- 1College of Plant Protection, Yangzhou University, Yangzhou, China
- 2College of Horticulture and Landscape Architecture, Yangzhou University, Yangzhou, China
Introduction: Brown planthopper (BPH), Nilaparvata lugens Stål (Hemiptera: Delphacidae), is a major rice pest causing significant damage to rice throughout the world. Intensive pesticide usage often causes resistance in these seasonal pests, mainly through the modulation of antioxidant machinery. The superoxide dismutase (SOD) gene family is known for regulating BPH response to pesticides.
Methods: In the present study, we identified eight NlSOD genes from the NCBI using the BLASTP program. The bioinformatics analysis includes a phylogenetic tree, conserved domain, motifs, gene ontology (GO) analysis, Kyoto encyclopedia of genes and genomes (KEGG) pathways, and protein-protein interaction, highlighting the distinctive functional elements of NlSOD genes.
Results and discussion: Additionally, the NlSOD genes showed expression in all developmental stages of BPH. Under three sugars (glucose, sucrose, and trehalose) treatment, the respective upregulation of NlSOD8, NlSOD6, and NlSOD2 was noted. The NlSOD1 induced significantly under jinggamycin (JGM) deduced its potential as a key regulator of BPH response to the pesticide. Our study has provided detailed knowledge of the NlSOD gene family in-silico analysis and the defensive response to insecticide and high sugar of BPH. We hope the results of this research will help to shed light on the resistance of BPH towards insecticide toxicity and high sugar and help to control it more efficiently.
1 Introduction
Aerobic organisms thrive on oxygen through mitochondrial machinery in order to sustain life. Some of these oxygen molecules are converted into reactive oxygen species (ROS) during metabolic processes (Atalay et al., 2020). Exposure of tissues and cells to radiation (ultraviolet rays) and chemical agents also produces ROS (Lee et al., 2019; Qureshi et al., 2021b). ROS, including superoxide anion (O2−) and hydrogen peroxide (H2O2), react nonspecifically with various substances and damage cells (Hermes-Lima and Zenteno-Savı́n, 2002; Fukai and Ushio-Fukai, 2011). Aerobic organisms have antioxidant enzymes that scavenge or remove ROS to prevent such oxidative damage. SOD (EC 1.15.1.1) is one such antioxidant enzyme that catalyzes the dismutation of superoxide anions into molecular O2 and H2O2. Thus, SOD breaks down superoxide anions generated in cells. It plays an important role in protecting organisms from oxidative damage (Valentine and Hart, 2003a; Fukai and Ushio-Fukai, 2011). SOD is a metalloenzyme constituting three types: cytosolic Cu and Zn-containing (Cu/Zn-SOD), mitochondrial Mn-containing (Mn-SOD), and extracellular (Ec-SOD) SOD (Okado and Fridovich, 2001; Yamamoto et al., 2005).
The SOD gene family provides the first line of defense against ROS primarily generated by aerobic metabolism (Parker et al., 2004). SOD enzymes transform negatively charged oxygen molecules into hydrogen peroxide, which catalase then destroys. Not only are ROS extremely harmful to cellular components, but research is increasingly identifying ROS as one of the primary mediators of aging processes (Sohal et al., 2002; Hasty et al., 2003; Sharif et al., 2022). This protective role of SOD is prevalent in both eukaryotic and prokaryotic organisms (Sharif et al., 2022). Thus, it’s quite possible that some major SOD gene families are also implicated in insect body plans (Bordo et al., 1994; Zelko et al., 2002).
There are three basic families of animal SOD enzymes with two possible metal configurations in the active sites using either Mn or Cu and Zn ions (Bordo et al., 1994). The Mn-SOD is restricted to the inner matrix of the mitochondria (Okado-Matsumoto and Fridovich, 2001) and is very different in structure from the Cu/Zn-SOD. Of the two Cu/Zn-SOD, one gene product is primarily localized in the cytoplasm (Crapo et al., 1992), with some also occurring in the mitochondrial intermembrane space (Okado-Matsumoto and Fridovich, 2001). This gene is usually referred to as the cytoplasmic Cu/Zn-SOD. The Cu/Zn-SOD is predominantly extracellular, although it has also been shown to be membrane-bound (Folz et al., 1997; Fujii et al., 1998). When this second SOD, termed extracellular Cu/Zn-SOD, is secreted rather than routed to the membrane, it can be reabsorbed and transported inside the nucleus, where it protects genomic DNA (Ookawara et al., 2002) and slows telomere shortening (Von Zglinicki, 2002; Serra et al., 2003). The extracellular Cu/Zn-SOD gene is distinguished from the cytoplasmic Cu/Zn-SOD gene by having an N-terminal signal cleavage peptide that routes the extracellular Cu/Zn-SOD for secretion (Folz et al., 1997; Fujii et al., 1998).
In recent decades, rice consumption has increased globally. A statistical analysis of rice consumption showed a high increase. In the 2008/2009 crop year, 437.18 million metric tons (MMTs) of rice were consumed; compared to 2021/2022, 509.87 MMTs worldwide (Shahbandeh, 2021). Although the rice crop gets affected by a number of insect pests, however, BPH is one of the most destructive insects of rice (Hu et al., 2014). Rice is directly affected by BPH infestations, which also disseminate several viral pathogens, such as the rice grassy stunt virus (RGSV) and rice black-streaked dwarf virus (RBSDV) (Zhou et al., 2008; Nguyen et al., 2015). Host plant resistance is the best approach for mitigating BPH; however, pesticide measures are at the top of the list because of their broad availability and expeditious results (Jena et al., 2006). JGM is a fungicide utilized for the treatment of rice fields for the disease Rhizoctonia solani, which leads to rice sheath blight (Jiang et al., 2012). BPH’s flight ability, body weight, thermo-tolerance, protein and lipid contents, and reproduction are all reportedly impacted by the JGM (Ge et al., 2019; Ahmad et al., 2022b).
The pesticide applications on pest population notoriety remain an important research area. The JGM fungicide, a product of Streptomyces var. jinggangen, is used for controlling rice sheath blight R. solani commercially used in BPH-threatened areas in China (Peng et al., 2014). JGM is commonly applied two to three times in the rice growing season at a commercial level of 125–175 g.a.i. ha-1 through the foliar spraying method. As experienced with insecticides, the JGM sprayed leaf enhances the BPH survival rate and fecundity (Wu et al., 2001; Jiang et al., 2012). JGM is generally overused due to its low toxicity and low impact on biodiversity. SOD studies in insects are comparatively rare compared to plants and mammals. However, some of the genes have been studied in insects, such as SOD1 in B. mori (Yamamoto et al., 2005a) and Hyphantria cunea (Yamamoto et al., 2007), as well as SOD2 in B. mori and NlSOD1 in N. lugens (Yamamoto and Yamaguchi, 2022). In the current study, we have systematically characterized the NlSOD gene family in BPH. Further, the gene structure, conserved motifs, protein-protein interactions, KEGG enrichment pathways, GO analysis, and the expression analysis of SOD genes in response to high sugar treatment and JGM have been studied. We believe our work will contribute to future functional studies regarding pesticide resistance in BPH and minimize the overuse of toxic pesticides on rice crops.
2 Materials and methods
2.1 Identification of NLSOD gene family members in N. lugens
To obtain insights into the NlSOD gene family, the protein sequences of N. lugens were retrieved from the publically available National Center for Biotechnology (NCBI) (http://www.ncbi.nlm.nih.gov/cdd/) and eight NlSOD genes were identified. For the phylogenetic relationship of the NlSOD gene family, 8 NlSOD, Drosophila melanogaster (3 DmSOD), Laodelphax striatellus (3 LsSOD), Scirpophaga incertulas (4 SiSOD), Spodoptera frugiperda (4 SfSOD) proteins sequences were downloaded from the NCBI online database. Furthermore, To avoid the possible loss of a single NlSOD protein due to the fact of a missing domain, a local BLASTP with a 1E-5 cutoff was performed. The Conserved Domain Database (CDD) of the NCBI (http://www.ncbi.nlm.nih.gov/cdd/) (accessed on 14 January 2023), SMART database (http://smart.embl-heidelberg.de/) (accessed on 18 January 2023) (Schultz et al., 1998), Inter Pro Scan program (https://www.ebi.ac.uk/interpro/) (accessed on 18 January 2023), and Scan Prosite (https://prosite.expasy.org/scanprosite/) (accessed on 20 January 2023) was used to confirm the NlSOD gene family-specific domains. Additionally, the physiochemical properties of the NlSOD proteins in the studied species were discovered using the ExPASy online server (http://web.expasy.org/protparam/) following the same protocol as the previous study of Sohail et al. (2022).
2.2 NlSOD gene family phylogenetic relationship, conserved domain, motif distribution, and GO analysis
To obtain detailed knowledge regarding the evolutionary relationship of the NlSOD gene family in N. lugens with the counter species. Herein, we investigated the phylogenetic relationships of the NlSOD gene family members of N. lugens with the developed genome D. melanogaster and the other three insects species (L. striatellus, S. incertulas, and S. frugiperda). Firstly, all the mentioned insect’s SOD amino acid sequences were downloaded from NCBI and were then aligned through ClustalW software (version 2.1) (http://www.genome.jp/tools/clustalw/) (accessed on 13 February 2023) following the default parameters to examine the evolutionary relationships among the sequences and construct the maximum likelihood (ML) phylogenetic tree using MEGA7 (version 7.0) (Kumar et al., 2016). For the reliability of clades, bootstrap with 1,000 replicates was used.
Domain architecture of identified NlSOD proteins was performed by using the full-length protein sequence to CDD NCBI (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) (accessed on 23 February 2023) and downloaded hit data subjected to DOG software (version 2.0) (visualization of protein domain structure) for domains visualization and finally for better graphical representation the Microsoft PowerPoint 365 was used.
Furthermore, the conserved motifs of the NlSOD gene family were predicted using the MEME online server (Version 4.12.0) (http://meme-suite.org/) (accessed on 23 February 2023) with the default settings. The details of the top 5 predicted motifs were obtained from the MEME suite and then cross-checked with NCBI-CDD (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) (accessed on 23 February 2023). Finally, the motif distribution was drawn via Microsoft PowerPoint 365 software.
The obtained NlSOD protein sequences were implied in the online server “CELLO2GO” (http://cello.life.nctu.edu.tw/cello2go/alignment.php) to determine the predicted functions, such as molecular functions, biological processes, and cellular components. Finally, the GO classifications were recovered using GraphPad Prism (Version 9.4.1) (GraphPad Software, Inc., LA Jolla, CA, United States) was used for graphical representation.
