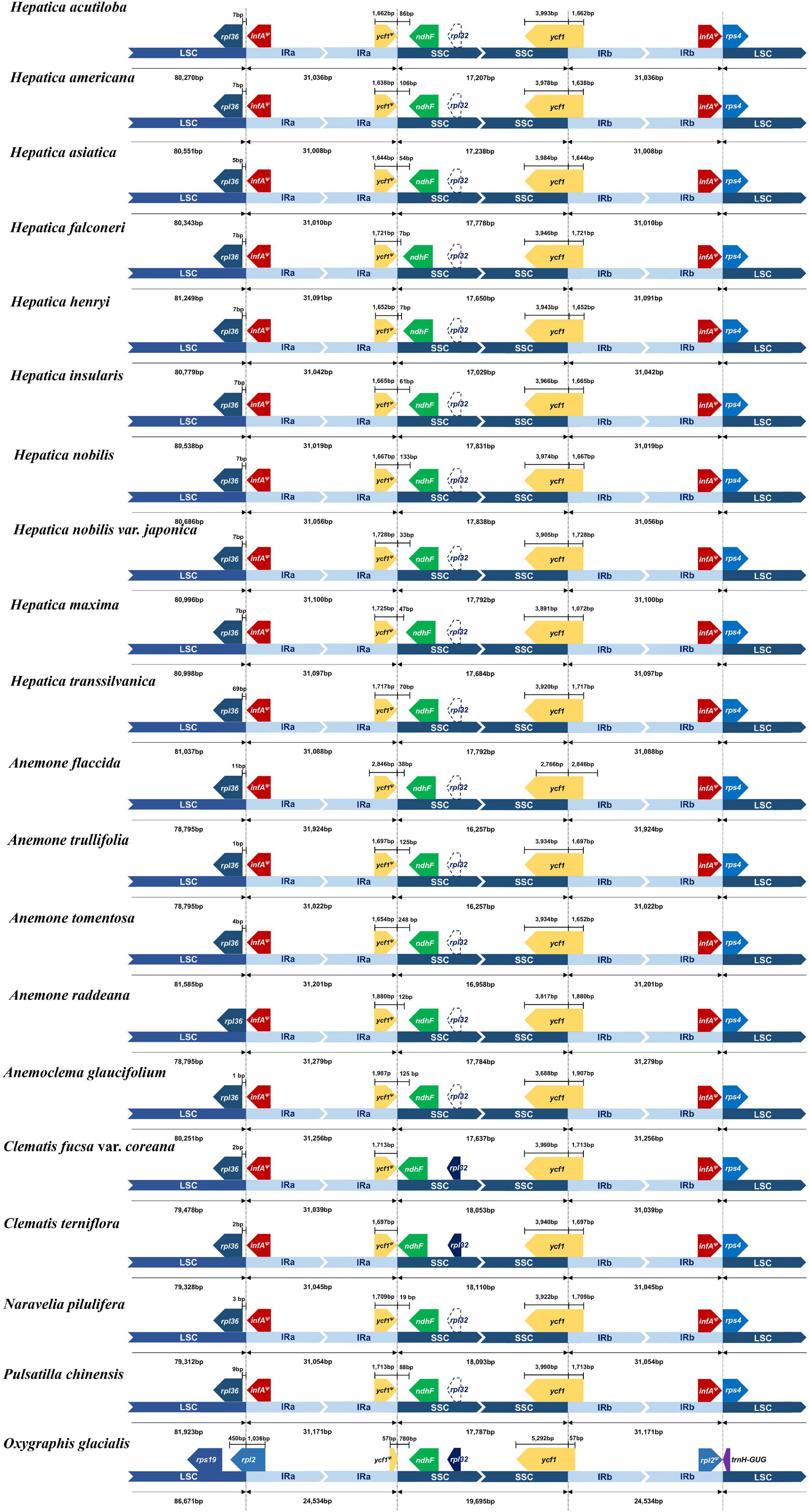
Figure 2. Comparison of the large single-copy region, inverted repeats, and small single-copy region boundaries within tribe Anemoneae.
The dN/dS ratios of most PCGs were less than 1 for all Hepatica species and greater than 1 for rpl20 in H. acutiloba (1.6113), H. americana (1.6113), and H. falconeri (3.5576). The photosynthesis apparatus genes (pet, psa, and psb), ATP synthase gene (atp), and RNA polymerase gene (rpo) had low dN/dS ratios (≤0.5), while atpF and petL in H. falconeri had higher dN/dS ratios (0.7456 and 0.7391, respectively) than in other Hepatica species. The RNA processing gene (matK) and NADH dehydrogenase gene (ndh) showed moderate dN/dS ratios (≤0.67). ndhH and ndhJ had low dN/dS ratios (<0.039). Ribosomal protein genes (rps and rpl) had a wide range of dN/dS ratios (0–3.5576). Most of the rps and rpl genes had moderate dN/dS ratios, and some genes (rps7, rps8, rps11, rps12, rps19, rpl23, and rpl36) had a ratio of 0. The dN/dS of rpl22 was 1.1592 in H. transsilvanica (Supplementary Figure 3 and Supplementary Table 3).
