- UOSD SAFU, Department of Research, Diagnosis and Innovative Technologies, IRCCS-Regina Elena National Cancer Institute, Rome, Italy
Preclinical animal models are valuable tools to improve treatments of malignant diseases, being an intermediate step of experimentation between cell culture and human clinical trials. Among different animal models frequently used in cancer research are mouse and, more recently, zebrafish models. Indeed, most of the cellular pathways are highly conserved between human, mouse and zebrafish, thus rendering these models very attractive. Recently, several transgenic reporter mice and zebrafishes have been generated in which the luciferase reporter gene are placed under the control of a promoter whose activity is strictly related to specific cancer cellular processes. Other mouse models have been generated by the cDNA luciferase knockin in the locus of a gene whose expression/activity has increased in cancer. Using BioLuminescence Imaging (BLI), we have now the opportunity to spatiotemporal visualize cell behaviors, among which proliferation, apoptosis, migration and immune responses, in any body district in living animal in a time frame process. We provide here a review of the available models to visualized cancer and cancer-associated events in living animals by BLI and as they have been successful in identifying new stages of early tumor progression, new interactions between different tissues and new therapeutic responsiveness.
Bioluminescence Imaging Technique – Bli
In vivo bioluminescence imaging, BLI, represents an interesting current and future new approach to molecular imaging. It allows imaging of internally generated light linked to specific physiological and/or pathological cellular processes in living small animals. This non-invasive technique, allows quantification in the same animal of spatial and temporal progression of the process of interest and identification of animal-to-animal variations (Signore et al., 2010).
BioLuminescence imaging in living animals takes advantage of luciferase reporter genes as internal sources of light. Usually, using tissue specific promoters, animals are engineered to express luciferase gene in a specific tissue and/or in a particular cellular process. If the activity of the promoter is strictly dependent on a single protein, this approach allows to followed in vivo the transcription activity of that specific protein. Another strategy involves the cloning of the luciferase cDNA in the locus of the gene whose expression you want to follow in time. In contrast to the above startegy, in this case is visualized the expression and not the activity of the protein of interest. Both these strategies enable real-time non-invasive imaging of several biological processes (Figure 1). In addition, with the advent of CRISPR/Cas technology, it will be possible from now to insert the luciferase gene in the locus of interest in a manner much easier than in the past. Until now, the animal model that has been most used to study via BLI cellular processes is the mouse model, however, zebrafish models for BLI have been recently described, too.
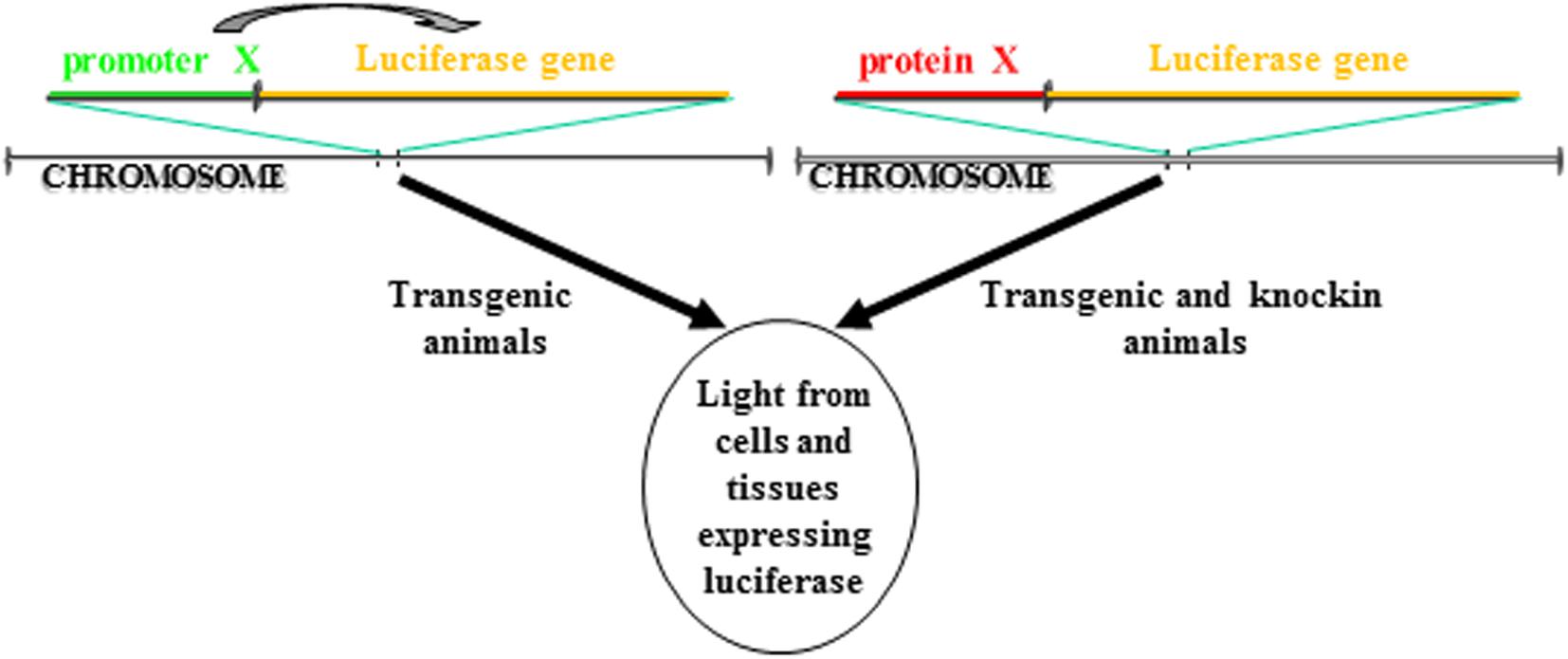
Figure 1. Different strategies to make a luciferase-based transgenic animal. One strategy take advantage of the transgenesis of cassettes in which the luciferase gene transcription is regulated by a promoter. The luciferase will be expressed only in cells in which the promoter is active. In another strategy, the cassette for transgenesis can include a fusion protein between the luciferase and the gene of interest. The cassette can be either knockin into the locus of the gene coding for the protein so that the cassette is under the transcription control of the endogenous promoter or the protein of interest can be directly inserted in frame into the locus. In both cases, the luciferase will be expressed only in cells in which the protein is expressed.
The most common reporter gene useful for BLI is the firefly (Photinus pyralis) luciferase, a heat-unstable enzyme with a half-life of approximately 2h thus useful to study dynamics of biological processes. Animals do not produce the substrate for the light producing process, the luciferin, thus giving an excellent signal-to-noise ratio, as there is virtually no background in the animal tissues. Upon intraperitoneal (i.p.) injection the luciferin distributes throughout the mouse rapidly and it passes across blood-tissue barriers including placenta (Lipshutz et al., 2001; Ciana et al., 2003). Likewise in mice, in zebrafish models, luciferin can be injected intraperitoneally or simply dissolved in aquarium water, letting the fish swimming (Chen et al., 2013). In organs expressing luciferase and in the presence of oxygen and ATP as a source of energy, the luciferin become oxyluciferin by a reaction catalyzed by luciferase and coupled with light emission (Figure 2). In addition to the firefly, many other luciferases are available for application of in vivo BLI. Among them, the most widely used are the sea pansy Renilla reniformis, the click beetle Pyrophorus plagiophthalamus, the marine copepod Gaussia princeps and the recently developed deep-sea shrimp derived NanoLuc. The substrate for the first two is d-luciferin while for Gaussia is coelenterazine and for NanoLuc a novel coelenterazine analog, furimazine (for a detailed review on available luciferase genes see Mezzanotte et al., 2017). The animals are imaged with cooled charge-coupled device (CCD) cameras mounted within a light-tight box in which the anesthetized animals are placed. The camera is accompanied with computer software for image data acquisition and analysis. The software converts electron signals into numerical values. The data are quantified by region-of-interest analysis, measuring photon flux from bioluminescence (Figure 1). The majority of the devices produce 2D images but a device has been developed that allows a 3D diffuse luminescence tomography (DLIT) that takes into account the scattering and absorption of light in tissue and provides an estimate of the 3D location and brightness of the light-emitting sources (Signore et al., 2010; Thompson et al., 2013). The sensitivity of the technique is influenced by many parameters. One crucial issue is the depth of the luciferase-labeled cells within the body. Upon administration of the luciferin substrate to the transgenic animals, the light emitted by luciferase is able to penetrate tissue depths however light intensity decreases 10-fold each centimeter of tissue depth (Signore et al., 2010). In addition BLI is decreased by pigmentation of fur. Removing fur or alternatively breeding mice into an albino background, can be useful (Signore et al., 2010). In the context of zebrafish models, due to the small size of fishes that reaches 4 cm in length and 0,5 cm of thickness and the absence of fur, difficulties about tissue penetration are slight, owing to the shorter distance through which the emitted light has to travel to reach body surface. The sensitivity of BLI is also influenced by the strength of the promoter used to drive luciferase expression as well as the number of incorporated transgenes per cells. Finally, CCD cameras used are also an important variable that modulates the sensitivity of this technique.