2.3 Protein interaction network construction
The online server String (version 11.5) (https://string-db.org) designed by the Swiss Institute of Bioinformatics (accessed on 25 February 2023) was used for the protein-protein interaction (PPI), and the network was recovered using the NlSOD1 protein as our reference protein. The obtained results were based on the available sources in the string database, including text mining co-expression, determined, co-occurrence, intersection, curated databases, and fusion (Szklarczyk et al., 2015). The pathway enrichment analysis was also conducted by searching for NlSOD genes in the NCBI database’s online pathway enrichment tool.
2.4 Insect rearing, stress treatment, and expression profiling of the NlSOD genes
The insects BPH and the rice verity Ninjing4 were used in the study, obtained from China National Rice Research Institute (Hangzhou, China). The Ninjing4 rice variety, known to be preferred because of no resistance to BPH infestations, was used for rearing. Initially, the BPH colonies were reared on the rice seedlings in cement tanks covered with fine mesh outdoors (i.e., in natural conditions) for 6 months (i.e., April to October) and overwintered in lab-controlled conditions.
First, we soaked the seeds for 24 h in a water dip plastic tray with a standard size of one-quarter (60 cm H_100 cm W_200 cm L) in lab standard conditions of 26°C ± 2°C with the 16 h L:8 h D in relative humidity of 80% ± 10% in the ecological laboratory of Yangzhou University. The germinated seeds were transferred to cement tanks covered with fine mesh in an outdoor natural environment and were grown until the six-leaf seedling stage. The seedlings were then transferred into plastic pots (dimensions = R ¼ 16 cm). Finally, the stress treatments proceeded at the tillering stage (40 ± 2, 40 ± 4, and 40 ± 8 days) following the same procedure as Ahmad et al. (2022a). Furthermore, the JGM technical grade of 61.7% used in this study was obtained from the Qianjiang Biochemistry Co., Ltd. (Haining, Zhejiang, China). Following the protocol in a previous report by Ahmad et al. (2022a), the two hundred parts per million (PPM) solution was Tween 20 obtained from the Sinopsin Group Chemical Reagent Company (Shanghai, China). The fungicide was then sprayed on the rice seedlings, following the procedure in a previous study (Ge et al., 2010).
The rice seeds were sown in standard cement tanks containing standard rice-growing soil at the Yangzhou University experimental field under natural outdoor conditions (Ge et al., 2013). Seedlings bearing six leaves were transplanted into plastic pots (R ¼ 8 cm) with four hills per pot, three plants per hill, and grown under natural conditions for experiments. Rice plants were pulled from the pots (at about 25 days), keeping the root system intact. The plants were gently washed without damaging the roots, then cultured in plastic containers (D ¼ 12 cm and H ¼ 25 cm) containing rice hydroponic liquid solution obtained from International Rice Research Institute (Los Banos, Laguna, Philippines). The pH of the hydroponic solution was maintained at 5.0 by daily adjustments with 1 mol/L HCl and 1 mol/L NaOH. The root systems were immersed in the hydroponic solution, and the solution was replaced every 3 days to ensure sufficient plant nutrition. Experimental rice plants (n ¼ 3) were transferred into a 1% glucose, sucrose, and trehalose hydroponic solution after a week following the method of Lin et al. (2018), and controls (n ¼ 3) were cultured in normal hydroponic solution (without sugar). Thirty-third-instar nymphs were transferred onto rice plants in the 1% glucose, sucrose, and trehalose hydroponic solution and covered with 60 mesh nylon net, and the BPH nymphs infestations for two, four and 8 days were collected and snap frozen in liquid nitrogen then and stored in −80°C degree till further use. Finally, the stored samples were then used for qRT-PCR analysis, and the specific primers used in the study are listed in (Supplementary Table S1).
2.5 Expression profiling of the NlSOD genes in N. lugens
BPH was introduced on the forty-day-old (40 ± 2) to plastic buckets having rice plants, and samples were collected at 2, 4, and 8 days after infestation. First, the BPH samples were snap-frozen in liquid nitrogen and then stored at −80°C degrees till further experiments. Furthermore, total RNA was extracted from the samples using an RNA extraction kit (Vazyme, Nanjing, China). First, the free DNA was removed using DNase I, and the concentration and purity were measured using a NanoDrop 1,000 spectrophotometer (Thermo Fisher Scientific, Rockford, IL, United States). Also, the integrity was checked using 1.5% agarose gel electrophoresis. Finally, the resulting complementary DNA (cDNA) was used as a template for quantitative real-time PCR (qRT-PCR) analysis using SYBR Green real-time PCR master mix (Vazyme, Nanjing, China). The qRT-PCR assays were performed in triplicate using a real-time PCR system (Bio-Rad, Hercules, CA, United States) following the manufacturer’s protocol.
Furthermore, the 2 μL aliquots of cDNA were amplified by qPCR in 20 μL reaction volumes using the SYBR Premix Ex Taq™ II (TaKaRa, Dalian, China). The cDNAs were amplified at 95°C for 2 min, followed by 35 cycles of 10 s at 95°C, then for 30 s, and at 72°C for 30 s, with a final extension step of 72°C for 10 min in a CFX96 real-time PCR system (Bio-Rad Co., Ltd., CA, United States). The mRNA amounts of all genes were separately quantified with the stable expression of the constitutive reference gene, actin. The specific primers are listed in (Supplementary Table S1). After amplification, the target gene cycle threshold (Ct) values were normalized to the reference gene (actin) by the 2−ΔΔCT method (Livak and Schmittgen, 2001). The data’s mean values of three biologically independent replicates were used for the final graphs.
2.6 Statistical analysis
The data presented in this paper were analyzed using the SPSS software (version 25.0, SPSS Inc., Chicago, IL, United States) for statistical analysis (ANOVA), statistical significance, and a 95% confidence interval (p ≤ 0.05). The data were analyzed and expressed as the mean ± standard deviation (SD) of three biologically independent replicates in all measured parameters, and finally, GraphPad Prism (Version 9.4.1) (GraphPad Software, Inc., LA Jolla, CA, United States) was used for graphical representation.
3 Results
3.1 Identification and sequence analysis of NlSOD genes
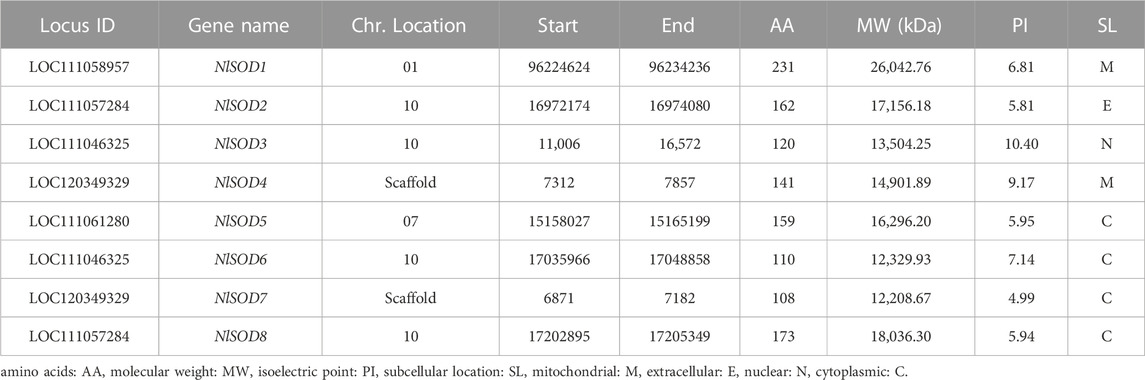
The present study retrieved 8 NlSOD genes, and the nomenclature was given (NlSOD1-NlSOD8) based on the three functional domains and chromosomal positions. Among the eight NlSOD gene family members, the NlSOD1 contains two functional domains, namely, Sod_Fe_N and Sod_Fe_C; the NlSOD4 and NlSOD7 exhibit single domains (Supplementary Figure S1). Moreover, the five members of the SOD genes family (NlSOD2, NlSOD3, NlSOD5, NlSOD6, and NlSOD8) were found with the Sod_Cu conserved domain. The accession number and annotations of the conserved domains are listed in supplementary data (Supplementary Table S2).
Following the deep scan, eight NlSOD genes were isolated from the NCBI online database; the detailed information is listed in Table 1. Among them, NlSOD1 and NlSOD4 resided in the mitochondrial, NlSOD2 extracellular, and NlSOD3 nuclear, whereas NlSOD5, NlSOD6, NlSOD7 and NlSOD8 resided in cytoplasmic. Other features of NlSOD proteins such as locus ID, chromosomal coordinates, molecular weight, chemical properties, and isoelectric point (PI) are tabulated.
3.2 Phylogenetic relationship and motif patterns of SOD genes in N. lugens
To obtain the evolutionary relationship of the NlSOD gene family, 8 NlSOD proteins with 4 other insects’ species, including 3 DmSOD, 3 LsSOD, 4 SiSOD and 4 SfSOD protein sequences were aligned using ClustalX software. After alignment, an ML phylogenetic tree was constructed using MEGA7 software (version 7.0). Finally, the phylogenetic tree was displayed using subscription-based iTOL: Interactive Tree Of Life (https://itol.embl.de/) (Letunic and Bork, 2021). The phylogenetic analysis clustered the SOD proteins into three subgroups such as SODI, SODII, and SODIII, and the number of SOD proteins was counted in each subgroup, whereas subgroup I accounted for 2 proteins and subgroup II and subgroup III with 3 NlSOD proteins (Figure 1), following the same procedure of our previous report by Ahmad et al. (2022b).