Chloroplast Genome Rearrangements and Gene Loss
Nine LCBs identified through whole-genome alignments were shared by all members of tribe Anemoneae and Oxygraphis (Figure 3 and Supplementary Table 4). In Anemoneae, gene order is conserved within Hepatica and similar to Anemone, Pulsatilla, and Anemoclema. In comparison to Oxygraphis, six rearrangement events were detected in Anemoneae: three inversions (LCB1, LCB2, and LCB4) and three relocations (LCB1, LCB5, and LCB6). Among six rearrangements, Hepatica shared three inversions with Anemone, Pulsatilla, and Anemoclema (LCB1, ∼1.2 kb, including rps4; LCB2, ∼9.1 kb, including trnH-GUG–rps16; LCB4, ∼49 kb, including trnG-UCC–ycf3) and two relocations (LCB1 and LCB5), whereas in Clematis including Naravelia, additional rearrangements, inversion of LCB4, and relocation of LCB5 and LCB6 (∼4.6 kb, including trnL-UAA–ndhC) were identified (Figure 3).

Figure 3. Whole-genome alignment of tribe Anemoneae. Each locally collinear block (LCB) is color-coded and represents a syntenic region. Blocks below the horizontal center line represent inversions relative to the reference (Oxgraphis glacialis). The height of the colored region with a block reflects the average sequence identity relative to the reference. The numbers indicate each LCB number.
We identified two pseudogenes (infA and rps16) and one gene loss (rpl32) in Hepatica. infA was a non-functional structure with a 3′ end truncated across the Anemoneae including Hepatica. The length of the residual infA sequence ranged from 75 to 77 bp (Supplementary Figure 4B). Within Anemoneae, only Hepatica was missing a functional rps16; exon 1 of the gene was present and conserved in all of the Anemoneae; however, 150 bp of intron and exon 2 were deleted across Hepatica species (Supplementary Figure 4A). The rpl32, which is located between ndhF and trnL-UAG, has been completely lost in Hepatica and two Anemone (A. flaccida and A. trullifolia), whereas rpl32 of other Anemoneae was identified as a pseudogene except in Clematis fusca var. coreana (Supplementary Figures 4C, 5).
Phylogenetic Analyses
The total alignment length of the nucleotide dataset was 69,400 bp, and the optimal phylogenetic tree in ML analysis had a likelihood score of ln(L) = −151,170.677. The ML tree and Bayesian tree had similar topologies (Figure 4). Hepatica formed a monophyletic group and is sister to a clade of Anemone trullifolia and A. flaccida (BS/PP = 100/1.00). Anemoclema was sister to the Clematis + Naravelia clade (BS/PP = 100/1.00). Anemone was not monophyletic. A. trullifolia and A. flaccida are closely related to Hepatica, whereas A. tomentosa and A. raddeana form a sister clade to Pulsatilla; the clade consisting of Anemone + Pulsatilla is sister to the Clematis + Naravelia + Anemonclema lineage (BS/PP = 61/0.86). Among Hepatica species, H. falconeri is sister to the rest of the genus. H. asiatica and H. insularis were grouped as a clade with a high support value (BS/PP = 100/1.00). However, H. maxima is sister to H. nobilis with weak support (BS/PP = 62/–). H. nobilis var. japonica was grouped together with H. acutiloba and H. americana with moderate support (BS/PP = 65/.98). H. transsilvanica was sister to the H. nobilis and H. maxima clade.