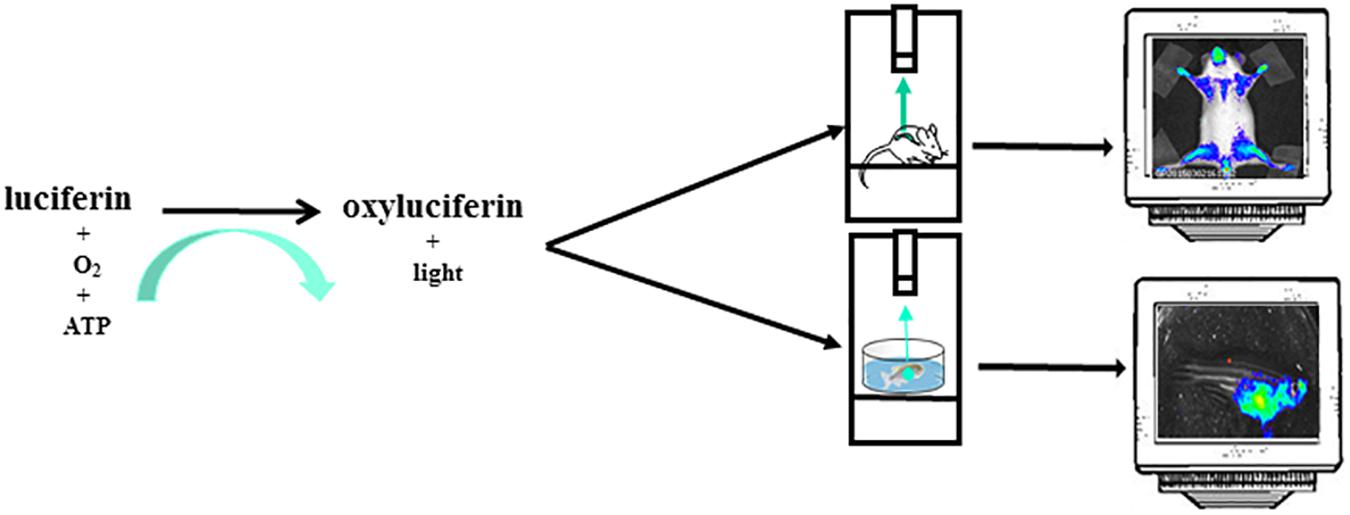
Figure 2. The instrument used for luminescence is composed of an ultra-sensitive cooled CCD camera linked to a dark box. In the dark box is present a shelf heated at 37°C where the mice, or the petri dish with zebrafishes, are positioned. The mice are euthanized trough a system delivering isoflurane or by i.p. injection of sedatives. For zebrafish imaging sessions the sedative is dissolved in the water. The CCD camera detects light coming from mice or zebrafishes. The images are collected and analyzed by a dedicated software. The mice image is referred to Mito-Luc model described in the text (Goeman et al., 2012). The zebrafish image represents a not already published bioluminescent model of proliferation (GP personal communication).
From an ethical point of view, the BLI technology offers significant values. In fact, the assay is performed on the same living animal through a series of non-invasive imaging sessions.
The most important thing is that this technique allows longitudinal experiments to be carried out without sacrificing mice at all times, thus reducing the number of animals needed.
Transgenic reporter mouse models for BLI
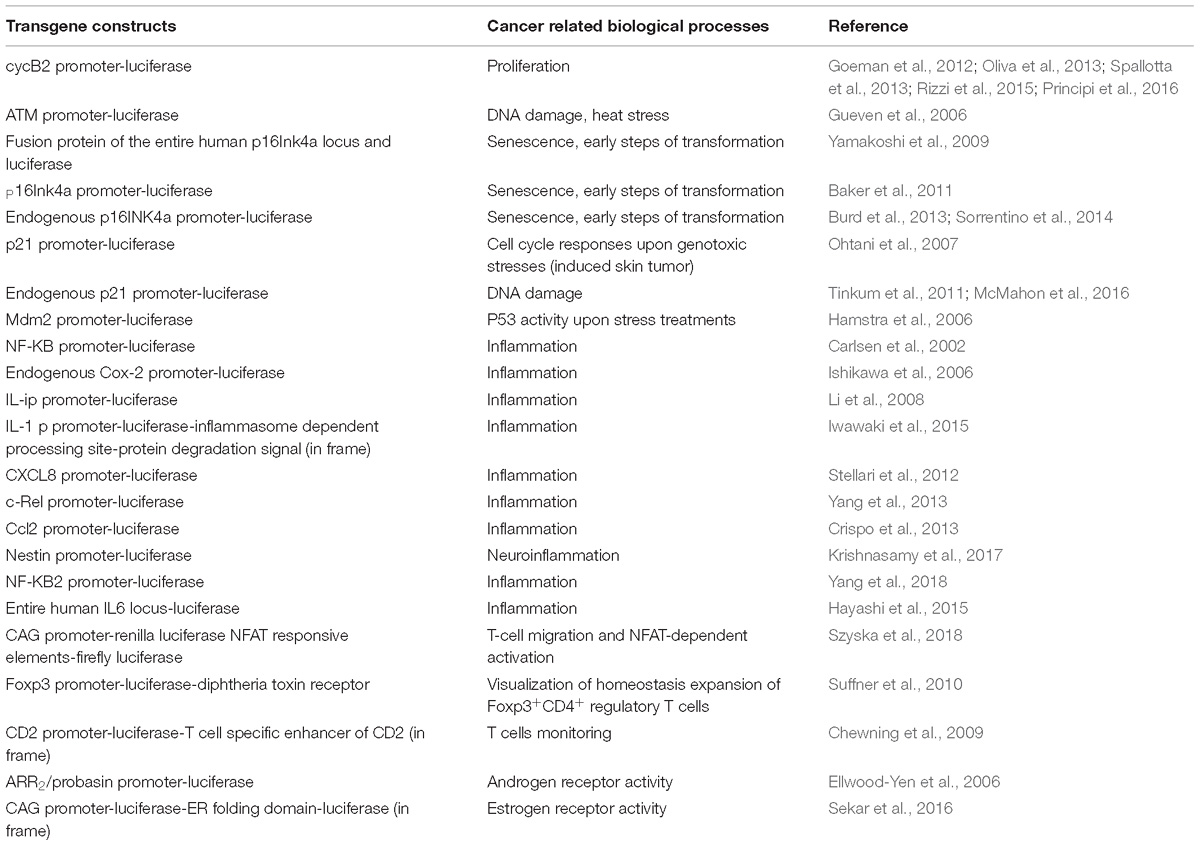
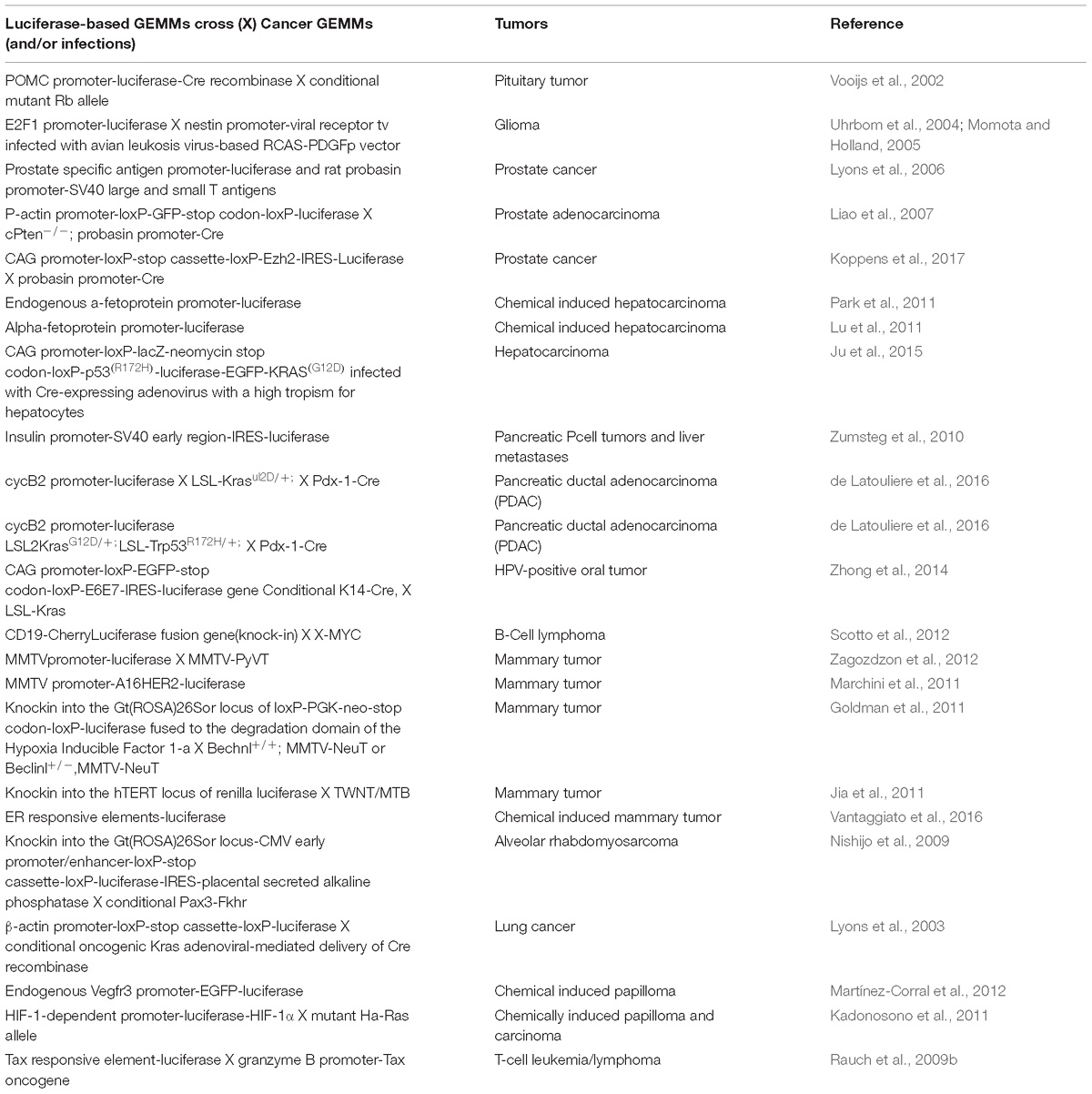
Non-invasive molecular imaging, BLI, is a powerful tool to study single molecular events in time in the physiological setting of a living organism. In the last decade, the application of this technique has been extended from static observation of anatomical structures to dynamic analysis of molecular events. When combined with cancer models, BLI is giving an unprecedented opportunity to investigate in the entire organism the molecular events leading to neoplastic development and progression. Several transgenic mouse models have been developed to visualize cancer growth and cancer-related cellular processes such as proliferation, tumor progression, inflammation and immune processes in living animals by BLI. These mice are not only useful for tracking healthy and disease processes in vivo, but also for testing the efficacy of therapeutic compounds. Although some models could be missed, the following list represents an update of transgenic models based on luciferase-dependent imaging generated so far. Tables 1, 2 summarize genetically engineered mouse models (GEMMs) described in this review. In Table 1 are reported GEMMs useful to illuminate cancer-related cellular processes while Table 2 reports GEMMs in which it is possible to image uptake, growth and progression of different cancers.
Reporter Mice of Cell Growth: Useful Tools for Oncology Research
Progression through each phase of the cell cycle is governed by cyclin/cyclin-dependent kinases (Cdks) complexes. We and others have contributed to demonstrate that the transcription of a plethora of cyclins and cdks, responsible for the cell cycle progression, is driven by the transcription factor NF-Y (Gurtner et al., 2017). Based on the rationale that the activity of NF-Y is restricted in vitro to proliferating cells, our group has generated a transgenic reporter mouse, called MITO-Luc (MITO stands for mitosis and Luc for luciferase), in which an NF-Y–dependent promoter controls luciferase expression. The model stems from a decade of research to identify an NF-Y-dependent cyclin B2 minimal promoter driving luciferase expression in all phases of the cell cycle. In these mice luciferase is expressed in each proliferating cell. Using BLI in these mice we visualize areas of physiological cell proliferation as well as regeneration processes in response to injury demonstrating a role of NF-Y activity on hepatocyte proliferation during liver regeneration. MITO-Luc reporter mice would be useful to study the involvement of genes in cell proliferation and aberrant proliferation in disease pathogenesis. They will be also useful to study the effect of new anti/pro-proliferative drugs and assessment of their efficacy both on target and non-target tissues. We have demonstrated that MITO-Luc mouse model is suitable to characterize the effects of small molecules acting on proliferation of bone marrow (BM) and spleen cells in time course experiments in intact animals (Goeman et al., 2012; Oliva et al., 2013; Spallotta et al., 2013; Rizzi et al., 2015; Principi et al., 2016).