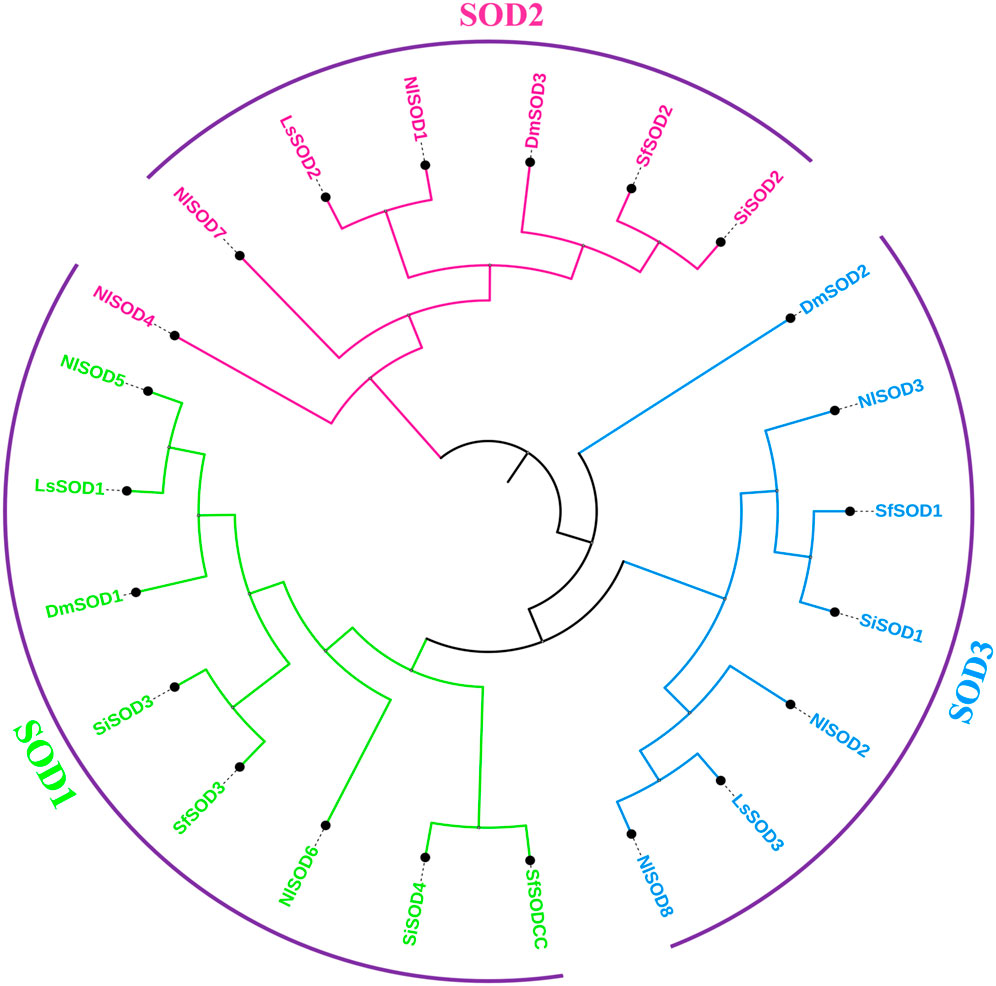
FIGURE 1. Schematic representation of the phylogenetic analysis of the SOD gene family: the phylogenetic tree was generated using the amino-acid sequences of selected SODs via the neighbor-joining (NJ) tree method. All N. lugens SODs, D. melanogaster, L. striatellus, S. incertulas, and S. frugiperda, with their counterparts, were classified into three subgroups, and the final tree was displayed using iTOL.
A total of eighteen conserved motifs were discovered using the MEME online server (Version 5.4.1) (https://meme-suite.org/meme/tools/meme) (accessed on 26 February 2023) reported by Bailey et al. (2009), and they were found to be appropriate for explaining the NlSOD gene’s structure (Figure 2). Among the eight NlSOD genes, the NlSOD2 and NlSOD8 were counted with four motifs, followed by NlSOD5 with three motifs, and NlSOD1 and NlSOD3 exhibited two motifs. Finally, the three NlSOD genes such as NlSOD4, NlSOD6, and NlSOD7, were recorded for a single motif.
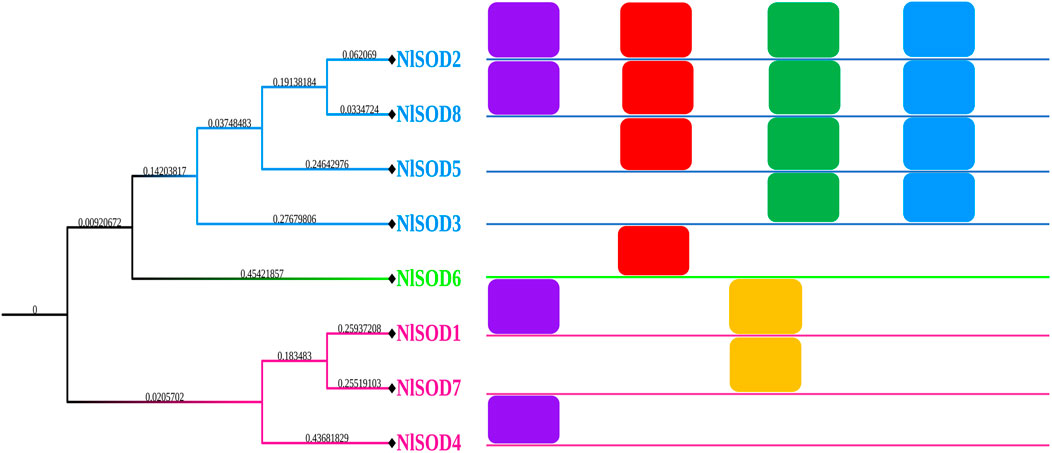
FIGURE 2. Schematic representation of the conserved motifs of NlSOD genes in N. lugen. The phylogenetic tree was recovered using Mega (V. 7.0), and then the motifs were combined using Microsoft PowerPoint 365.
3.3 Go analysis and KEGG pathways enrichment classification
NlSOD genes GO analysis showed various key functional predictions. For instance, the biological, molecular, and cellular processes following the same as our previous study by Ahmad et al. (2022b). In the biological processes category, the NlSOD genes are mainly involved in the regulation of stress-responsive with a high percentage of 22% and 20% in the biosynthetic process, whereas the other functions such as cell pigmentation, cell differentiation, and other environmental stimuli via hormonal and metabolic modulation were also observed (Supplementary Figure S2). As stated in the molecular processes category, the NlSOD genes were also found to regulate oxidoreductase activity, ion binding, protein binding, and enzyme binding activity. A large number of NlSOD genes reside in the intercellular region by up to 26%, followed by cytoplasm, and plasma membrane, whereas some members are localized to extracellular space and plasma membrane.
KEGG pathway enrichment analysis is an improtant tool for understanding the molecular framework ignited by a cluster of genes. For instance, in our study, the NlSOD genes showed their involvement in different KEGG pathways. Amongst them, the FoxO signaling pathway was regulated by seven percent of NlSOD genes, followed by the MAPK signaling pathway, which accounted for five percent of genes in their regulatory machinery. The chemical carcinogenesis-reactive oxygen species signaling pathway, which is moderately influenced by three percent of NLSOD genes in response to JGM stress and serves as the first line of defense was also found in the analysis. Additionally, the longevity regulating pathway, which is key to insect lifespan, was also fine-tuned by NlSOD genes. A series of disease-related pathways such as prion, parkinson, and huntington could rely on the transcriptional of NlSOD genes. The KEGG pathway analysis recorded the lipid and peroxisomes pathways (Figure 3).
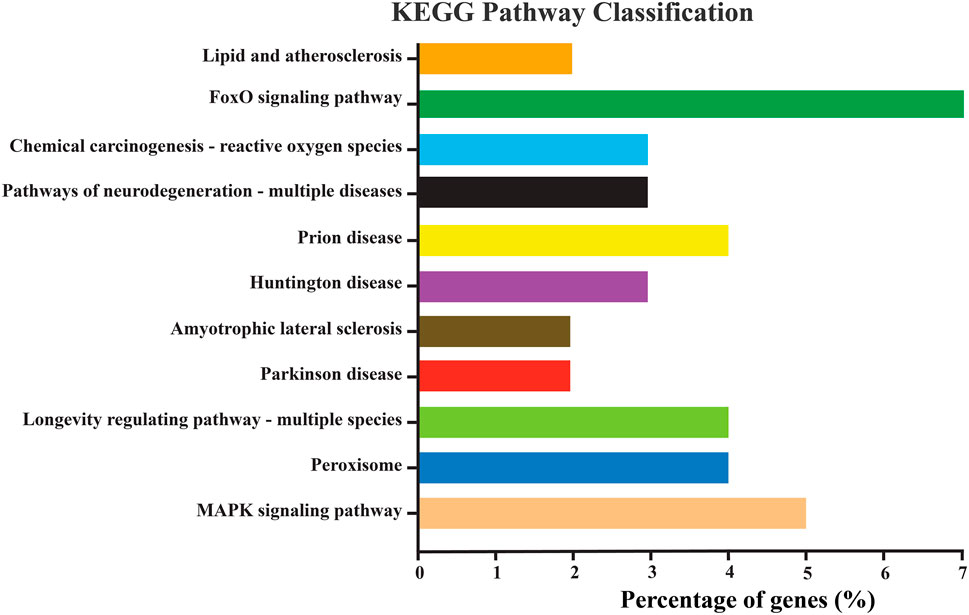
FIGURE 3. Represents the KEGG pathway enrichment analysis of differentially expressed genes of N. lugens. KEGG pathways with (p-value < 0.01, log2 ≥ 5, ≤–5). The Y-axis indicates the KEGG pathways, and the X-axis indicates the percentage of genes in each pathway. The percentage in the X-axis is DEGs significantly enriched in the relevant pathways.
3.4 NlSOD proteins interactive network analysis
The detailed information of our reference protein NlSOD1 protein interactive partners were initially searched in the online server FlyBase (https://flybase.org/reports/FBgn0032350) (accessed on 27 February 2023) and was confirmed by STRING: functional protein association networks (https://cn.string-db.org/) and the integrative protein partners network was programmed. The detailed information and accessions are tabulated in (Supplementary Table S3). The NlSOD1 protein plays a crucial role in BPH growth and development and is also highly stress-responsive. Numerous other crucial proteins are also found to be highly interactive with our reference NlSOD1 protein. (Supplementary Figure S3). For instance, the SOD1 protein interacted highly with CAT, Prx5, PHGPx, GCLC, and CG15116. Almost similar binding was also observed for SOD2 protein with other crucial proteins such as TAM and MRPS29, which is extensively studied on the stress/diseases side. Finally, the SOD3 protein was found with high interaction with all the proteins named above, revealing the potential role of the NlSOD protein as crucial in the developmental processes and highly stress-responsive.