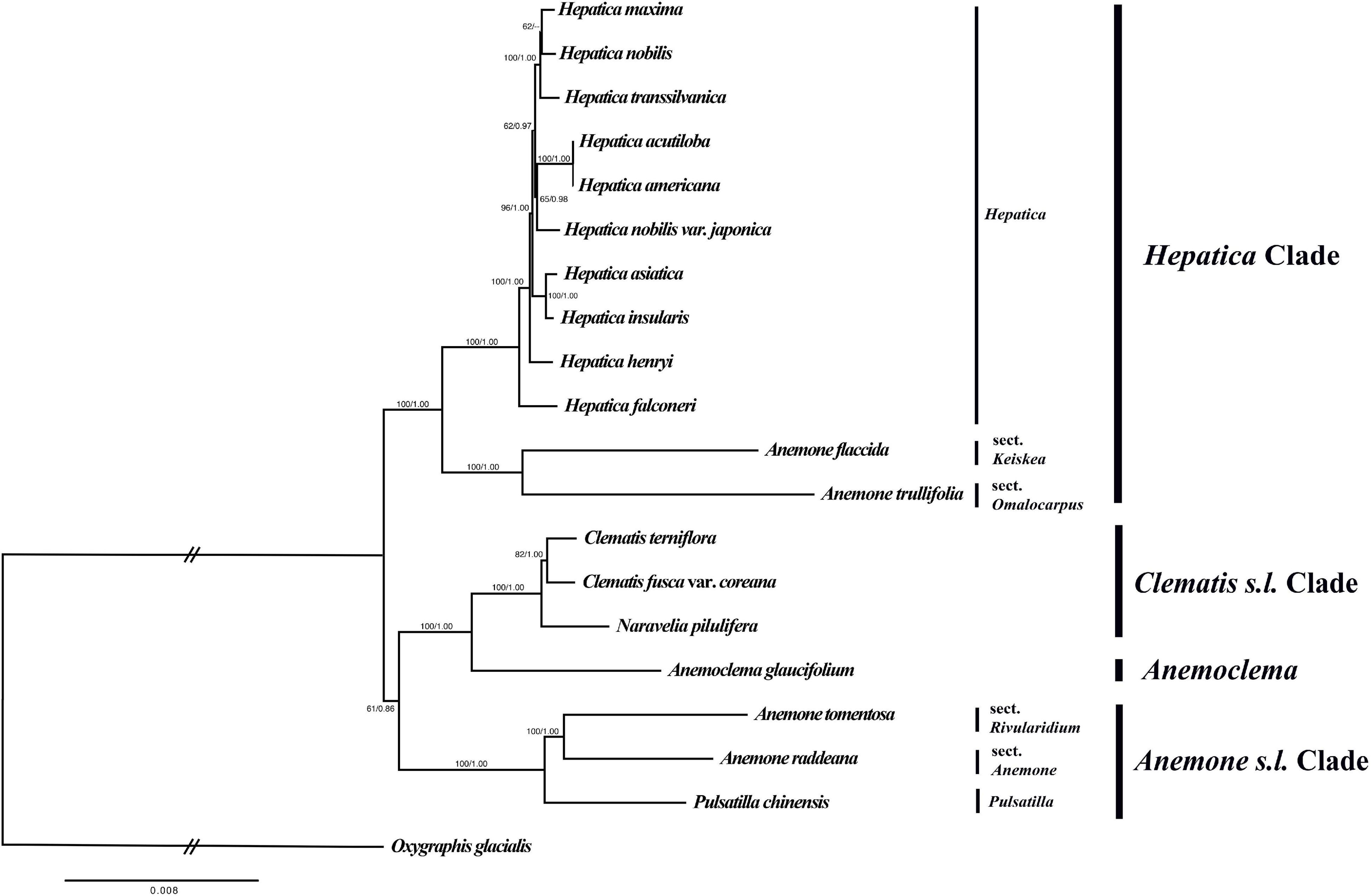
Figure 4. Phylogenetic tree reconstruction of 20 taxa using maximum likelihood based on the concatenated sequence of 76 PCGs. Numbers above the branches indicate bootstrap value and posterior probabilities.
Discussion
Comparative Characteristics of cp Genome for Hepatica and Its Implication
When compared to other closely related taxa, Hepatica has fewer PCGs (76 genes) than other genera (77–78 genes) because of pseudogenization or gene loss of infA, rps16, and rpl32 (Zhai et al., 2019). The loss or pseudogenization of three genes (rps16, rpl32, and infA) in the Ranunculaceae cp genome seems to be the result of parallel evolution (Zhai et al., 2019). The infA was pseudogenized by truncation, and only 77 bp of the 5′ end of the sequence is remaining in the cp genomes of Hepatica and other Anemoneae species. Although pseudogenization of infA appeared in several genera of Ranunculaceae, truncation of infA was found in only the Anemoneae lineage (Supplementary Figure 4). Usually, infA is located in the LSC region in Ranunculaceae, whereas infA of Anemoneae is located on the end of IR/LSC boundaries (Figure 2). Thus, it is suggested that IR expansion into the LSC region leads to the truncation of infA within Anemoneae lineages. The rps16 was identified as a pseudogene by deletion of the second exon and intron. The rps16 pseudogene was also found in only Hepatica among the Anemoneae lineage. The existence of the rps16 pseudogene provides additional molecular evidence that Hepatica is monophyletic. Pseudogenization or gene loss of rps16 has been reported in various lineages, such as Medicago (Fabaceae) and Populus (Downie and Palmer, 1992; Ueda et al., 2008), and some Ranunculaceae with the loss of complete sequence or frameshift deletion (Zhai et al., 2019; Park et al., 2020), Draba (Brassicaceae), and Lobularia (Brassicaceae) with deletion of the first exon or deletion of the second exon and intron (Roy et al., 2010), and Veratrum (Melanthiaceae) with deletion of the second exon and intron (Do and Kim, 2017). The phylogenetic distribution of the rpl32 gene loss shows two patterns: (1) a complete loss of all sequences across the Hepatica clade and (2) pseudogenization with partial sequences or a frameshift across Clematis s.l. + Anemone s.l. clade except Clematis. Meanwhile, both rpl32 pseudogenes and intact genes appeared in Clematis (Liu et al., 2018a, b; He et al., 2019; Zhai et al., 2019). Therefore, rpl32 seems to have undergone a gradual gene loss through deletion. The gene loss of rpl32 has been reported within several lineages of Ranunculaceae (Park et al., 2015; Zhai et al., 2019; Park and Park, 2020). Park et al. (2015) suggested that the reduction of the ndhF and trnL intergenic spacer (IGS) region is associated with the loss or pseudogenization of rpl32. In this study, however, we could not find an affinity between gene loss and length variation of ndhF and trnL IGS.