Cyclin-Cdk complexes are negatively regulated by Cdk-inhibitors (CKIs) among which p16INK4a and p21Cip1. Apart from their capacity to arrest the cell cycle they have been shown to participate in an increasing number of cellular processes. p16INK4a limits cell-cycle progression and promotes cellular senescence in response to stress such as oncogene activation, while p21Cip1 is mainly involved in growth arrest, quiescence and induction of differentiation.
In 2006 Guaven and colleagues described a mouse model useful to visualize the transcriptional control of ATM, a gene whose mutations lead to a pleiotropic phenotype, including a predisposition to develop malignancies. In these animals the luciferase gene is under the control of the murine Atm promoter. Using this tool the authors demonstrated for the first time that ionizing radiation and heat stress induce ATM promoter in different mouse tissues (Gueven et al., 2006).
To monitor senescence and early steps of transformation transgenic reporter systems driven by fragments of the p16INK4a promoter (Yamakoshi et al., 2009; Baker et al., 2011), have been generated. Yamakoshi and colleagues engineered mice to express a fusion protein of the entire human p16Ink4a locus and luciferase. Baker and colleagues generated transgenic mice carrying a cassette in which a 2,617-bp fragment of the p16Ink4a gene promoter, that is transcriptionally active in senescent cell, drives transcription of luciferase gene. Burd and colleagues targeted luciferase gene into the translational start site of the endogenous p16INK4a locus employing a targeted knock-in strategy concerning in the cloning of luciferase gene into the p16INK4a open reading frame preserving known cis-regulatory elements (Burd et al., 2013). In these three systems the authors have been able to monitor senescence and the earliest steps of tumorigenesis suggesting that p16INK4a activation is a common characteristic of early steps of carcinogenesis. Interestingly, the knock-in model has been also employed to study the influence of several environmental agents on aging (Sorrentino et al., 2014).
To monitor cell cycle responses upon genotoxic stresses, reporter models useful to spatio-temporally monitor p21Cip1 activity have been generated. In the model generated by Ohtani et al., 2007, luciferase gene transcription is driving by a p21 promoter fragment. The authors induced skin tumor formation and by time-course BLI experiments they observed that the expression of p21Cip1 oscillates over the time (Ohtani et al., 2007). Later, in 2011 and 2016 two models have been described in which, using a knock-in strategy, the expression of luciferase is placed under the control of endogenous p21 promoter (Tinkum et al., 2011; McMahon et al., 2016). The treatments of these mice with several DNA-damaging agents, including ionizing radiation lead to a temporary light induction. Interestingly, Tinkum and colleagues crossed their mice with p53 null mice, demonstrating that p53 was required for the p21-dependent light. Indeed, both exogenous (Ohtani et al., 2007) and endogenous (Tinkum et al., 2011) p21 promoters are under the control of p53, making these models ideal for monitoring not only p21Cip1 but p53 activation in vivo, too.
p53 is a key mediator of cell cycle arrest of stressed cells. Two different reporter models have been generated to track p53 transcriptional activity by BLI. In these mice luciferase gene is transcribed by a p53-dependent promoter, mdm2 and Puma (Hamstra et al., 2006; Briat and Vassaux, 2008). In both papers it is possible to temporal evaluate p53 activity upon stress treatments: ionizing radiation and chemically induced DNA damage, respectively. In view of the major role that p53 plays in normal and transformed cells, these mice represent a powerful tool to predict, map and characterize side effects of drugs helping in the design of new therapeutic strategies.
Making Tumors Bright
Genetically engineered mouse models of human cancer are a useful tool to investigate molecular mechanisms of tumorigenesis (Tuveson and Hanahan, 2011). The utility of GEMMs in preclinical research may improve even further when they are combined with non-invasive bio-imaging techniques. GEMMs carrying bioimaging reporter systems that tag specific cellular processes or molecular events observed in human cancers are useful tools for early detection of cancer as well as for understanding of cancer initiation, immune system roles, tumor angiogenesis, invasion and cancer therapy. In the past years several different GEMMs have been generated in which cancer growth can be followed by BLI. These models represent several big killer cancers among which pancreatic, prostate, brain, breast and liver cancer.