3.5 Tissue-specific expression analysis of NlSOD genes in developmental stages
Tissue-specific expression analysis unfolded the potential role of NlSOD genes in the development of BPH (Figure 4). For instance, dominant expression was observed in the fifth instral stage of NlSOD6 up to 14 folds, followed by NlSOD2 in the fifth instral stage, 1 day after emergence (1A) and 2A up to six folds expression. Furthermore, the NlSOD4 showed moderate transcription in 2A, and NlSOD8 displayed the five folds expression in 1A. However, the lower transcription was recorded in NlSOD1, NlSOD5, and NlSOD7, suggesting that the NlSOD gene family has a potential role in the developmental stages of N. lugens (Figure 4). Notably, the lowest transcription was observed by overall NlSOD genes in the first to fourth instral stage, excluding NlSOD3 and NlSOD7, which have a moderate expression pattern in the first-fourth instral development stage. Among all the developmental stages, the NlSOD genes showed high expression in the fifth instral stage; for a detailed description and a graphical representation, see the appended data in the supplementary file (Supplementary Figure S4).
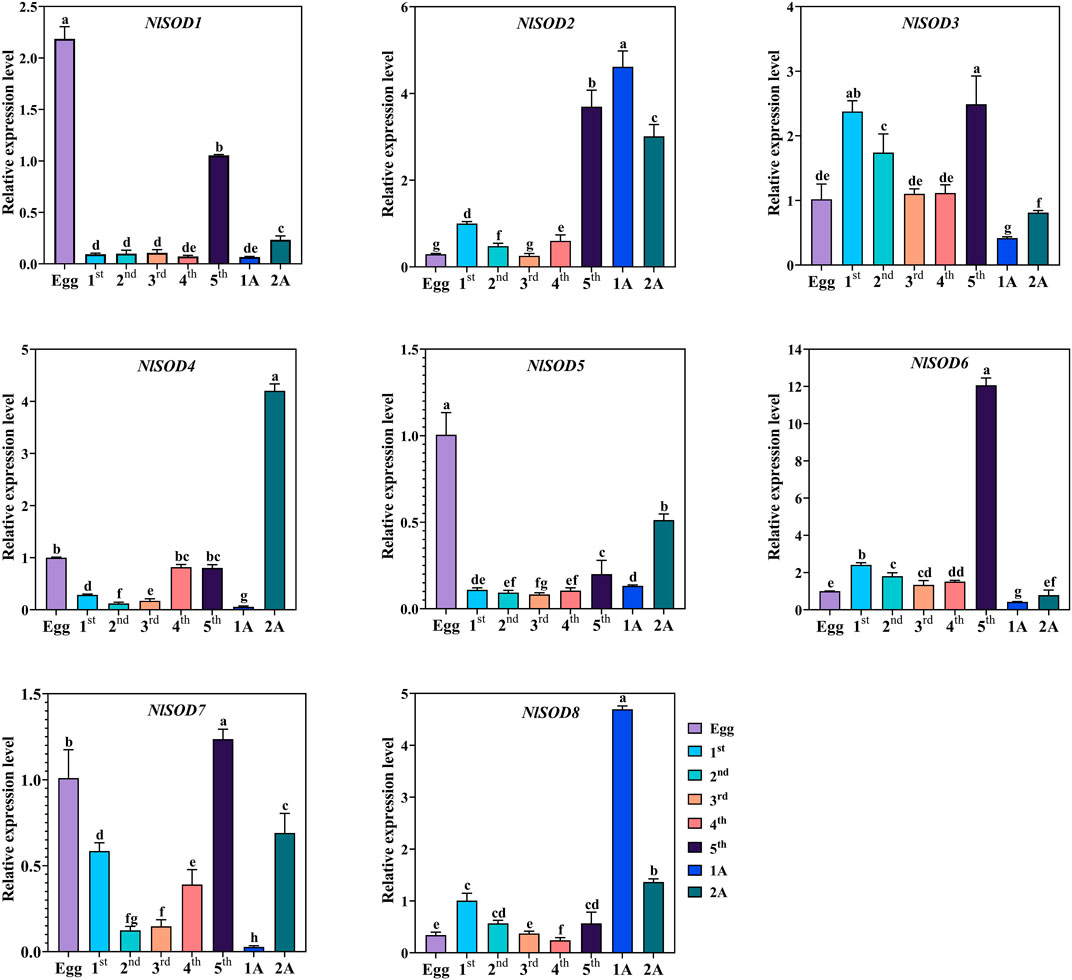
FIGURE 4. Expression analysis of NlSOD genes in developmental stages. 1A represents 1 day after emergence (1 DAE) and 2 DAE. Histogram bars indicate expression, and error bars show means ± SEM. Bars annotated with different lowercase letters are statistically significant at p < 0.05 (Tukey test).
We have confirmed the tissues specific expression of NlSOD genes in four major tissues (head, midgut, ovary, and fat body) at 2 days after emergence (2DAE) by qRT-PCR. The results showed that most of the NlSOD genes have the dominant expression mostly in the head and ovary followed by the fat body and midgut (Figure 5). In particular, the high expression of NlSOD5 up to 10 folds, followed by NlSOD1 with five folds in the ovary. Moreover, the moderate transcriptional pattern was shown by most of the NlSOD genes, such as the NlSOD2, NlSOD3, NlSOD4, NlSOD6, and NlSOD8, with 1.5 folds in different tissues. A single NlSOD family member NlSOD7 displayed lower transcription in almost all tested tissues (Figure 5).
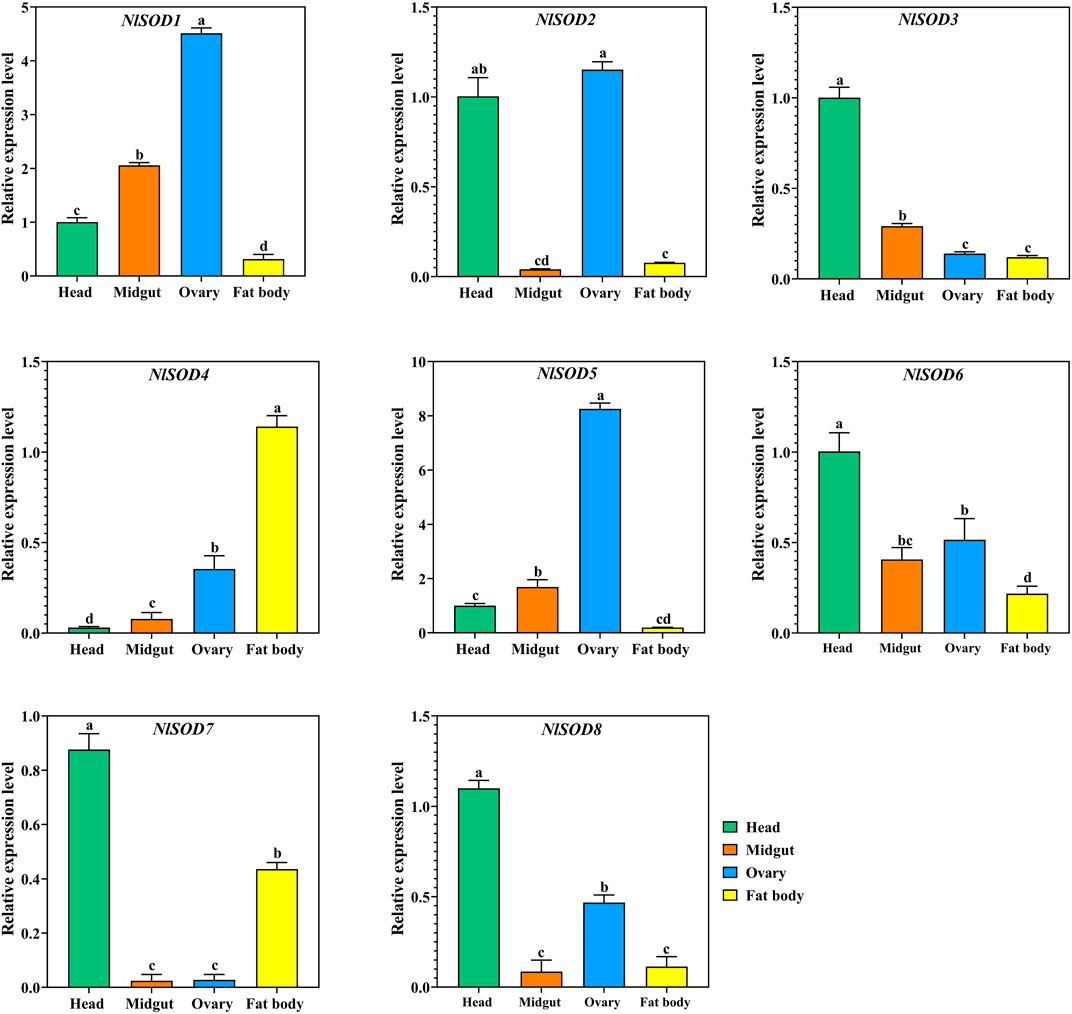
FIGURE 5. Expression analysis of NlSOD genes in selected segments/tissues. Histogram bars indicate expression, and error bars show means ± SEM. Bars annotated with different lowercase letters are statistically significant at p < 0.05 (Tukey test).
3.6 Expression analysis of the NlSOD genes under JGM treatment
We performed qRT-PCR analysis to validate the NlSOD genes expressions in response to JGM spraying treatment (Figure 6). Our study revealed that NlSOD genes, such as NlSOD1 and NlSOD5, displayed upregulated expressions at 2 days (2D) and 4D time points. Followed by NlSOD2 and NlSOD8 had moderate transcription, and the rest of the four genes, such as NlSOD3, NlSOD4, NlSOD6, and NlSOD7, had the lowest transcription patterns. These results suggest that the JGM spraying application induces the NlSOD genes and enhances BPH’s immunity regulations against JGM.