Non-functional genes in chloroplast are often associated with functional transfer to the nucleus, such as rpl32 in Salicaceae and Ranunculaceae (Ueda et al., 2008; Park et al., 2015, 2020; Zhai et al., 2019), rps16 in Medicago, Salicaceae, Thalictrum, and Delphinineae (Ueda et al., 2008; Park et al., 2015, 2020), and infA in Arabidopsis, Glycine, Solanum, and Mesembryanthemum (Millen et al., 2001). However, further investigations that search for transferred genes in nuclear transcriptomes are needed to resolve the fate of missing cp genes.
Structural rearrangements in the chloroplast genomes have been reported in a variety of seed plants, including a 50-kb inversion in Papilionoideae (Doyle et al., 1996), a 22-kb inversion in Asteraceae (Kim et al., 2005), a 42-kb inversion in Abies (Tsumura et al., 2000), a 21-kb inversion in Jasmineae (Lee et al., 2007), and multiple inversions in Passiflora (Shrestha et al., 2019). We characterized a highly conserved genome structure across Anemoneae including Hepatica except for the Clematis + Naravelia lineage (Figure 3 and Supplementary Figure 1).
Although Hepatica cp genomes have an identical structure to those in related taxa, the structural variation compared with Oxygraphis could indicate an evolutionary history around the tribal level.
The phylogenetic distribution of arrangements suggests that three inversions (LCB1, LCB2, and LCB4) and two relocations (LCB1 and LCB5) occurred in the early Anemoneae. On the other hand, the rearrangements in LCB4, LCB5, and LCB6 occurred independently in the Clematis + Naravelia lineage (Supplementary Figures 5, 6). Repeat analysis identified 30-bp repeats in the flanking regions of LCB4 and LCB5 in Anemoneae, thus suggesting that these inversions may have been repeat-mediated. Based on these results, the structural rearrangement of Hepatica is assumed to have occurred via the following four inversions: (1) inversion of LCB1 to LCB5, (2) inversion of LCB4 and LCB, (3) inversion of LCB2 to LCB5, and (4) the inverted LCB2 (Supplementary Figures 5, 6). The Clematis + Naravelia lineage underwent two additional inversions: inversion of LCB4 to LCB6 and inversion of LCB5 and LCB6. The rearrangements in Anemoneae have been reported (Hoot and Palmer, 1994; Liu et al., 2018b; Park and Park, 2020) as we observed four to six inversion events. In addition, the phylogenomic results suggest that the cp genome structure of the ancestor of Anemoneae might be similar to those of Hepatica, Anemone, Anemoclema, and Pulsatilla (Supplementary Figures 5, 6).
The synonymous (dS) and non-synonymous (dN) substitution rate ratios are valuable for understanding molecular evolution (Drouin et al., 2008). A dN/dS ratio >1, <1, and = 1 indicates positive selection, negative selection, and neutral selection, respectively. Nucleotide substitution rate analyses in the Hepatica cp genome revealed that most cp genes are under negative selection (<1). rpl20 and rpl22 had significantly high dN/dS (>1) in H. falconeri, H. americana, H. acutiloba, and H. transsilvanica. The rpl20 gene in H. falconeri had a particularly high dN/dS ratio (3.5576). Based on this, we presume that natural selection pressure was applied to maintain the protein translation system.
Phylogenetic Relationships
Ulbrich (1906) suggested that Hepatica is divided into two sections based on the crenate lobe: sect. Hepatica with an entire lobe (H. acutiloba, H. americana, H. asiatica, H. falconeri, H. insularis, H. maxima, H. nobilis, and H. nobilis. var. japonica) and sect. Angulosa with a crenate lobe (H. henryi, H. nobilis var. pubescens, and H. transsilvanica). Our phylogenetic tree does not support this classification.
Thomson (1852) described H. falconeri as a species of Anemone; however, uncertainty remains about its generic position in Anemone or Hepatica (Ogisu et al., 2002). Although the leaf shape of H. falconeri resembles that of Anemone, the morphology of the involucral bracts, pistils, and achenes and the karyotype are closer to H. nobilis (Ogisu et al., 2002). According to our study, H. falconeri is an early branching species (Figure 4) that features the rps16 pseudogene, which is only found in the Hepatica lineage. Thus, our data support H. falconeri as falling into the genus Hepatica.
Among Asian Hepatica, H. asiatica is sister to H. insularis. Interestingly, H. maxima, a species endemic to Uleung Island, South Korea, is sister to European Hepatica (H. nobilis and H. transsilvanica) rather than Asian Hepatica. Previous studies suggested that H. maxima originated from populations of H. asiatica (Pfosser et al., 2011). However, in contrast with previous results, our phylogenetic analysis shows that H. maxima is close to H. nobilis.