The pioneer example of bright tumor in mice is that of pituitary tumor development. Vooijs and colleagues (Vooijs et al., 2002) took advantage of animals carrying one conditional mutated Rb allele. They crossed these mice with mice expressing Cre recombinase and luciferase gene under the control of a pituitary specific promoter. These mice endogenously develop pituitary bright tumors. These mice are a powerful tool to study cancer prevention and treatment using anticancer agents that interfere with the Rb pathway.
To measure brain tumor growth non-invasively, Ef-luc transgenic mice expressing luciferase gene under the control of the E2F1 promoter have been crossed GEMM of human gliomas. The E2F1 promoter is autoregulatory during late G1 and S phase of the cell cycle being inhibited by E2F-RB during early G1. Ef-luc mouse line crossed with the N–tv-a mouse strain expressing the viral receptor tv-a from the nestin promoter. Upon infection with a viral vector expressing RCAS-PDGFB, oligodendrogliomas have been induced in these mice and the authors were able to follow development of these tumors by BLI. This system has been used to follow early stages of brain tumorigenesis and to examine where tumors initiate (Uhrbom et al., 2004; Momota and Holland, 2005).
Several murine models have been developed to real-time monitoring of prostate cancer growth based on the use of a specific prostate promoter. In one model luciferase is under the control of the PSA promoter, whose activity is confined to the epithelial cells of the prostate. In these animals the expression of SV40 large T antigene is targeted to prostate using the rat probasin promoter. In these mice prostate lesions rise progressing from hyperplasia through to high-grade lesions that eventually metastasize. The authors demonstrated that these mice are useful to follow the tumor response to androgen ablation in a long timeframe (Lyons et al., 2006). In another example a composite promoter fragment from PSA promoter has been used to generate the PSA-E2/P-luc transgenic mice. In these mice the luciferase is mainly expressed in the ventral prostate lobes. This model, when bred unto the TRAMP prostate cancer background allows not only the uptake but also the progression of distant soft-tissue metastasis of prostate cancer (Seethammagari et al., 2006). On the basis of these data it is reasonable to assume that if the transgenic mouse model PSA-Luc were crossed with transgenic models that develop prostate cancer it would be possible to visulalize the onset, growth and progression of prostate cancer through BLI. The other model take advantage of the Cre/loxP technology, inactivating Pten alleles and activating luciferase and GFP expression in the prostate epithelial cells. Interestingly, although Cre expression is under an androgen-dependent promoter, once the genes undergo recombination neither the Pten allele nor luciferase requires androgen for expression thus making this model useful for monitoring growth, regression, or relapse of cancer irrespective of hormonal manipulations (Liao et al., 2007). On the other end, a model useful to image androgen receptor signaling is the transgenic mouse ARR2 Pb-Lux that expresses luciferase specifically in the prostate in an androgen-dependent fashion (Ellwood-Yen et al., 2006). Recently, a mouse model has been generated in which it is possible to visualize prostate tumor growth and metastatic progression over time. In this mouse model the Polycomb Group protein EZH2, whose overexpression is associated with poor prognosis in prostate cancer, and luciferase gene are transcribed together. The prostate-specific expression of the transgene is mediated by crossing of these mice with Probasin-Cre carrying mice (Koppens et al., 2017).
In 2011 Lu and colleagues described the generation of a knock in mice in which a thymidine kinase and luciferase reporter genes were placed under the transcriptional control of the endogenous alpha-fetoprotein (AFP) promoter. In the same year a transgenic mouse expressing luciferase under control of AFP promoter has been described (Park et al., 2011). Both models have been demonstrated to be useful to monitor chemical induced hepatocarcinoma by BLIs (Lu et al., 2011). Recently, a GEMM has been developed in which, following Cre-mediated DNA excision, the two oncogenes p53R172H and KRASG12D, are expressed in a single open reading frame with luciferase and GFP genes. In these mice, systemic injection of Cre encoding adenovirus induce the expression of the four transgenes in the liver leading to the development of bright liver tumors The bioluminescent signals correlates with tumor sizes, demonstrating the utility of this model for BLI of liver tumor development (Ju et al., 2015).
With the purpose to monitor pancreatic T-cell tumors non-invasively by BLI, Zumsteg and colleagues in the 2010 developed mice in which the expression of a bicistronic mRNA coding for large T antigen and luciferase occurs specifically in β-cells of Langerhans islets. In these mice the authors were able to illuminate tumor progression as well as lymph node and liver metastases (Zumsteg et al., 2010).
Genetically engineered mouse models designed to recapitulate genetic and pathologic aspects of pancreatic ductal adenocarcinoma (PDAC) are the LSL-KrasG12D/+;Pdx-1-Cre (KC) and LSL-KrasG12D/+;LSL-Trp53R172H/+;Pdx-1-Cre (KPC) mice, in which the Cre-recombinase, transcribed by the pancreas-specific Pdx-1 promoter, leads to the expression of oncogenic mutant for of Kras alone or in combination with a mutant p53 protein, respectively (Hingorani et al., 2003, 2005). Our group has recently developed and characterized two novel mouse models, MITO-Luc-KC (MKC) and MITO-Luc-KPC (MKPC), obtained through intercross of KC and KPC with our MITO-Luc mouse model, engineered to express the luciferase reporter gene in cells undergoing active proliferation. In these mice we have had the opportunity to follow PDAC evolution in the living animal in a time frame process. Altogether, in vivo and ex vivo analyses demonstrate that MKC and MKPC mouse models are powerful tools for visualizing PDAC development in terms of proliferation in the entire living animal in a spatio-temporal manner. Most notably, these results also demonstrate that, in these mouse models, it is possible to identify early steps of pancreatic carcinogenesis non-invasively in living animals in which proliferation events take place before tumor appearance (de Latouliere et al., 2016). From a pharmacological point of view, this opens the possibility to design therapeutic protocols with a more precise timing than those using the tumor palpability as a starting point, a parameter of low specificity and sensitivity.
To track responses to small molecules, Zhong and collegues developed and HPV-positive oral tumor mouse model in which HPV oncogenes E6, E7, mutant kras as well as a luciferase reporter (iHPV-Luc) are conditionally express in the epithelial cells upon a tamoxifen-regulated Cre recombinase system. In these transgenic mice tamoxifen treatment resulted in oral tumor development the development of which can be easily monitored by BLI (Zhong et al., 2014).
To achieve bioluminescence in B-cell lymphoma Scotto and colleagues have developed a transgenic mouse carrying CherryLuciferase fusion protein targeted to one allele of the CD19 locus crossed with GEMM of an aggressive lymphoma due to a chromosomal translocation of the c-myc gene (Scotto et al., 2012).
For BLI mammary tumor visualization and monitoring, a mouse strain expressing luciferase and Polyoma Virus middle T antigen in mammary glands under control of MMTV promoter (Zagozdzon et al., 2012) has been generated. Similarly, Marchini and colleagues, to examine the ability of Δ16HER2, a splice variant of HER2 gene, to induce transformation of mammary epithelium and to monitor Δ16HER2-mediated tumorigenesis in alive mice, developed a mouse strain in which transcription of luciferase and Δ16HER2 are under control of MMTV promoter (Marchini et al., 2011). Goldman and colleagues in the 2011, developed a transgenic mouse model in which regions of hypoxia are imaged via BLI thanks to the expression of the oxygen-dependent degradation domain of the Hypoxia Inducible Factor 1-α gene in frame with luciferase gene. These mice have been cross-breed with transgenic mice developing mammary tumors (MMTV-neu and Beclin1 +/−), and tumor growth were longitudinally tracked over several weeks and in response to treatments (Goldman et al., 2011). Based on this proof-of-principle the authors hypothesized that this GEMM could serve as a platform for non-invasive imaging of solid tumors in mice. An interesting model has been generated useful to image the transcriptional regulation of human TERT expression. These mice carry a 160-kb transgenic bacterial artificial chromosome containing the human TERT locus and Renilla luciferase cassette downstream human TERT promoter (Jia et al., 2011). This model has been crossed with mice overexpressing Wnt1 oncogene by a Dox-dependent promoter activated via mammary-specific expression of reverse tetracycline transactivator. Mammary adenocarcinomas developed consistently in female mice and imaging of human TERT transcription has been followed in mammary gland during tumorigenesis. To determine the dynamics of estrogen receptor activity, GEMM expressing a luciferase reporter gene under the control of estrogen receptor responding elements has been generated. This study has been the pioneer in the field of transgenic luciferase-reporter animals models and, using young mice before sex hormones production and/or ovariectomized adult mice, it highlights, for the first time, the importance of hormone-independent activation of the estrogen receptor (Ciana et al., 2003). This model has been later used to demonstrate the applicability of BLI to the study of ER signaling during breast transformation. The authors induced sporadic mammary cancers in these mice with DMBA (9,10-dimethyl 1,2-benzanthracene) and imaged estrogen receptors activity by BLI from early stages to palpable tumors in the breast (Vantaggiato et al., 2016). Recently, a GEMM expressing an ERα folding-biosensor has been developed. In these mice it is possible to evaluate the impact of diverse estrogenic ligands and to study the estrogen induced carcinogenesis in vivo (Sekar et al., 2016).