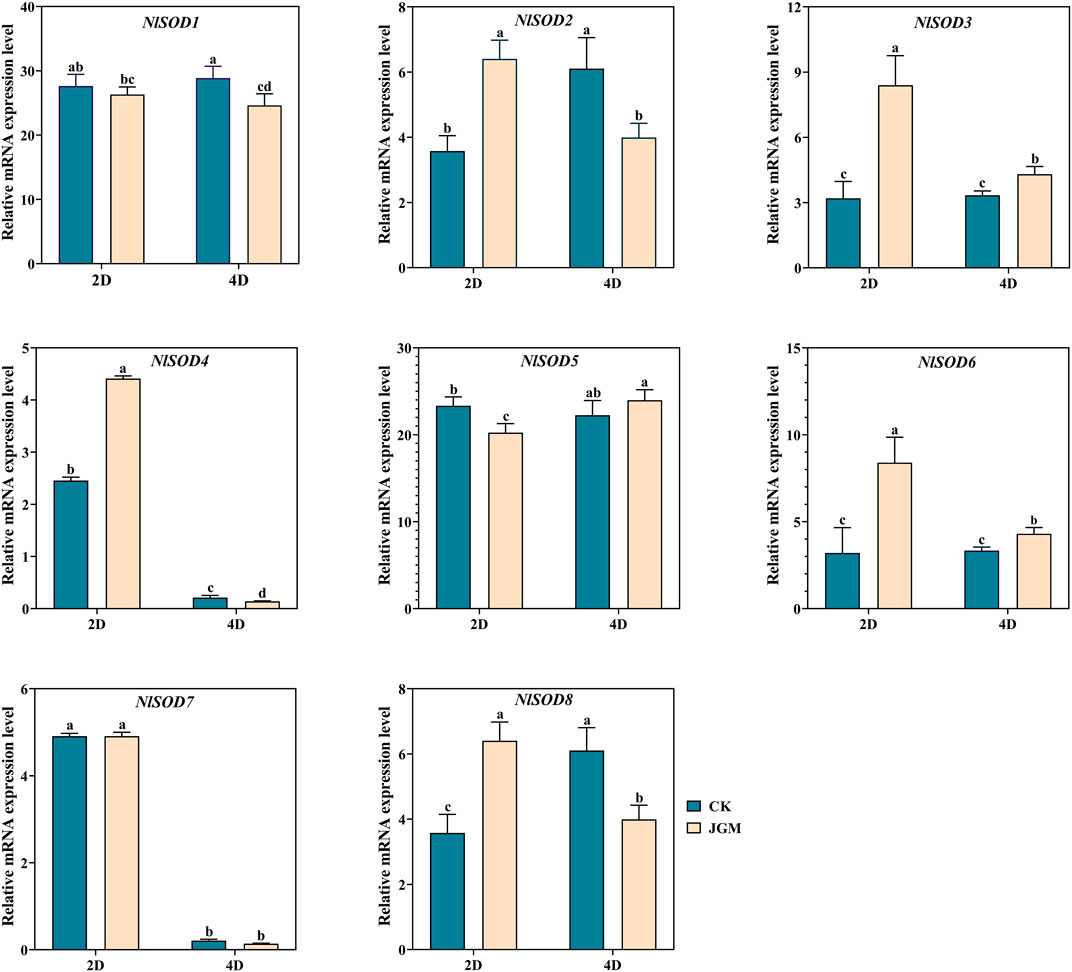
FIGURE 6. Expression analysis of NlSOD genes in response to Jinggangmycin 2 days (2D) and 4D after treatment. The histogram bars indicate expression, and the error bars show means ± SEM. Bars annotated with different lowercase letters are statistically significant at p < 0.05 (Tukey test).
3.7 Expression analysis of the NlSOD genes in response to glucose, sucrose, and trehalose hydroponic treatment
The qRT-PCR analysis revealed the potential role of the NlSOD gene family. Among the 8 NlSOD genes, the NlSOD6 gene displayed dominant expression of fifteen folds at 24h, followed by NlSOD2 and NlSOD3 with five and four folds expressions under glucose, respectively. Whereas the rest of the genes, such as NlSOD1, NlSOD4, NlSOD5, and NlSOD6, displayed comparatively low transcription at 3 h and 6h, while NlSOD7 showed a moderate expression of 2.5 folds at 24 h (Figure 7).
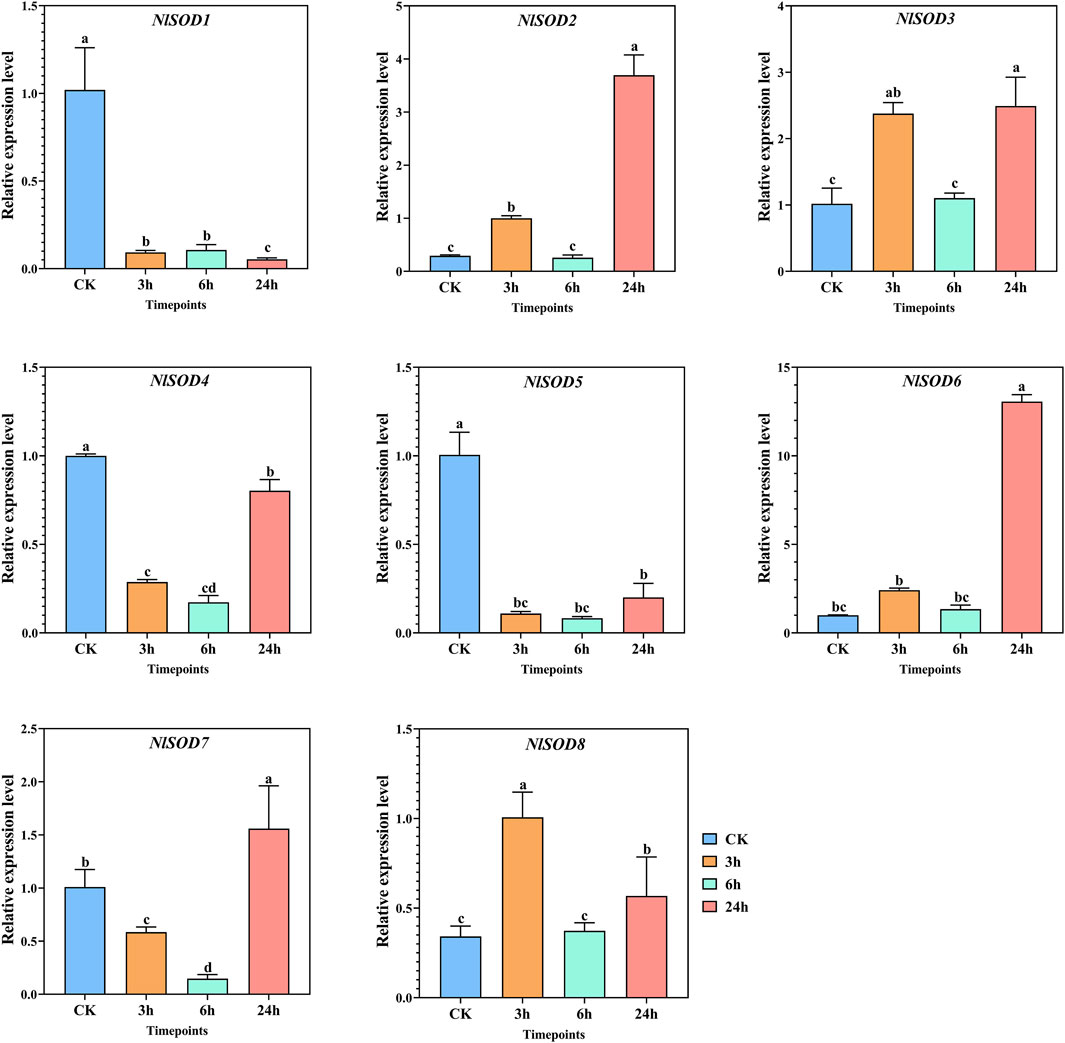
FIGURE 7. Expression analysis of NlSOD genes in response to glucose at 3h, 6h, and 24h. The histogram bars indicate expression, and the error bars show means ± SEM. Bars annotated with different lowercase letters are statistically significant at p < 0.05 (Tukey test).
Sugar transporters may offer a new potential target for controlling insect pests (Kikuta et al., 2010). Herein, the expression of NlSOD genes in BPH fed on rice plants treated with sucrose hydroponics was investigated. Similar to glucose, NlSOD genes were triggered under sucrose treatment at almost all the time points (Figure 8). Furthermore, the NlSOD6 displayed a dominant expression of 15 folds at 24 h. Followed by NlSOD2 and NlSOD8 of 5 folds expression at 24h, whereas moderate expression was observed by NlSOD5 and NlSOD7 of 3 folds at 24h, and the lowest transcription was observed by NlSOD3 of 1.3 folds at 3 h. Seven of the eight NlSOD genes showed an upregulated expression, and only a single gene, such as the NlSOD1, displayed a relatively downregulated expression at 3, 6 and 24 h. The results suggested that most of the NlSOD genes are highly responsive to sucrose and could work in concert with metabolism to tailor developmental and defense processes.

FIGURE 8. Expression analysis of NlSOD genes in response to sucrose at 3h, 6h, and 24h. The histogram bars indicate expression, and the error bars show means ± SEM. Bars annotated with different lowercase letters are statistically significant at p < 0.05 (Tukey test).
Gene expression analysis was conducted to understand the role of NlSOD genes of BPH fed with rice plants treated with trehalose hydroponic solution. Furthermore, the eight NlSOD genes showed upregulated expression patterns under trehalose hydroponic treatment. In which the NlSOD6 was observed with the dominant expression of 15 folds at 24 h. Followed by NlSOD8 of 6 folds expression at 24h, whereas the four NlSOD genes such as NlSOD1, NlSOD2, NlSOD4 and NlSOD7, showed moderate expression of 5 folds at 24 h. Contrastingly, the NlSOD5 and NlSOD3 were observed with the lowest transcription of 4 and 3 folds, respectively. The expression analysis of NlSOD genes under trehalose showed upregulated expression patterns and might finetuning the development of BPH and provide defense on a cellular basis.
4 Discussion
When insects suffer from environmental stresses such as extreme temperatures and insecticide toxicities, the ROS are spawned (Gao X. L. et al., 2013; Gao X. M. et al., 2013). Metallo enzyme SOD is the most effective intracellular enzymatic antioxidant, ubiquitous in all aerobic organisms and subcellular compartments prone to ROS-mediated oxidative stress. It eliminates extremely toxic ROS, such as O2, and reduces the possibility of hydroxyl radical OH generation through a metal-catalyzed reaction of the Haber-Weiss type (Fridovich, 1978). Mn-SOD is considered a general stress-responsive element, and several internal and external cues, including pesticide toxicity at the transcriptional and translational levels, can substantially impact its expression profiles (Zelko et al., 2002; Cho et al., 2006). The SOD enzymes are a group of highly conserved stress response enzymes that function as molecular chaperones assisting in protein refolding and degradation in stressed cells (Mayer and Bukau, 2005; Yamamoto et al., 2005d; Yamamoto et al., 2007). However, only a little is known to date about oxidative stress induced by JGM pesticides and sugars (glucose, sucrose, trehalose) in BPH. In the current study, the SOD gene family in BPH was characterized, and the JGM and sugar protective effect of protein was investigated for the first time.