On the contrary, H. nobilis var. japonica, an endemic to Japan, is phylogenetically close to the North American Hepatica. H. nobilis var. japonica was previously classified as H. acutiloba before Nakai (1937b), who identified it as a variety of H. nobilis based on the shapes of its lobes and bracts. In contrast, Zonneveld (2010) demonstrated that H. nobilis var. japonica is very similar to H. asiatica in genome size and geographically separated from Europe. He also suggested that H. nobilis var. japonica should be treated as a subspecies of H. asiatica (Zonneveld, 2010). Our phylogenetic analysis shows that H. nobilis var. japonica needs to be elevated to species level rather than treated as a subspecies of H. asiatica. H. nobilis var. japonica is closer to North American Hepatica than it is to H. nobilis. However, we could not include H. nobilis var. pubescens, a Japanese endemic, in this study. To evaluate the classification position of H. nobilis var. japonica, the relationship between the two Japanese endemics should be investigated further.
In this study, the Hepatica is sister to A. flaccida (sect. Keiskea) and A. trullifolia (sect. Omalocarpus), whereas Pulsatilla is sister to A. raddeana (sect. Anemone) and A. tomentosa (sect. Rivularidium). The Pulsatilla + Anemone clade is close to Clematis (including Naravelia) and Anemoclema (Figure 4). These results are similar to those based on another plastid dataset (Jiang et al., 2017). However, Liu et al. (2018b) found that the Hepatica + sect. Omalocarpus clade was sister to Clematis + Anemoclema. Although the topological incongruence was found previously, Anemoneae was divided into three major clades in common. The first clade is subgenus Anemonidium of genus Anemone including Anemonidium, Omalocarpus, Keiskea, and Hepatica. The second clade is the subgenus Anemone of genus Anemone including Anemone, Barneoudia, Knowltonia, Oreithales, Pulsatilla, and Pulsatilloides. The last clade is Anemonclema and Clematis s.l., including Archiclematis, Clematis, and Naraverilia. Based on the nrITS and atpB-rbcL dataset, phylogenetic analyses recovered the monophyly of Anemone s.l. (Hoot et al., 2012; Jiang et al., 2017), whereas five plastid datasets (atpB-rbcL, matK, psbA-trnQ, rbcL, and rpoB-trnC) revealed the paraphyly of Anemone s.l. (Jiang et al., 2017; Liu et al., 2018b; in this study). According to our study, Anemone s.l. is paraphyletic, and our result did not support the classification by Hoot et al. (2012), which placed Hepatica into Anemone. Thus, the subgenus Anemonidium needs to be separated as an independent genus, Hepatica, as suggested by Jiang et al. (2017) and Liu et al. (2018b).
Conclusion
In this study, we sequenced and analyzed the cp genome of nine species of Hepatica. The comparative analyses revealed distinct characters of Hepatica cp genomes. Even though the cp genome size, genome structure, and gene contents of Hepatica were similar to those of other Anemoneae cp genomes, the IR expansion and gene loss or pseudogene demonstrate the evolutionary history of the genus and its relatives. We resolved the monophyly of Hepatica and found that some taxa need to be reassessed in tribe Anemoneae. Our results supported that H. nobilis var. japonica is not closely related to H. nobilis and indicated that this taxon needs to be promoted to species level. We identified that Anemone s.l. was paraphyletic and recommended that Anemone s.l. should be reclassified.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
KP and SP conceived and designed the experiments and modified the manuscript. KP performed the experiments, analyzed the data, and prepared a draft of the manuscript and figures. Both authors read and approved the final manuscript.
Funding
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2015R1D1A3A01020009).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Akhtar H. Malik at University of Kashmir (CBI) for kindly providing H. falconeri samples.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.638580/full#supplementary-material
Footnotes
- ^ http://www.phyzen.com
- ^ http://lowelab.ucsc.edu/tRNAscan-SE/
- ^ https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi
- ^ http://ogdraw.mpimp-golm.mpg.de/
- ^ http://tree.bio.ed.ac.kr/softwar/figtree/
References
Allen, G., Flores-Vergara, M., Krasynanski, S., Kumar, S., and Thompson, W. (2006). A modified protocol for rapid DNA isolation from plant tissues using cetyltrimmethylammonium bromide. Nat. Protoc. 1, 2320–2325. doi: 10.1038/nprot.2006.384
Chang, C., Lin, H., Lin, I., Chow, T., Chen, H., Chen, W., et al. (2006). The chloroplast genome of Phalaenopsis aphrodite (Orchidaceae): comparative analysis of evolutionary rate with that of grasses and its phylogenetic implications. Mol. Biol. Evol. 23, 279–291. doi: 10.1093/molbev/msj029
Darling, A., Mau, B., Blattner, F., and Perna, N. (2004). Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14, 1394–1403. doi: 10.1101/gr.2289704
Do, H. D. K., and Kim, J.-H. (2017). A dynamic tandem repeat in monocotyledons inferred from a comparative analysis of chloroplast genomes in Melanthiaceae. Front. Plant Sci. 8:693. doi: 10.3389/fpls.2017.00693
Downie, S., and Palmer, J. (1992). “Use of chloroplast dna rearrangements in reconstructing plant phylogeny,” in Molecular Systematics of Plants, eds P. S. Soltis, D. E. Soltis, and J. J. Doyle (Boston, MA: Springer). doi: 10.1007/978-1-4615-3276-7_2
Doyle, J., Doyle, J., Ballenger, J., and Palmer, J. (1996). The distribution and phylogenetic significance of a 50-kb chloroplast DNA inversion in the flowering plant family Leguminosae. Mol. Phylogenet. Evol. 5, 429–438. doi: 10.1006/mpev.1996.0038
Drouin, G., Daoud, H., and Xia, J. (2008). Relative rates of synonymous substitutions in the mitochondrial, chloroplast and nuclear genomes of seed plants. Mol. Phylogenet. Evol. 49, 827–831. doi: 10.1016/j.ympev.2008.09.009
Ehrendorfer, F. (1995). Evolutionary trends and patterns in the Anemoninae. Plant Syst. Evol. Suppl. 9, 283–293.