Luciferase activity measured by BLI in entire living animals allow a spatio-temporal imaging of its expression. However, it is a semi-quantitative measure being possible only a relative measure between the different individuals and/or the different treatments. Nishijo et al. (2009) developed a bicistronic Cre/LoxP reporter mouse strain in which semiquantitative, spatially expression of luciferase is coupled with quantitative expression of a serum biomarker, the human placental secreted alkaline phosphatase (SeAP). These mice have been crossed with an inducible Pax7-CreER mouse reporter line, allowing a muscle specific expression of luciferase and SeAP and with a mouse model of rhabdomyosarcoma. In this contest they investigate correlations between tumor volume, luciferase signal and serum SeAP levels, demonstrating that they have an higher precision than the traditional pre-clinical models (Nishijo et al., 2009).
To image spontaneously arising tumor burden, Lyons and colleagues developed a ubiquitously expressed conditional (Cre/Lox) luciferase transgene. In these mice it is possible to visualize cells that have undergone Cre-mediated recombination in alive animals. The mice have been crossed with transgenic mice in which a conditional oncogenic Kras expressed in lung epithelial cells induces non-small cell lung cancer and the authors demonstrate that, in this contest, longitudinal non-invasive visualization of lung tumorigenesis by BLI is feasible (Lyons et al., 2003).
An interest GEMM is represented by the Vegfr3 mouse model, where an EGFP-luciferase fusion protein, is expressed under transcriptional control of the endogenous Vegfr3 promoter. In these mice Martinez-Corral and colleagues were able to track tumor-induced lymphangiogenesis in the context of papillomas induced by dimethylbenzanthracene/12-O-tetradecanoylphorbol-13-acetate (DMBA/TPA) (Martínez-Corral et al., 2012). Similar to the GEMM developed by Goldman and collegues, an HRE/ODD-luciferase mouse model consisting of a HIF-1-dependent promoter and a cDNA encoding luciferase fused to the sequence of the ODD domain of human HIF-1α has been developed (Kadonosono et al., 2011). These mice were crossed with mice carrying a Ha-Ras mutated allele. which show a higher sensitivity to carcinogens than conventional mice. Treatment of these mice with N-methyl-N-nitrosourea (MNU), leads to bioluminescent tissues contained papillomas and carcinomas.
Imaging Cancer-Associated Inflammation and Immune Response in Mice
Among different transgenic models developed to spatio-temporally image inflammation in living animals, the most used is the one generated by Carlsen et al., 2002. In these mice expression of luciferase is driven by NF-kB response elements. Playing NF-KB a major role in physiological and pathological inflammation, these mice are a very useful tool to visualize inflammation in intact animals. They were used to image inflammation in any body district associated with several inputs among them pro- and anti-inflammatory treatments including stressors (Carlsen et al., 2004; Vykhovanets et al., 2008; Vykhovanets et al., 2012; Young et al., 2014), b-islet and cardiac transplantation (Roth et al., 2006; Ma et al., 2008), myocardial infarction (Tillmanns et al., 2006), Escherichia Coli infection of mammary glands (Notebaert et al., 2008), autoimmunity (Zangani et al., 2009) and high fat diet (Vykhovanets et al., 2011). Other tools to image chronic and acute inflammation are two knock-in mice in which luciferase gene is cloned in the endogenous cyclooxygenase-2 locus (Ishikawa et al., 2006). In other transgenic mouse lines useful to image inflammation, transcription of luciferase gene is driven by promoters of genes involved in inflammation processes, such as a Smad2/3-dependent promoter (Lin et al., 2005), human IL-1beta (Li et al., 2008; Iwawaki et al., 2015), CXCL8 (Stellari et al., 2012), c-Rel (Yang et al., 2013) ccl20 (Crispo et al., 2013), nestin (Krishnasamy et al., 2017) and NFkb2 (Yang et al., 2018) promoters. A sensitive GEMM useful to monitor inflammation has been generated integrating luciferase reporter gene in a bacterial artificial chromosome containing sequences of the human interleukin 6 gene (Hayashi et al., 2015). In all these animal models, systemic inflammation has been monitored in a variety of healthy and damaged tissues. However, any of these interest models has yet been studied in the contest of cancer-related processes.
Few studies visualize inflammation processes related to cancer. One interest study by Rauch and colleague have employed a mouse model of spontaneous lymphoma in which inflammation associated to malignant transformation is coupled with light emission. They crossed transgenic mice in which luciferase transcription is regulated by HTLV-1 LTR promoter with mice expressing Tax oncogene under the human granzyme B promoter in activated T cells and natural killer cells. These mice develop leukemia and lymphoma and the authors were able to discover an already unknown inflammation step preceding tumorigenesis (Rauch et al., 2009b). Using this model the authors were also able to demonstrate that inflammatory stimulus leads to the development of lymphoma (Rauch et al., 2009a).
Although bioimaging of immune response is a challenging item and many efforts have been made to date to visualize in vivo the immune response to antigens relatively few transgenic animal models have been produced. To study adoptive T-cell therapy efficacy in the presence of large solid tumors a transgenic mouse models that allow monitoring T-cell activation in response to cancer has been generated. In this model renilla luciferase is under control of a constitutive promoter while firefly luciferase transcription is driven by the NFAT promoter thus allowing the concomitant analysis of T-cell migration and NFAT-dependent activation. The T-cells isolated from these mice are useful for longitudinal studies have been employed to study the kinetics of T-cell activation upon immune reconstitution of myelo ablated hosts in the presence of two tumor models (Szyska et al., 2018). Suffner and colleagues in 2010 generated transgenic animals in which it is possible to visualize homeostasis expansion of Foxp3+CD4+ regulatory T cells (Tregs). These cells play a major role in maintaining self-tolerance and limiting immune responses to pathogens. In these mice the promoter of Foxp3 gene transcribes luciferase and the diphtheria toxin receptor. By BLI the authors demonstrated that Tregs were mainly located in lymphoid organs. Treg depletion upon diphtheria toxin treatment was also monitored by BLI, and longitudinal studies showed that the Treg compartment was recovered to its original size due to homeostatic proliferation of the remaining Tregs (Suffner et al., 2010). To study T cell dynamics following antigen encounter, a GEMM has been generated using a human CD2 mini-gene to drive luciferase transcription on T cell compartment. To real time image antigen-specific CD4+ T cell kinetics, the authors crossed these mice with OVA-specific CD4 TCR transgenic mice and analyzed CD4+ T cell antigen-specific responses. They were able in this system to compare kinetics and magnitude of clonal expansion/contraction in lymph nodes and tissue sites of antigen injections (Chewning et al., 2009). Although the two last models have not been used to follow cancer–associated immune response, they definitely represent useful tools for future studies in immuno-oncology research.
Real-Time Imaging of Gene Expression: Light Insights From Zebrafish
Despite 450 million years of evolutionary distance, cell and molecular pathways that govern signaling, proliferation, differentiation, and apoptosis are highly conserved between human and zebrafish, thus Zebrafish models represent a fundamental tool to improve treatments of malignant disease, as an intermediate experimental step between cell culture based assays and human clinical trials. Contrary to what has been published for mouse models, there are still few transgenic zebrafish models that exploit BLI by driving the expression of luciferase reporter gene under the control of a regulatory sequence or embedded on endogenous loci. Due to the optical transparency, defective or pathological phenotypes can be analyzed in the whole-mount embryos using fluorescent proteins that allows the study of the disease processes. On the other hand, the detection of fluorescent proteins in living adult animals is particularly tricky, due to the presence of the no-transparent skin of adult zebrafish that avoids to detect signals from an internal organ and restricts the observation on tissues near the body surface. Multiple advantages of bioluminescence overcome fluorescence detection in living adult animals among which the lack of background noise signal and the fact that the wavelength of the light emitted from luciferase-expressing cells is enough to penetrate all tissues in small animals.