4.1 SOD genes are widely distributed in class insecta
A BLASTP search demonstrated that the LsSOD gene family and the NlSOD gene family were closely related, with the LsSOD genes having a higher proportion of matched sequences, indicating that the NlSOD gene is more diverged compared to other SOD genes. Moreover, the phylogenetic analysis also confirmed this relationship (Umasuthan et al., 2012). Both insects are members of the same insect family, the Hemiptera (Delphacidae), and share shorter N-terminal sequences, which strongly suggests their taxonomic association (Zelko et al., 2002). The phylogenetic tree analysis demonstrated that Mn-SOD and Cu/Zn-SOD may have shared a common ancestor and that Mn-SOD may have undergone more evolutionary change than Cu/Zn-SOD. Additionally, the Cu/Zn-SOD clade was divided into the Cu/Zn-SOD and Cu/Zn-SOD subgroups (Figure 1), with the latter showing greater divergence; these two protein sub-families may develop through gene replication as previously reported by Liu et al. (2015).
Members of the SOD gene family are highly conserved, and our results also uncover this high homology. The current study identified highly inducible SOD genes and shared close similarities with other insect species. Comparing the amino acid sequence of NlSOD with its counterparts from Lepidopteran insect species, including L. striatellus, S. incertulas, S. frugiperda, and D. melanogaster, the NlSOD showed high similarity with L. striatellus (Figure 1).
Domains are a protein’s distinct functional and/or structural units (Marchler-Bauer et al., 2015). Usually, they are responsible for a particular function or interaction, contributing to the overall role of a protein (Sigrist et al., 2010). The current study found that NlSOD proteins exhibit three functional domains (Supplementary Figure S1). The SOD_Fe_N functional domain is reportedly strongly associated with innate immunity, which is essential for survival organisms, for instance, in the survival and stress response in Chlamys farreri (Wang et al., 2018). Similarly, we also found the SOD_Fe_N, Sod_Fe_C, and Sod_Cu domains, which might help the BPH withstand the JGM and high sugar.
Additionally, usually, they are responsible for a particular function or interaction; because of these domains in NlSOD proteins, the NlSOD candidate proteins (Supplementary Figure S3) are found in high interactions with other functional proteins. Moreover, sequence motifs are short, recurring patterns in DNA that are presumed to have a biological function. Often, they indicate sequence-specific binding sites for proteins such as nucleases and transcription factors. Others are involved in important processes at the RNA level, including ribosome binding, mRNA processing (splicing, editing, polyadenylation), and transcription termination (D'Haeseleer, 2006). Herein, we also found the five motifs in the NlSOD genes (Figure 2) to explain the gene structure and influence their role in development and stress response, which needs further study.
4.2 NLSOD genes play an essential role in N. lugens growth and development
In prior investigations, it was discovered that SOD genes were crucial not only for growth and development but also for stress response and other vital processes throughout various tissues and organs (Okado-Matsumoto and Fridovich, 2001; Zelko et al., 2002). In the present study, we found that the SOD was also distributed in all the tested tissues, such as in the head, midgut, ovary, and fat body (Figure 5). Additionally, the SOD genes showed transcriptional activity in the BPH egg, first to fifth instral nymph, 1 day after emergence (1A) and 2A (Figure 4), suggesting that this transcript may be involved in stress sensing and signaling transduction.
Moreover, there were alterations in the NlSOD gene expression levels throughout distinct tissues. However, the fact that the midgut and fat body exhibited the greatest expression implies that it may be contributing to development. The midgut includes the malpighian tubule, which plays an important role in detoxifying and eliminating toxins (Beyenbach et al., 2010). And fat body is one of the prime sites for antioxidant enzymes (Yamamoto et al., 2005b; Lee et al., 2005; Yamamoto et al., 2005). Our findings are consistent with those of Glossina morsitans (Munks et al., 2005) and Agrilus planipennis (Rajarapu et al., 2011), in which the midgut and fat body showed a substantial upregulation of SOD genes. The hindgut and fat body’s upregulation of SOD suggests that these two organs served as the primary sites for the production of antioxidant enzymes and provided resistance to oxidative stress.
4.3 NlSOD genes regulate the BPH immune response to pesticide toxicity and high sugar
Aerobic organisms constantly consume oxygen, which the mitochondria utilize to produce the energy required to sustain life (Atalay et al., 2020). Some of these oxygen molecules may convert into reactive oxygen species (ROS) during metabolic processes (Qureshi et al., 2021a). Exposure of tissues and cells to ultraviolet radiation and chemical agents generates ROS such as superoxide anion (O2−) and hydrogen peroxide (H2O2) and causes oxidative damage to cells (Hermes-Lima and Zenteno-Savı́n, 2002; Fukai and Ushio-Fukai, 2011; Lee et al., 2019). SOD gene family is an essential antioxidant that catalyzes the dismutation of superoxide anions into molecular O2 and H2O2, thereby protecting cellular components from extreme oxidative stress impairments (Valentine and Hart, 2003b; Fukai and Ushio-Fukai, 2011). Insects have developed tolerance to the insecticides JGM and sugars as a consequence of their widespread application (Matsumura et al., 2008). In the present study, the qRT-PCR analysis showed that most of the NlSOD gene expressions were induced after JGM treatment. For instance, the NlSOD1 and NlSOD5 displayed dominant expressions (Figure 6). Following that, under the sugar (glucose, sucrose and trehalose) treatment, the NlSOD6 and NlSOD2 were observed with high transcription (Figures 7–9) and might be involved in regulating BPH immune response to JGM and high sugar. However, the rest of the NlSOD genes displayed mixed expression patterns under the JGM and high sugar treatment and cannot be ruled out from functional studies. These genes may work redundantly and therefore rely on each other transcriptional changes. Interactive protein studies would be useful to confirm the statement of whether these amino acids have homodimeric or heterodimeric interactions. Additionally, our results provide a theoretical platform for pesticide regulatory bodies.
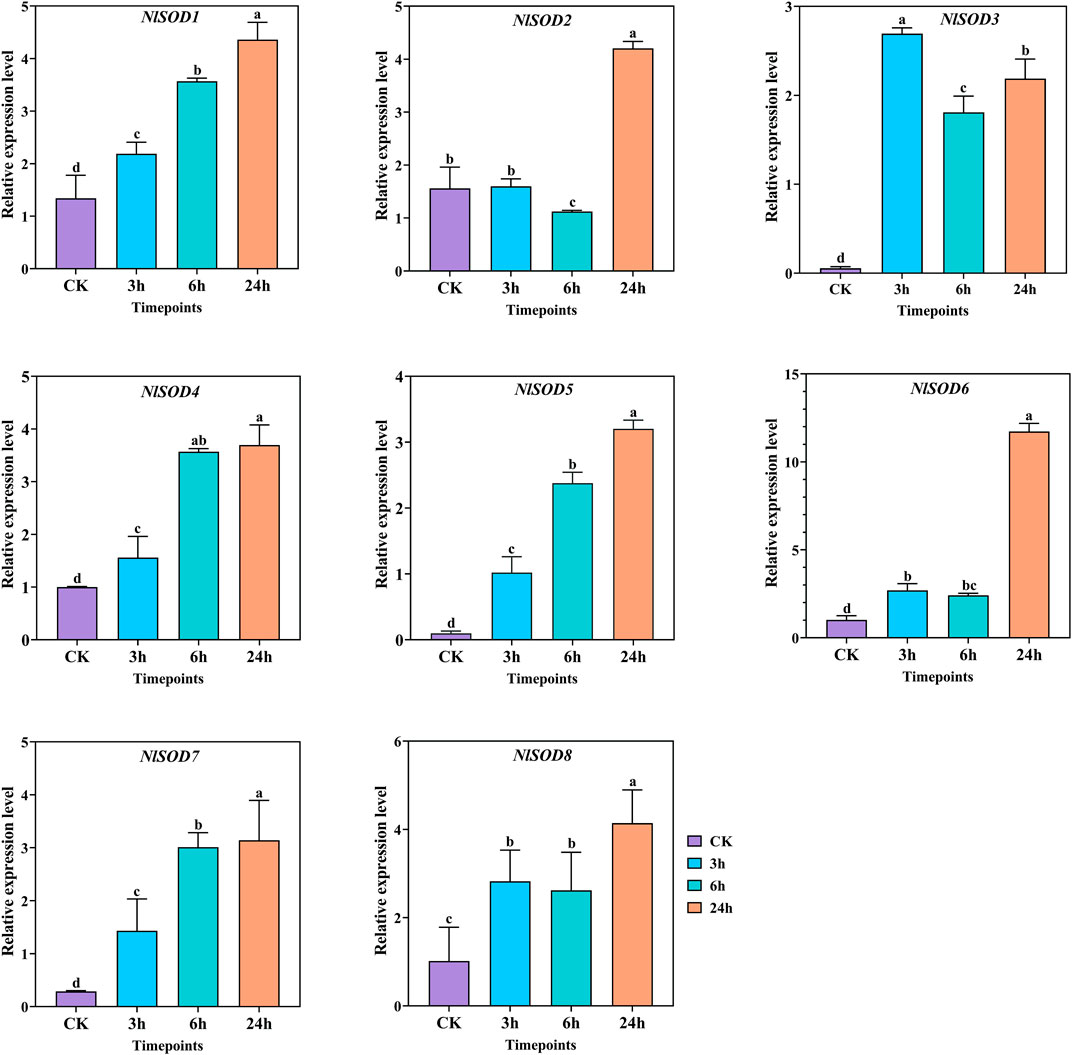
FIGURE 9. Expression analysis of NlSOD genes in response to trehalose at 3h, 6h, and 24h. The histogram bars indicate expression, and the error bars show means ± SEM. Bars annotated with different lowercase letters are statistically significant at p < 0.05 (Tukey test).