Ehrendorfer, F., and Samuel, R. (2001). Contributions to a molecular phylogeny and systematics of Anemone and related genera (Ranunculaceae-Anemoniae). Acta Phytotaxonomica Sin. 39, 293–307.
Frazer, K., Pachter, L., Poliakov, A., Rubin, E., and Dubchak, I. (2004). VISTA: computational tools for comparative genomics. Nucleic Acids Res. 32(Suppl. 2), W273–W279. doi: 10.1093/nar/gkh458
He, J., Yao, M., Lyu, R. D., Lin, L. L., Liu, H. J., Pei, L. Y., et al. (2019). Structural variation of the complete chloroplast genome and plastid phylogenomics of the genus Asteropyrum (Ranunculaceae). Sci. Rep. 9:15285. doi: 10.1038/s41598-019-51601-2
Hiroe, M. (1957). A cytoxanomical study on Anemone hepatica L. (Ranunculaceae) of Japan. Bot. Mag. Tokyo 70, 4–7. doi: 10.15281/jplantres1887.70.4
Hoot, S. (1995). Phylogenetic relationships in Anemone (Ranunculaceae) based on DNA restriction site variation and morphology. Plant Syst. Evol. 9(Suppl. 9), 285–300. doi: 10.1007/978-3-7091-6612-3
Hoot, S., Meyer, K., and Manning, J. C. (2012). Phylogeny and reclassification of Anemone (Ranunculaceae), with an emphasis on austral species. Syst. Bot. 37, 139–152. doi: 10.1600/036364412X616729
Hoot, S., and Palmer, J. (1994). Structural rearrangements, including parallel inversions, within the chloroplast genome of Anemone and related genera. J. Mol. Evol. 38, 274–281. doi: 10.1007/BF00176089
Jalas, J., and Suominen, J. (1976). “Atlas florae Europeae3,” in Salicaceae to Balanophoraceae, eds J. Jalas and J. Suominen (Helsinki: Committee for Mapping the Flora of EuropeJ).
Jansen, R., and Ruhlman, T. (2012). “Plastid genomes of seed plant,” in Genomics of Chloroplasts and Mitochondira, eds R. Bock and V. Knoop (Dordrecht: Springer), 103–126.
Jiang, M., Chen, H., He, S., Wang, L., Chen, A. J., and Liu, C. (2018). Sequencing, characterization, and comparative analyses of the plastome of Caragana rosea var. rosea. Int. J. Mol. Sci. 19:1419. doi: 10.3390/ijms19051419
Jiang, N., Zhou, Z., Yang, J., Zhang, S., Guan, K., Tan, Y., et al. (2017). Phylogenetic reassessment of tribe Anemoenae (Ranunculaceae): non-monophyly of Anemone s.l. revealed by plastid datasets. PLoS One 12:e0174792. doi: 10.1371/journal.pone.0174792
Johansson, J. T. (1999). There larege inversions in the chloroplast genomes and one loss of the chloroplast gene rps16 suggest an early evolutionary split in the genus Adonis (Ranunculaceae). Plant Syst. Evol. 218, 133–143. doi: 10.1007/BF01087041
Katoh, K., and Standley, D. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kim, K., Choi, K., and Jansen, R. (2005). Two chloroplast DNA inversions originated simultaneously during the early evolution of the sunflower family (Asteraceae). Mol. Biol. Evol. 22, 1783–1792. doi: 10.1093/molbev/msi174
Kurita, M. (1955). Chromosome studies in Ranunculaceae. I. Karyotypes of the subtribed Anemoninae. Rep. Biol. Inst. Ehime Univ. 1, 1–10.