Up today the BLI technology has been mainly applied to zebrafish tumor xenograft model, using luciferase labeled cancer cells to track tumor growth in vivo and to test the efficacy of antitumor and antiangiogenic compounds (Letrado et al., 2018). Noteworthy Chen’s group in the 2013, for the first time, has applied BLI methodology in a transgenic zebrafish model. They found that luciferase activity in adult zebrafish can be reproducibly monitored in tissues of freely swimming animals in a non-invasive manner. Although melanin absorbs light, the photons emitted from inner tissues do not show signal attenuation by the melanophores (Chen et al., 2013). Both ubiquitous and tissue-specific luciferase-based transgenic lines has been described. In the ubiquitous lines, ubiquitin or β-actin2 promoters drive widespread luciferase expression. On the contrary, in the tissue specific lines, cryaa or cmlc2 promoters drive luciferase expression specifically in the lens and cardiomyocytes, respectively. Interestingly, in the cmlc2-luc line, authors were able to demonstrate that luciferase-dependent imaging is useful to estimate muscle quantity in the inner organ, the heart, and to longitudinally follow cardiac regeneration after injury.
A second example of BLI methodology applied to a transgenic zebrafish model comes from the paper of Astuti and colleagues in the 2017. The authors generated a zebrafish strain that ubiquitously expressed luciferase gene under control of the ubiquitin promoter. Hematopoietic stem/progenitor cells have been collected from marrow contained in the kidney of adult zebrafish of this strain and it has been transplanted in an irradiated non luciferase expressing zebrafish strain. In this model system it is possible to track by BLI, hematopoietic cell homing function, immediately after the hematopoietic cell transplantation and using this model they have been able to demonstrate a positive role of ergosterol, a vitamin D2 precursor, in the homing of hematopoietic cells (Astuti et al., 2017).
In the same year, another zebrafish model has been generated in which an NF-κB promoter fragment drives expression of luciferase and GFP. Inflammation treatments of these animals, such as TNFα, induces a dose-dependent luciferase signal in live transgenic embryos demonstrating that this models is a valuable tool for studying NF-κB signaling in a spatiotemporal resolution manner (Kuri et al., 2017).
Drawbacks of Luciferase-Mediated Imaging
As described so far, the use of BLI in preclinical studies certainly has many advantages. However, when interpreting the results, it is important to take into account several shortcomings. BLI is a semiquantitative method and all results are relative to each other that’s why it’s not easy to standardize the experiments. The BLI signal could not be directly proportional to cell number since both endogenous and exogenous factors can impact the various steps of the luciferase/substrate reaction and could lead to misinterpretations. It is important to have in mind that the amount of luciferases, luciferin and cofactors such as ATP can be different in different cells/body districts (Fan and Wood, 2007). Half-life and stability of different luciferases can vary between few hours to several days. In case of use of secreted luciferases it’s important to take into account that some circulating components of blood and urine can also affect the enzyme stability (Czupryna and Tsourkas, 2011). The technique is restricted to small animals. Indeed, the depth and the location of the luciferase-expressing cells in the body can impact on the BLI signal. The administration route of the substrate is also critical for BLI. After i.p. or i.v. injections d-luciferin has different biodistribution being more homogeneous among tissues when the substrate is administered i.v. However, i.p administration yielded a prolonged organ uptake of the substrate (Berger et al., 2008). Light quenching is mediated by pigmented molecules (e.g., hemoglobin) and depends to the depth of the source of the signal. Hair and fur can scatter and attenuate light signal. The black fur color of the animals can attenuate the light and the signal is lower in black compared to white mice (Edinger et al., 2002). Although the high sensitivity of BLI is due to its low background, light emission by substrates such as coelenterazine, in the absence of the enzyme, can increase the luminescence background.
Conclusion and Future Perspectives
It has been evident for over a century that cancer is a systemic disease but the participation of the host macro-environment in tumor development and progression is only beginning to be appreciated. Recently, several emerging evidences highlight the temporal and spatial activation of hematopoiesis and immune response during cancer progression. Indeed cancer tissue- and lymphoid-related circuits promote reciprocal interplay with unexpected complexity. For example, it has been described that luminal breast cancer (LBC) cells establish a systemic macro-environment that supports outgrowth of otherwise-indolent disseminated tumors. LBCs secrete cytokines that activate BMCs (Kuznetsov, 2012). This cascade results in growth of adenocarcinoma and is abolished when BMCs activation is inhibited by an anti-inflammatory compound such as aspirin. These findings highlight the systemic macro-environment as an important component of disease progression (Becker, 2014). However, the crosstalk between macro-environment and tissue target of disease is still not yet widely known and there are still important unanswered questions. The lack of this knowledge are primarily due to the lack of human and animal models in which it is possible to follow tumor evolution in a time frame process.
BLI reporter animals are going to revolutionize the way biological processes, among which cancer, can be studied. Signals are generated with a fast kinetics, enabling the visualization of molecular events in real time, not only on the target organs but in all body districts allowing the visualization of the communicative reprogramming occurring in the entire animal. The non-invasive nature of the model allows investigations of physiological events from embryos to adult animals, as well as longitudinal studies of disease progression and/or drug response. The ability to monitor individual animals throughout longitudinal studies without sacrificing them significantly reduce the number of animals required in each experiment. It will also be possible to detect even sligth changes due to variability among individuals. Although they are already giving important results on basic and applied cancer research, we can anticipate that in the future they will be increasingly used in translational cancer research, finding application among the others in drug discovery development and toxicology.
Author Contributions
IM, LdL, AG, and GP conceived and designed the review. GP wrote the manuscript. All authors read and approved the final manuscript.
Funding
This work has been partially funded by intramural IRE Internal Projects and by AIRC (MFAG 11752) to AG and by AIRC (IG-2012 n. IG13234) to GP.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work is dedicated to the memory of Daniela Bona. We are grateful to Gabriele Toietta for graphic support.
References
Astuti, Y., Kramer, A. C., Blake, A. L., Blazar, B. R., Tolar, J., Taisto, M. E., et al. (2017). A Functional bioluminescent zebrafish screen for enhancing hematopoietic cell homing. Stem Cell Rep. 8, 177–190. doi: 10.1016/j.stemcr.2016.12.004
Baker, D. J., Wijshake, T., Tchkonia, T., LeBrasseur, N. K., Childs, B. G., van de Sluis, B., et al. (2011). Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature 479, 232–236. doi: 10.1038/nature10600
Becker, J. C. (2014). Tumor-educated myeloid cells: impact the micro- and macroenvironment. Exp. Dermatol. 23, 157–158. doi: 10.1111/exd.12241
Berger, F., Paulmurugan, R., Bhaumik, S., and Gambhir, S. S. (2008). Uptake kinetics and biodistribution of 14C-D-luciferin—a radiolabeled substrate for the firefly luciferase catalyzed bioluminescence reaction: impact on bioluminescence based reporter gene imaging. Eur. J. Nucl. Med. Mol. Imag. 35, 2275–2285. doi: 10.1007/s00259-008-0870-6
Briat, A., and Vassaux, G. (2008). A new transgenic mouse line to image chemically induced p53 activation in vivo. Cancer Sci. 99, 683–688. doi: 10.1111/j.1349-7006.2008.00742.x
Burd, C. E., Sorrentino, J. A., Clark, K. S., Darr, D. B., Krishnamurthy, J., Deal, A. M., et al. (2013). Monitoring tumorigenesis and senescence in vivo with a p16(INK4a)-luciferase model. Cell 152, 340–351. doi: 10.1016/j.cell.2012.12.010