5 Conclusion
In the present study, we identified eight NlSOD genes from NCBI and classified them into three subgroups based on their functional domain and structural properties. Furthermore, the qRT-PCR analysis revealed that NlSOD genes play an essential role in BPH growth and development by expression in all tested tissues. The NlSOD genes were also found responsive to JGM and sugar stress. Among them, the NlSOD6 gene displayed dominant expression and could be involved in regulating BPH response to JGM and sugar. The interactive protein analysis disclosed that these NlSOD, in concert with other stress markers, is crucial for BPH response to external stimuli. Stress control is a complex mechanism, and our study demonstrated the potential role of NlSOD genes in modulating BPH development. These myriad molecules could be used in future molecular work to manipulate BPH pesticide resistance.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author contributions
LG and TaW designed and supervised the research; AS, HZ, and QD conducted the experimental work. TiW and SZ contributed to the preparation of biological materials. AS and HZ analyzed the data. AS wrote the main manuscript, and LG and TaW revised the manuscript. All authors have read and agreed to publish the current version of the manuscript.
Funding
The authors are thankful to the National Natural Science Foundation of China (32072415) and the Key Research and Development Plan of Jiangsu Province (BE2022345) for providing financial support.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2023.1197395/full#supplementary-material
References
Ahmad, S., Zhang, J., Wang, H., Zhu, H., Dong, Q., Zong, S., et al. (2022a). The phosphoserine phosphatase alters the free amino acid compositions and fecundity in Cyrtorhinus lividipennis Reuter. Int. J. Mol. Sci. 23, 15283. doi:10.3390/ijms232315283
Ahmad, S., Zhu, H. W., Chen, Y., Xi, C. Y., Shah, A. Z., and Ge, L. Q. (2022b). Comprehensive bioinformatics and expression analysis of the TLP gene family revealed its role in regulating the response of Oryza sativa to Nilaparvata lugens, Laodelphax striatellus, and jinggangmycin. Agronomy 12, 1297. doi:10.3390/agronomy12061297
Atalay, S., Dobrzyńska, I., Gęgotek, A., and Skrzydlewska, E. (2020). Cannabidiol protects keratinocyte cell membranes following exposure to UVB and hydrogen peroxide. Redox Biol. 36, 101613. doi:10.1016/j.redox.2020.101613
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). Meme suite: Tools for motif discovery and searching. Nucleic acids Res. 37, W202–W208. doi:10.1093/nar/gkp335
Beyenbach, K. W., Skaer, H., and Dow, J. A. (2010). The developmental, molecular, and transport biology of Malpighian tubules. Annu. Rev. Entomology 55, 351–374. doi:10.1146/annurev-ento-112408-085512
Bordo, D., Djinović, K., and Bolognesi, M. (1994). Conserved patterns in the Cu,Zn superoxide dismutase family. J. Mol. Biol. 238, 366–386. doi:10.1006/jmbi.1994.1298
Cho, Y. S., Choi, B. N., Kim, K. H., Kim, S. K., Kim, D. S., Bang, I. C., et al. (2006). Differential expression of Cu/Zn superoxide dismutase mRNA during exposures to heavy metals in rockbream (Oplegnathus fasciatus). Aquaculture 253, 667–679. doi:10.1016/j.aquaculture.2005.05.047
Crapo, J. D., Oury, T., Rabouille, C., Slot, J. W., and Chang, L. Y. (1992). Copper,zinc superoxide dismutase is primarily a cytosolic protein in human cells. Proc. Natl. Acad. Sci. U. S. A. 89, 10405–10409. doi:10.1073/pnas.89.21.10405
D'haeseleer, P. (2006). What are DNA sequence motifs? Nat. Biotechnol. 24, 423–425. doi:10.1038/nbt0406-423
Folz, R. J., Guan, J., Seldin, M. F., Oury, T. D., Enghild, J. J., and Crapo, J. D. (1997). Mouse extracellular superoxide dismutase: Primary structure, tissue-specific gene expression, chromosomal localization, and lung in situ hybridization. Am. J. Respir. Cell Mol. Biol. 17, 393–403. doi:10.1165/ajrcmb.17.4.2826
Fridovich, I. (1978). The biology of oxygen radicals. Science 201, 875–880. doi:10.1126/science.210504
Fujii, M., Ishii, N., Joguchi, A., Yasuda, K., and Ayusawa, D. (1998). A novel superoxide dismutase gene encoding membrane-bound and extracellular isoforms by alternative splicing in Caenorhabditis elegans. DNA Res. 5 1, 25–30. doi:10.1093/dnares/5.1.25
Fukai, T., and Ushio-Fukai, M. (2011). Superoxide dismutases: Role in redox signaling, vascular function, and diseases. Antioxid. Redox Signal. 15, 1583–1606. doi:10.1089/ars.2011.3999
Gao, X. L., Li, J. M., Wang, Y. L., Jiu, M., Yan, G. H., Liu, S. S., et al. (2013). Cloning, expression and characterization of mitochondrial manganese superoxide dismutase from the Whitefly, Bemisia tabaci. Int. J. Mol. Sci. 14, 871–887. doi:10.3390/ijms14010871
Gao, X. M., Jia, F. X., Shen, G. M., Jiang, H. Q., Dou, W., and Wang, J. J. (2013). Involvement of superoxide dismutase in oxidative stress in the oriental fruit fly, Bactrocera dorsalis: Molecular cloning and expression profiles. Pest Manag. Sci. 69, 1315–1325. doi:10.1002/ps.3503
Ge, L. Q., Huang, L. J., Yang, G. Q., Song, Q. S., Stanley, D., Gurr, G. M., et al. (2013). Molecular basis for insecticide-enhanced thermotolerance in the Brown planthopper Nilaparvata lugens stål (hemiptera:delphacidae). Mol. Ecol. 22, 5624–5634. doi:10.1111/mec.12502
Ge, L. Q., Wang, L. P., Zhao, K. F., Wu, J. C., and Huang, L. J. (2010). Mating pair combinations of insecticide-treated male and female Nilaparvata lugens stal (Hemiptera: Delphacidae) planthoppers influence protein content in the male accessory glands (MAGs) and vitellin content in both fat bodies and ovaries of adult females. Pesticide Biochem. Physiology 98, 279–288. doi:10.1016/j.pestbp.2010.06.019
Ge, L. Q., Zheng, S., Gu, H. T., Zhou, Y. K., Zhou, Z., Song, Q. S., et al. (2019). Jinggangmycin-induced UDP-glycosyltransferase 1-2-like is a positive modulator of fecundity and population growth in Nilaparvata lugens (stål) (Hemiptera: Delphacidae). Front. physiology 10, 747. doi:10.3389/fphys.2019.00747
Hasty, P., Campisi, J., Hoeijmakers, J., Van Steeg, H., and Vijg, J. (2003). Aging and genome maintenance: Lessons from the mouse? Science 299, 1355–1359. doi:10.1126/science.1079161
Hermes-Lima, M., and Zenteno-Savı́n, T. (2002). Animal response to drastic changes in oxygen availability and physiological oxidative stress. Comp. Biochem. Physiol. Part - C: Toxicol. Pharmacol. 133, 537–556. doi:10.1016/s1532-0456(02)00080-7
Hu, G., Lu, F., Zhai, B. P., Lu, M. H., Liu, W. C., Zhu, F., et al. (2014). Outbreaks of the Brown planthopper Nilaparvata lugens (stål) in the yangtze river delta: Immigration or local reproduction? PLos one 9, e88973. doi:10.1371/journal.pone.0088973
Jena, K. K., Jeung, J. U., Lee, J. H., Choi, H. C., and Brar, D. S. (2006). High-resolution mapping of a new Brown planthopper (BPH) resistance gene Bph18(t) and marker-assisted selection for BPH resistance in rice (Oryza sativa L), Theor. Appl. Genet. 112, 288–297. doi:10.1007/s00122-005-0127-8
Jiang, L. B., Zhao, K. F., Wang, D. J., and Wu, J. C. (2012). Effects of different treatment methods of the fungicide jinggangmycin on reproduction and vitellogenin gene (nlvg) expression in the Brown planthopper Nilaparvata lugens stål (Hemiptera: Delphacidae). Pesticide Biochem. physiology 102, 51–55. doi:10.1016/j.pestbp.2011.10.009
Kikuta, S., Kikawada, T., Hagiwara-Komoda, Y., Nakashima, N., and Noda, H. (2010). Sugar transporter genes of the Brown planthopper, Nilaparvata lugens: A facilitated glucose/fructose transporter. Insect Biochem. Mol. Biol. 40, 805–813. doi:10.1016/j.ibmb.2010.07.008
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi:10.1093/molbev/msw054
Lee, K. S., Ryul Kim, S., Sook Park, N., Kim, I., Dong Kang, P., Hee Sohn, B., et al. (2005). Characterization of a silkworm thioredoxin peroxidase that is induced by external temperature stimulus and viral infection. Insect Biochem. Mol. Biol. 35, 73–84. doi:10.1016/j.ibmb.2004.09.008
Lee, W., Yang, H.-W., Cha, S.-H., Han, E. J., Shin, E.-J., Han, H.-J., et al. (2019). “Radio-protective effects of Loliolus beka gray meat consisted of a plentiful taurine against damages caused by gamma ray irradiation,” in Taurine 11 (Germany: Springer), 729–738. doi:10.1007/1978-1981-1013-8023-1005_1063
Letunic, I., and Bork, P. (2021). Interactive tree of life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–w296. doi:10.1093/nar/gkab301
Lin, X., Xu, Y., Jiang, J., Lavine, M., and Lavine, L. C. (2018). Host quality induces phenotypic plasticity in a wing polyphenic insect. Proc. Natl. Acad. Sci. U. S. A. 115, 7563–7568. doi:10.1073/pnas.1721473115
Liu, H., He, J., Chi, C., and Gu, Y. (2015). Identification and analysis of icCu/Zn-SOD, Mn-SOD and ecCu/Zn-SOD in superoxide dismutase multigene family of Pseudosciaena crocea. Fish Shellfish Immunol. 43, 491–501. doi:10.1016/j.fsi.2015.01.032