Lee, H., Jansen, R., Chumley, T., and Kim, K. (2007). Gene relocations within chloroplast genomes of Jasminum and Menodora (Oleaceae) are due to multiple, overlapping inversions. Mol. Biol. Evol. 24, 1161–1180. doi: 10.1093/molbev/msm036
Lehtonen, S., Christenhusz, M., and Falck, D. (2016). Sensitive phylogenetics of Clematis and its position in Ranunculaceae. Bot. J. Linn. Soc. 182, 825–867. doi: 10.1111/boj.12477
Li, M., Yang, B., Chen, Q., Zhu, W., Ma, J., and Tian, j. (2016). The complete chloroplast genome sequence of Clematis terniflora DC. (Ranunculaceae). Mitochondrial DNA A DNA Mapp. Seq. Anal. 27, 2470–2472. doi: 10.3109/19401736.2015.1033702
Librado, P., and Rozas, J. (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452. doi: 10.1093/bioinformatics/btp187
Liu, H., Ding, C., He, J., Cheng, J., Pei, L., and Xie, L. (2018a). Complete chloroplast genomes of Archiclematis, Naravelia and Clematis (Ranunculaceae), and their phylogenetic implications. Phytotaxa 343, 214–226. doi: 10.11646/phytotaxa.343.3.2
Liu, H., He, J., Ding, C., Lyu, R., Pei, L., Cheng, J., et al. (2018b). Analysis of complete chloroplast genomes of Anemoclema, Anemone, Pulsatilla, and Hepatica revealing structural variations among genera in Tribe Anemoneae (Ranunculaceae). Front. Plant Sci. 9:1097. doi: 10.3389/fpls.2018.01097
Lohse, M., Drechsel, O., Kahlau, S., and Bock, R. (2013). OrganellarGenomeDRAW–a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41, W575–W581. doi: 10.1093/nar/gkt289
Lou, R., Liu, B., Xie, Y., Huang, W., Yuan, J., He, G., et al. (2012). SOAPdeonvo2: an empirically improved memory-efficient shrot-read de novo assembler. Gigascience 1:18. doi: 10.1186/2047-217X-1-18
Lowe, T., and Chan, P. (2016). tRNAscan-SE On-line: search and contextual analysis of transfer RNA genes. Nucleic Acids Res. 44, W54–W57. doi: 10.1093/nar/gkw413
Meusel, H., Jäger, E., and Weinert, S. (1965). Vergleichende Chorologie der Zentraleuropäischen Flora 1. Jena: Gustav Fischer.
Meyer, K., Hoot, S., and Arroyo, M. (2010). Phylogenetic affinities of South American Anemone (Ranunculaceae), including the endemic segregate genera, Barneoudia and Oreithales. Int. J. Plant Sci. 171, 323–331. doi: 10.1086/650153
Mikeda, O., Kita, K., Handa, T., and Yukawa, T. (2006). Phylogenetic relationships of Clematis (Ranunculaceae) based on chloroplast and nuclear DNA sequences. Bot. J. Linn. Soc. 152, 153–168. doi: 10.1111/j.1095-8339.2006.00551.x
Millen, R., Olmstead, R., Adams, K., Palmer, J., Lao, N., Heggie, L., et al. (2001). Many parallel losses of infA from chloroplast DNA during angiosperm evolution with multiple independent transfers to the nucleus. Plant Cell 13, 645–658. doi: 10.2307/3871412
Ogisu, M., Awan, M. R., Mabuchi, T., and Mikanagi, Y. (2002). Morphology, phenology and cytology and Hepatica falconeri in Pakistan. Kew Bull. 57, 943–953. doi: 10.2307/4115724
Palmer, J. (1985). Comparative organization of chloroplast genomes. Annu. Rev. Genet. 19, 325–354. doi: 10.1146/annurev.ge.19.120185.001545
Palmer, J. (1991). “Plastid chromosomes: structure and evolution,” in The Molecular Biology of Plastids. Cell Culture and Somatic Cell Genetics of Plants, ed. R. G. Hermann (Vienna: Springer), 5–53. doi: 10.1016/b978-0-12-715007-9.50009-8
Park, K., and Park, S. J. (2016). Complete chloroplast genome of Clematis fusca var. coreana (Ranunculaceae). Mitochondrial DNA A DNA Mapp. Seq. Anal. 27, 4056–4058. doi: 10.3109/19401736.2014.1003841
Park, S., An, B., and Park, S. J. (2020). Recurrent gene duplication in the angiosperm tribe Delphinieae (Ranunculaceae) inferred from intracellular gene transfer events and heteroplasmic mutations in the mutations in the plasmid matK gene. Sci. Rep. 10:2720. doi: 10.1038/s41598-020-59547-6
Park, S., Jansen, R., and Park, S. J. (2015). Complete plastome sequence of Thalictrum coreanum (Ranunculaceae) and transfer of the rpl32 gene to the nucleus in the ancestor of the subfamily Thalictroideae. BMC Plant Biol. 15:40. doi: 10.1186/s12870-015-0432-6
Park, S., and Park, S. J. (2020). Large-scale phylogenomics reveals ancient introgression in Asian Hepatica and new insights into the origin of the insular endemic Hepatica maxima. Sci. Rep. 10:16288. doi: 10.1038/s41598-020-73397-2
Patel, R., and Jain, M. (2012). NGS QC toolkit for quality control of next generation sequencing data. PLoS One 7:e30619. doi: 10.1371/journal.pone.0030619
Pfosser, M., Sun, B.-Y., Stuessy, T., Jang, C.-G., Guo, Y.-P., Kim, T., et al. (2011). Phylogeny of Hepatica (Ranunculaceae) and origin of Hepatica maxima Nakai endemic to Ullung Island, Korea. Stapfia 95, 16–27.