Carlsen, H., Alexander, G., Austenaa, L. M., Ebihara, K., and Blomhoff, R. (2004). Molecular imaging of the transcription factor NF-κB, a primary regulator of stress response. Mutat. Res. 13, 199–211. doi: 10.1016/j.mrfmmm.2004.02.024
Carlsen, H., Moskaug, J., Fromm, S. H., and Blomhoff, R. (2002). In vivo imaging of NF-kappa B activity. J. Immunol. 168, 1441–1446. doi: 10.4049/jimmunol.168.3.1441
Chen, C. H., Durand, E., Wang, J., Zon, L. I., and Poss, K. D. (2013). Zebraflash transgenic lines for in vivo bioluminescence imaging of stem cells and regeneration in adult zebrafish. Development 140, 4988–4997. doi: 10.1242/dev.102053
Chewning, J. H., Dugger, K. J., Chaudhuri, T. R., Zinn, K. R., and Weaver, C. T. (2009). Bioluminescence-based visualization of CD4 T cell dynamics using a T lineage-specific luciferase transgenic model. BMC Immunol. 10:44. doi: 10.1186/1471-2172-10-44
Ciana, P., Raviscioni, M., Mussi, P., Vegeto, E., Que, I., Parker, M. G., et al. (2003). In vivo imaging of transcriptionally active estrogen receptors. Nat. Med. 9, 82–86. doi: 10.1038/nm809
Crispo, M., Van, Maele L, Tabareau, J., Cayet, D., Errea, A., Ferreira, A. M., et al. (2013). Transgenic mouse model harboring the transcriptional fusion ccl20-luciferase as a novel reporter of pro-inflammatory response. PLoS One 8:e78447. doi: 10.1371/journal.pone.0078447
Czupryna, J., and Tsourkas, A. (2011). Firefly luciferase and RLuc8 exhibit differential sensitivity to oxidative stress in apoptotic cells. PLoS One 6:e20073. doi: 10.1371/journal.pone.0020073
de Latouliere L, Manni, I., Iacobini, C., Pugliese, G., Grazi, G. L., Perri, P., et al. (2016). A bioluminescent mouse model of proliferation to highlight early stages of pancreatic cancer: a suitable tool for preclinical studies. Ann. Anat. 207, 2–8. doi: 10.1016/j.aanat.2015.11.010
Edinger, M., Cao, Y. A., Hornig, Y. S., Jenkins, D. E., Verneris, M. R., Bachmann, M. H., et al. (2002). Advancing animal models of neoplasia through in vivo bioluminescence imaging. Eur. J. Cancer 38, 2128–2136. doi: 10.1016/S0959-8049(02)00410-0
Ellwood-Yen, K., Wongvipat, J., and Sawyers, C. (2006). Transgenic mouse model for rapid pharmacodynamic evaluation of antiandrogens. Cancer Res. 66, 10513.31–10516.31. doi: 10.1158/0008-5472.CAN-06-1397
Fan, F., and Wood, K. V. (2007). Bioluminescent assays for high-throughput screening. Assay Drug Dev. Technol. 5, 127–136. doi: 10.1089/adt.2006.053
Goeman, F., Manni, I., Artuso, S., Ramachandran, B., Toietta, G., Bossi, G., et al. (2012). Molecular imaging of nuclear factor-Y transcriptional activity maps proliferation sites in live animals. Mol. Biol. Cell 23, 1467–1474. doi: 10.1091/mbc.E12-01-0039
Goldman, S. J., Chen, E., Taylor, R., Zhang, S., Petrosky, W., Reiss, M., et al. (2011). Use of the ODD-luciferase transgene for the non-invasive imaging of spontaneous tumors in mice. PLoS One 6:e18269. doi: 10.1371/journal.pone.0018269
Gueven, N., Fukao, T., Luff, J., Paterson, C., Kay, G., Kondo, N., et al. (2006). Regulation of the atm promoter in vivo. Genes Chromo. Cancer 45, 61.14–71.14. doi: 10.1002/gcc.20267
Gurtner, A., Manni, I., and Piaggio, G. (2017). NF-Y in cancer: Impact on cell transformation of a gene essential for proliferation. Biochim. Biophys. Acta Gene Regul. 1860, 604–616. doi: 10.1016/j.bbagrm.2016.12.005
Hamstra, D. A., Bhojani, M. S., Griffin, L. B., Laxman, B., Ross, B. D., and Rehemtulla, A. (2006). Real-time evaluation of p53 oscillatory behavior in vivo using bioluminescent imaging. Cancer Res. 66, 7482–7489. doi: 10.1158/0008-5472.CAN-06-1405
Hayashi, M., Takai, J., Yu, L., Motohashi, H., Moriguchi, T., and Yamamoto, M. (2015). Whole-body in vivo monitoring of inflammatory diseases exploiting human interleukin 6-luciferase transgenic mice. Mol. Cell. Biol. 35, 3590–3601. doi: 10.1128/MCB.00506-15
Hingorani, S. R., Petricoin, E. F., Maitra, A., Rajapakse, V., King, C., Jacobetz, M. A., et al. (2003). Pre invasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell 4, 437–450. doi: 10.1016/S1535-6108(03)00309-X
Hingorani, S. R., Wang, L., Multani, A. S., Combs, C., Deramaudt, T. B., Hruban, R. H., et al. (2005). Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell 7, 469–483. doi: 10.1016/j.ccr.2005.04.023
Ishikawa, T. O., Jain, N. K., Taketo, M. M., and Herschman, H. R. (2006). Imaging cyclooxygenase-2 (Cox-2) gene expression in living animals with a luciferase knock-in reporter gene. Mol. Imag. Biol. 8, 171–187. doi: 10.1007/s11307-006-0034-7
Iwawaki, T., Akai, R., Oikawa, D., Toyoshima, T., Yoshino, M., Suzuki, M., et al. (2015). Transgenic mouse model for imaging of interleukin-1β-related inflammation in vivo. Sci. Rep. 5:17205. doi: 10.1038/srep17205
Jia, W., Wang, S., Horner, J. W., Wang, N., Wang, H., Gunther, E. J., et al. (2011). A BAC transgenic reporter recapitulates in vivo regulation of human telomerase reverse transcriptase in development and tumorigenesis. FASEB J. 25, 979–989. doi: 10.1096/fj.10-173989
Ju, H. L., Calvisi, D. F., Moon, H., Baek, S., Ribback, S., Dombrowski, F., et al. (2015). Transgenic mouse model expressing P53(R172H), luciferase, EGFP, and KRAS(G12D) in a single open reading frame for live imaging of tumor. Sci. Rep. 5:8053. doi: 10.1038/srep08053
Kadonosono, T., Kuchimaru, T., Yamada, S., Takahashi, Y., Murakami, A., Tani, T., et al. (2011). Detection of the onset of ischemia and carcinogenesis by hypoxia-inducible transcription factor-based in vivo bioluminescence imaging. PLoS One 6:e26640. doi: 10.1371/journal.pone.0026640
Koppens, M. A., Tanger, E., Nacerddine, K., Westerman, B., Song, J. Y., and van, Lohuizen M (2017). A new transgenic mouse model for conditional overexpression of the polycomb group protein EZH2. Transgenic Res. 26, 187–196. doi: 10.1007/s11248-016-9993-x
Krishnasamy, S., Weng, Y. C., Thammisetty, S. S., Phaneuf, D., Lalancette-Hebert, M., and Kriz, J. (2017). Molecular imaging of nestin in neuroinflammatory conditions reveals marked signal induction in activated microglia. J. Neuroinflamm. 14:45. doi: 10.1186/s12974-017-0816-7
Kuri, P., Ellwanger, K., Kufer, T. A., Leptin, M., and Bajoghli, B. (2017). A high-sensitivity bi-directional reporter to monitor NF-κB activity in cell culture and zebrafish in real time. J. Cell Sci. 130, 648–657. doi: 10.1242/jcs.196485
Kuznetsov, A. V. (2012). A model of axonal transport drug delivery: effects of diffusivity. Int J. Numer. Method Biomed. Eng. 28, 1083–1092. doi: 10.1002/cnm.2469
Letrado, P., de Miguel, I., Lamberto, I., Díez-Martínez, R., and Oyarzabal, J. (2018). Zebrafish: speeding up the cancer drug discovery process. Cancer Res. 78, 6048–6058. doi: 10.1158/0008-5472.CAN-18-1029
Li, L., Fei, Z., Ren, J., Sun, R., Liu, Z., Sheng, Z., et al. (2008). Functional imaging of interleukin 1 beta expression in inflammatory process using bioluminescence imaging in transgenic mice. BMC Immunol. 9:49. doi: 10.1186/1471-2172-9-49
Liao, C. P., Zhong, C., Saribekyan, G., Bading, J., Park, R., Conti, P. S., et al. (2007). Mouse models of prostate adenocarcinoma with the capacity to monitor spontaneous carcinogenesis by bioluminescence or fluorescence. Cancer Res. 67, 7525–7533. doi: 10.1158/0008-5472.CAN-07-0668
Lin, S. L., Chen, R. H., Chen, Y. M., Chiang, W. C., Lai, C. F., Wu, K. D., et al. (2005). Pentoxifylline attenuates tubulointerstitial fibrosis by blocking Smad3/4-activated transcription and profibrogenic effects of connective tissue growth factor. J. Am. Soc. Nephrol. 16, 2702–2713. doi: 10.1681/ASN.2005040435
Lipshutz, G. S., Gruber, C. A., Cao, Y., Hardy, J., Contag, C. H., and Gaensler, K. M. (2001). In utero delivery of adeno-associated viral vectors: intraperitoneal gene transfer produces long-term expression. Mol. Ther. 3, 284–292. doi: 10.1006/mthe.2001.0267
Lu, X., Guo, H., Molter, J., Miao, H., Gerber, L., Hu, Y., et al. (2011). Alpha-fetoprotein-thymidine kinase-luciferase knockin mice: a novel model for dual modality longitudinal imaging of tumorigenesis in liver. J. Hepatol. 55, 96–102. doi: 10.1016/j.jhep.2010.10.020
Lyons, S. K., Lim, E., Clermont, A. O., Dusich, J., Zhu, L., Campbell, K. D., et al. (2006). Noninvasive bioluminescence imaging of normal and spontaneously transformed prostate tissue in mice. Cancer Res. 66, 4701–4707. doi: 10.1158/0008-5472.CAN-05-3598
Lyons, S. K., Meuwissen, R., Krimpenfort, P., and Berns, A. (2003). The generation of a conditional reporter that enables bioluminescence imaging of Cre/loxP-dependent tumorigenesis in mice. Cancer Res. 63, 7042–7046.