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi:10.1006/meth.2001.1262
Marchler-Bauer, A., Derbyshire, M. K., Gonzales, N. R., Lu, S., Chitsaz, F., Geer, L. Y., et al. (2015). Cdd: NCBI's conserved domain database. Nucleic acids Res. 43, D222–D226. doi:10.1093/nar/gku1221
Matsumura, M., Takeuchi, H., Satoh, M., Sanada-Morimura, S., Otuka, A., Watanabe, T., et al. (2008). Species-specific insecticide resistance to imidacloprid and fipronil in the rice planthoppers Nilaparvata lugens and Sogatella furcifera in East and South-east Asia. Pest Manag. Sci. 64, 1115–1121. doi:10.1002/ps.1641
Mayer, M. P., and Bukau, B. (2005). Hsp70 chaperones: Cellular functions and molecular mechanism. Cell. Mol. life Sci. 62, 670–684. doi:10.1007/s00018-004-4464-6
Munks, R. J. L., Sant’anna, M. R. V., Grail, W., Gibson, W., Igglesden, T., Yoshiyama, M., et al. (2005). Antioxidant gene expression in the blood-feeding fly Glossina morsitans morsitans. Insect Mol. Biol. 14, 483–491. doi:10.1111/j.1365-2583.2005.00579.x
Nguyen, T. D., Lacombe, S., Bangratz, M., Ta, H. A., Vinh Do, N., Gantet, P., et al. (2015). P2 of Rice grassy stunt virus (RGSV) and p6 and p9 of Rice ragged stunt virus (RRSV) isolates from Vietnam exert suppressor activity on the RNA silencing pathway. Virus Genes 51, 267–275. doi:10.1007/s11262-015-1229-2
Okado, M. A., and Fridovich, I. (2001). Subcellular distribution of superoxide dismutases (SOD) in rat liver: Cu, Zn-SOD in mitochondria. J. Biol. Chem. 276, 38388–38393. doi:10.1074/jbc.M105395200
Okado-Matsumoto, A., and Fridovich, I. (2001). Subcellular distribution of superoxide dismutases (SOD) in rat liver: Cu,Zn-SOD in mitochondria. J. Biol. Chem. 276, 38388–38393. doi:10.1074/jbc.M105395200
Ookawara, T., Kizaki, T., Takayama, E., Imazeki, N., Matsubara, O., Ikeda, Y., et al. (2002). Nuclear translocation of extracellular superoxide dismutase. Biochem. Biophysical Res. Commun. 296, 54–61. doi:10.1016/s0006-291x(02)00804-5
Parker, J. D., Parker, K. M., and Keller, L. (2004). Molecular phylogenetic evidence for an extracellular Cu Zn superoxide dismutase gene in insects. Insect Mol. Biol. 13, 587–594. doi:10.1111/j.0962-1075.2004.00515.x
Peng, D., Li, S., Wang, J., Chen, C., and Zhou, M. (2014). Integrated biological and chemical control of rice sheath blight by Bacillus subtilis NJ-18 and jinggangmycin. Pest Manag Sci. 70, 258–263. doi:10.1002/ps.3551
Qureshi, S., Chandra, S., Chopra, D., Dubey, D., Jain, V., Roy, S. K., et al. (2021a). Nabumetone induced photogenotoxicity mechanism mediated by ROS generation under environmental UV radiation in human keratinocytes (HaCaT) cell line. Toxicol. Appl. Pharmacol. 420, 115516. doi:10.1016/j.taap.2021.115516
Qureshi, S., Chandra, S., Chopra, D., Dubey, D., Jain, V., Roy, S. K., et al. (2021b). Nabumetone induced photogenotoxicity mechanism mediated by ROS generation under environmental UV radiation in human keratinocytes (HaCaT) cell line. Toxicol. Appl. Pharmacol. 420, 115516. doi:10.1016/j.taap.2021.115516
Rajarapu, S. P., Mamidala, P., Herms, D. A., Bonello, P., and Mittapalli, O. (2011). Antioxidant genes of the emerald ash borer (Agrilus planipennis): Gene characterization and expression profiles. J. Insect Physiology 57, 819–824. doi:10.1016/j.jinsphys.2011.03.017
Schultz, J., Milpetz, F., Bork, P., and Ponting, C. P. (1998). SMART, a simple modular architecture research tool: Identification of signaling domains. Proc. Natl. Acad. Sci. U. S. A. 95 11, 5857–5864. doi:10.1073/pnas.95.11.5857
Serra, V., Von Zglinicki, T., Lorenz, M., and Saretzki, G. (2003). Extracellular superoxide dismutase is a major antioxidant in human fibroblasts and slows telomere shortening. J. Biol. Chem. 278, 6824–6830. doi:10.1074/jbc.M207939200
Shahbandeh, M. (2021). Total rice consumption worldwide from 2008/2009 to 2020/2021. New York, United States: Statista Inc., (in 1000 Metric Tons) ").
Sharif, R., Su, L., Chen, X., and Qi, X. (2022). Hormonal interactions underlying parthenocarpic fruit formation in horticultural crops. Hortic. Res. 9, uhab024. doi:10.1093/hr/uhab024
Sigrist, C. J., Cerutti, L., De Castro, E., Langendijk-Genevaux, P. S., Bulliard, V., Bairoch, A., et al. (2010). PROSITE, a protein domain database for functional characterization and annotation. Nucleic Acids Res. 38, D161–D166. doi:10.1093/nar/gkp885
Sohail, H., Noor, I., Nawaz, M. A., Ma, M., Shireen, F., Huang, Y., et al. (2022). Genome-wide identification of plasma-membrane intrinsic proteins in pumpkin and functional characterization of CmoPIP1-4 under salinity stress. Environ. Exp. Bot. 202, 104995. doi:10.1016/j.envexpbot.2022.104995
Sohal, R. S., Mockett, R. J., and Orr, W. C. (2002). Mechanisms of aging: An appraisal of the oxidative stress hypothesis. Free Radic. Biol. Med. 33, 575–586. doi:10.1016/s0891-5849(02)00886-9
Szklarczyk, D., Franceschini, A., Wyder, S., Forslund, K., Heller, D., Huerta-Cepas, J., et al. (2015). STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic acids Res. 43, D447–D452. doi:10.1093/nar/gku1003
Umasuthan, N., Bathige, S. D. N. K., Revathy, K. S., Lee, Y., Whang, I., Choi, C. Y., et al. (2012). A manganese superoxide dismutase (MnSOD) from Ruditapes philippinarum: Comparative structural- and expressional-analysis with copper/zinc superoxide dismutase (Cu/ZnSOD) and biochemical analysis of its antioxidant activities. Fish Shellfish Immunol. 33, 753–765. doi:10.1016/j.fsi.2012.06.024
Valentine, J. S., and Hart, P. J. (2003a). Misfolded CuZnSOD and amyotrophic lateral sclerosis. Proc. Natl. Acad. Sci. 100, 3617–3622. doi:10.1073/pnas.0730423100
Valentine, J. S., and Hart, P. J. (2003b). Misfolded CuZnSOD and amyotrophic lateral sclerosis. Proc. Natl. Acad. Sci. U. S. A. 100, 3617–3622. doi:10.1073/pnas.0730423100
Von Zglinicki, T. (2002). Oxidative stress shortens telomeres. Trends Biochem. Sci. 27, 339–344. doi:10.1016/s0968-0004(02)02110-2
Wang, M., Wang, B., Jiang, K., Liu, M., Shi, X., and Wang, L. (2018). A mitochondrial manganese superoxide dismutase involved in innate immunity is essential for the survival of Chlamys farreri. Fish Shellfish Immunol. 72, 282–290. doi:10.1016/j.fsi.2017.11.010
Wu, J.-C., Xu, J.-X., Yuan, S.-Z., Liu, J.-L., Jiang, Y.-H., and Xu, J.-F. (2001). Pesticide-induced susceptibility of rice to Brown planthopper Nilaparvata lugens. Entomologia Exp. Appl. 100, 119–126. doi:10.1046/j.1570-7458.2001.00854.x
Yamamoto, K., Kimura, M., Aso, Y., Banno, Y., and Koga, K. (2007). Characterization of superoxide dismutase from the fall webworm, Hyphantria cunea: Comparison of its properties to superoxide dismutase from the silkworm Bombyx mori. Appl. entomology zoology 42, 465–472. doi:10.1303/aez.2007.465
Yamamoto, K., and Yamaguchi, M. (2022). Characterization of a novel superoxide dismutase in Nilaparvata lugens. Archives insect Biochem. physiology 109, e21862. doi:10.1002/arch.21862
Yamamoto, K., Zhang, P., Banno, Y., Fujii, H., Miake, F., Kashige, N., et al. (2005a). Superoxide dismutase from the silkworm, Bombyx mori: Sequence, distribution, and overexpression. Biosci. Biotechnol. Biochem. 69, 507–514. doi:10.1271/bbb.69.507
Yamamoto, K., Zhang, P., He, N., Wang, Y., Aso, Y., Banno, Y., et al. (2005b). Molecular and biochemical characterization of manganese-containing superoxide dismutase from the silkworm, Bombyx mori. Comp. Biochem. Physiology Part B Biochem. Mol. Biol. 142, 403–409. doi:10.1016/j.cbpb.2005.09.002
Yamamoto, K., Zhang, P., He, N., Wang, Y., Aso, Y., Banno, Y., et al. (2005). Molecular and biochemical characterization of manganese-containing superoxide dismutase from the silkworm, Bombyx mori. Comp. Biochem. physiology 142, 403–409. doi:10.1016/j.cbpb.2005.09.002
Zelko, I. N., Mariani, T. J., and Folz, R. J. (2002). Superoxide dismutase multigene family: A comparison of the CuZn-SOD (SOD1), Mn-SOD (SOD2), and EC-SOD (SOD3) gene structures, evolution, and expression. Free Radic. Biol. Med. 33, 337–349. doi:10.1016/s0891-5849(02)00905-x
Keywords: superoxide dismutase, Nilaparvata lugens, jinggangmycin, glucose, sucrose, trehalose, bioinformatics
Citation: Sheraz A, Zhu H, Dong Q, Wang T, Zong S, Wang H, Ge L and Wu T (2023) The superoxide dismutase (SOD) genes family mediates the response of Nilaparvata lugens to jinggangmycin and sugar. Front. Physiol. 14:1197395. doi: 10.3389/fphys.2023.1197395
Received: 31 March 2023; Accepted: 25 April 2023;
Published: 10 May 2023.
Edited by:
Bin Tang, Hangzhou Normal University, ChinaReviewed by:
Kui Kang, Zunyi Normal University College, ChinaHaijun Xu, Zhejiang University, China
Hai-jian Huang, Ningbo University, China
Copyright © 2023 Sheraz, Zhu, Dong, Wang, Zong, Wang, Ge and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Linquan Ge, lqge@yzu.edu.cn; Tao Wu, wutao@yzu.edu.cn
†These authors have contributed equally to this work
 Ahmad Sheraz
Ahmad Sheraz Haowen Zhu1†
Haowen Zhu1† Linquan Ge
Linquan Ge