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D., Darling, A., Hohna, S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Roy, S., Ueda, M., Kadowaki, K., and Tsutsumi, N. (2010). Different status of the gene for ribosomal protein S16 in the chloroplast genome during evolution of the genus Arabidopsis and closely related species. Genes Genet. Syst. 85, 319–326. doi: 10.1266/ggs.85.319
Shishkin, B. K. (1937). “Ranales and Rhoeadales,” in Flora of the U.S.S.R, Vol. 7, ed. B. K. Shishkin (Moskva: Izdatel’stvo Akademii Nauk SSSR), 219–221.
Shrestha, B., Weng, M., Theriot, E., Gilbert, L., Ruhlman, T., Krosnick, S., et al. (2019). Highly accelerated rates of genomic rearrangemnets and nucleotide substitutions in plastid genomes of Passiflora subgeneus Decaloba. Mol. Phylogenet. Evol. 138, 53–64. doi: 10.1016/j.ympev.2019.05.030
Szczecińska, M., and Sawicki, J. (2015). Genomic resources of three Pulsatilla species reveal evolutionary hotspots, species-specific sites and variable plastid structure in the family Ranunculaceae. Int. J. Mol. Sci.16, 22258–22279. doi: 10.3390/ijms160922258
Tamura, M. (1995). “Angiospermae: ordnung Raanunculales, Fam. Ranunculaceae,” in Die Natürlichen Pflanzenfamilien, 2nd Edn. Vol. 17aIV, eds A. Engler, K. Prantl, and P. Hiepko (Berlin: Duncker & Humblot), 349–351.
Tillich, M., Lehwark, P., Pellizzer, T., Ulbricht-Jones, E., Fischer, A., Bock, R., et al. (2017). GeSeq –versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45, W6–W11.
Timmis, J., Ayliffer, M., Huang, C., and Martin, W. (2004). Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat. Rev. Genet. 5, 123–135. doi: 10.1038/nrg1271
Tsumura, Y., Suyama, Y., and Yoshimura, K. (2000). Chloroplast DNA inversion polymorphism in populations of Abies and Tsuga. Mol. Biol. Evol. 17, 1302–1312. doi: 10.1093/oxfordjournals.molbev.a026414
Ueda, M., Nishikawa, T., Fujimoto, M., Takanashi, H., Arimura, S., Tsutsumi, N., et al. (2008). Substitution of the gene for chloroplast RPS16 was assisted by generation of a dual targeting signal. Mol. Biol. Evol. 25, 1566–1575. doi: 10.1093/molbev/msn102
Wang, W. (1980). “Ranuculaceae(2),” in Flora Reipublicae Popularis Sinicae, Vol. 28, ed. W. T. Wang (Beijing: Science Press), 56–60.
Wang, W., Lu, A., Ren, Y., Endress, M., and Chen, Z. (2009). Phylogeny and classification of Ranunculales: evidence from four molecular loci and morphological data. Perspect. Plant Ecol. Evol. Syst. 11, 81–110. doi: 10.1016/j.ppees.2009.01.001
Woo, H., Kim, J., and Lee, N. (2002). Morphological and isozyme divergence in Korean Hepatica sensu stricto (Ranunculaceae). Plant Syst. Evol. 236, 33–44.
Xie, L., Wen, J., and Li, L. (2011). Phylogenetic analyses of Clematis (Ranunculaceae) based on sequences of nuclear ribosomal ITS and three plastid regions. Syst. Bot. 36, 907–921. doi: 10.1600/036364411X604921
Zhai, W., Duan, X., Zhang, R., Guo, C., Li, L., Xu, G., et al. (2019). Chloroplast genomic data provide new and robust insights into the phylogeny and evolution of the Ranunculaceae. Mol. Phylogenet. Evol. 135, 12–21. doi: 10.1016/j.ympev.2019.02.024
Zhang, Y., Kong, H., and Yang, Q. (2015). Phylogenetic relationships and taxonomic status of the monotype Chinese genus Anemoclema (Ranunculaceae). Plant Syst. Evol. 301, 1335–1344. doi: 10.1007/s00606-014-1160-3
Keywords: chloroplast genome, inversion, pseudogenization, phylogenetic analyses, gene loss, rearrangement
Citation: Park KT and Park S (2021) Phylogenomic Analyses of Hepatica Species and Comparative Analyses Within Tribe Anemoneae (Ranunculaceae). Front. Plant Sci. 12:638580. doi: 10.3389/fpls.2021.638580
Received: 07 December 2020; Accepted: 01 April 2021;
Published: 04 June 2021.
Edited by:
Carl J. Rothfels, University of California, Berkeley, United StatesReviewed by:
Samuli Lehtonen, University of Turku, FinlandKeir Wefferling, University of Wisconsin–Green Bay, United States
Copyright © 2021 Park and Park. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: SeonJoo Park, sjpark01@ynu.ac.kr
 Kyu Tae Park
Kyu Tae Park SeonJoo Park
SeonJoo Park