Ma, L., Xiang, Z., Sherrill, T. P., Wang, L., Blackwell, T. S., Williams, P., et al. (2008). Bioluminescence imaging visualizes activation of nuclear factor-kappaB in mouse cardiac transplantation. Transplantation 85, 903–910. doi: 10.1097/TP.0b013e318166cde1
Marchini, C., Gabrielli, F., Iezzi, M., Zenobi, S., Montani, M., Pietrella, L., et al. (2011). The human splice variant Δ16HER2 induces rapid tumor onset in a reporter transgenic mouse. PLoS One 6:e18727. doi: 10.1371/journal.pone.0018727
Martínez-Corral, I., Olmeda, D., Diéguez-Hurtado, R., Tammela, T., Alitalo, K., and Ortega, S. (2012). In vivo imaging of lymphatic vessels in development, wound healing, inflammation, and tumor metastasis. Proc. Natl. Acad. Sci. U.S.A. 109, 6223–6228. doi: 10.1073/pnas.1115542109
McMahon, M., Frangova, T. G., Henderson, C. J., and Wolf, C. R. (2016). Olaparib, monotherapy or with ionizing radiation, exacerbates dna damage in normal tissues: insights from a new p21 reporter mouse. Mol. Cancer Res. 14, 1195–1203. doi: 10.1158/1541-7786.MCR-16-0108
Mezzanotte, L., van’t Root, M., Karatas, H., Goun, E. A., and Löwik, C. W. G. M. (2017). In vivo molecular bioluminescence imaging: new tools and applications. Trends Biotechnol. 35, 640–652. doi: 10.1016/j.tibtech.2017.03.012
Momota, H., and Holland, E. C. (2005). Bioluminescence technology for imaging cell proliferation. Curr. Opin. Biotechnol. 16, 681–686. doi: 10.1016/j.copbio.2005.10.012
Nishijo, K., Hosoyama, T., Bjornson, C. R., Schaffer, B. S., Prajapati, S. I., Bahadur, A. N., et al. (2009). Biomarker system for studying muscle, stem cells, and cancer in vivo. FASEB J. 23, 2681–2690. doi: 10.1096/fj.08-128116
Notebaert, S., Carlsen, H., Janssen, D., Vandenabeele, P., Blomhoff, R., and Meyer, E. (2008). In vivo imaging of NF-kappaB activity during Escherichia coli-induced mammary gland infection. Cell. Microbiol. 6, 1249–1258. doi: 10.1111/j.1462-5822.2008.01123.x
Ohtani, N., Imamura, Y., Yamakoshi, K., Hirota, F., Nakayama, R., Kubo, Y., et al. (2007). Visualizing the dynamics of p21(Waf1/Cip1) cyclin-dependent kinase inhibitor expression in living animals. Proc. Natl. Acad. Sci. U.S.A 104, 15034–15039. doi: 10.1073/pnas.0706949104
Oliva, P., Roncoroni, C., Radaelli, E., Brunialti, E., Rizzi, N., De, Maglie M, et al. (2013). Global profiling of TSEC pro-liferative potential by the use of a reporter mouse for proliferation. Reprod. Sci. 20, 119–128. doi: 10.1177/1933719111431002
Park, J. H., Kim, K. I., Lee, Y. J., Lee, T. S., Kim, K. M., Nahm, S. S., et al. (2011). Non-invasive monitoring of hepatocellular carcinoma in transgenic mouse with bioluminescent imaging. Cancer Lett. 310, 53–60. doi: 10.1016/j.canlet.2011.06.013
Principi, E., Girardello, R., Bruno, A., Manni, I., Gini, E., Pagani, A., et al. (2016). Systemic distribution of single-walled carbon nanotubes in a novel model: alteration of biochemical parameters, metabolic functions, liver accumulation, and inflammation in vivo. Int. J. Nanomed. 11, 4299–4316. doi: 10.2147/IJN.S109950
Rauch, D., Gross, S., Harding, J., Bokhari, S., Niewiesk, S., Lairmore, M., et al. (2009a). T-cell activation promotes tumorigenesis in inflammation-associated cancer. Retrovirology 6:116. doi: 10.1186/1742-4690-6-116
Rauch, D., Gross, S., Harding, J., Niewiesk, S., Lairmore, M., Piwnica-Worms, D., et al. (2009b). Imaging spontaneous tumorigenesis: inflammation precedes development of peripheral NK tumors. Blood 113, 1493–1500. doi: 10.1182/blood-2008-07-166462
Rizzi, N., Manni, I., Vantaggiato, C., Delledonne, G. A., Gentileschi, M. P., Maggi, A., et al. (2015). In vivo imaging of cell proliferation for a dynamic, whole body, analysis of undesired drug effects. Toxicol. Sci. 145, 296–306. doi: 10.1093/toxsci/kfv056
Roth, D. J., Jansen, E. D., Powers, A. C., and Wang, T. G. (2006). A novel method of monitoring response to islet transplantation: bioluminescent imaging of an NF-kB transgenic mouse model. Transplantation 81, 1185–1190. doi: 10.1097/01.tp.0000203808.84963.13
Scotto, L., Kruithof-de, Julio M, Paoluzzi, L., Kalac, M., Marchi, E., Buitrago, J. B., et al. (2012). Development and characterization of a novel CD19CherryLuciferase (CD19CL) transgenic mouse for the preclinical study of B-cell lymphomas. Clin. Cancer Res 18, 3803–3811. doi: 10.1158/1078-0432.CCR-11-2588
Seethammagari, M. R., Xie, X., Greenberg, N. M., and Spencer, D. M. (2006). EZC-prostate models offer high sensitivity and specificity for noninvasive imaging of prostate cancer progression and androgen receptor action. Cancer Res. 66, 6199–6209. doi: 10.1158/0008-5472.CAN-05-3954
Sekar, T. V., Foygel, K., Massoud, T. F., Gambhir, S. S., and Paulmurugan, R. (2016). A transgenic mouse model expressing an ERα folding biosensor reveals the effects of bisphenol A on estrogen receptor signaling. Sci. Rep. 6:34788. doi: 10.1038/srep34788
Signore, A., Mather, S. J., Piaggio, G., Malviya, G., and Dierckx, R. A. (2010). Molecular imaging of inflammation/infection: nuclear medicine and optical imaging agents and methods. Chem. Rev. 110, 3112–3145. doi: 10.1021/cr900351r
Sorrentino, J. A., Krishnamurthy, J., Tilley, S., Alb, J. G. Jr., Burd, C. E., and Sharpless, N. E. (2014). p16INK4a reporter mice reveal age-promoting effects of environmental toxicants. J. Clin. Invest. 124, 169–173. doi: 10.1172/JCI70960
Spallotta, F., Cencioni, C., Straino, S., Nanni, S., Rosati, J., Artuso, S., et al. (2013). A nitric oxide-dependent cross-talk between class I and III histonedeacetylases accelerates skin repair. J. Biol. Chem. 288, 11004–11012. doi: 10.1074/jbc.M112.441816
Stellari, F. F., Franceschi, V., Capocefalo, A., Ronchei, M., Facchinetti, F., Villetti, G., et al. (2012). In vivo imaging of transiently transgenized mice with a bovine interleukin 8 (CXCL8) promoter/luciferase reporter construct. PLoS One 7:e39716. doi: 10.1371/journal.pone.0039716
Suffner, J., Hochweller, K., Kühnle, M. C., Li, X., Kroczek, R. A., Garbi, N., et al. (2010). Dendritic cells support homeostatic expansion of Foxp3+ regulatory T cells in Foxp3.LuciDTR mice. J. Immunol. 184, 1810–1820. doi: 10.4049/jimmunol.0902420
Szyska, M., Herda, S., Althoff, S., Heimann, A., Russ, J., D’Abundo, D., et al. (2018). A transgenic dual-luciferase reporter mouse for longitudinal and functional monitoring of t cells in vivo. Cancer Immunol. Res. 6, 110–120. doi: 10.1158/2326-6066.CIR-17-0256
Thompson, L., Dias, H. G., and Campos, T. P. (2013). Dosimetry in brain tumor phantom at 15 MV3D conformal radiation therapy. Radiat. Oncol. 8:168. doi: 10.1186/1748-717X-8-168
Tillmanns, J., Carlsen, H., Blomhoff, R., Valen, G., Calvillo, L., Ertl, G., et al. (2006). Caught in the act: in vivo molecular imaging of the transcription factor NF-kappaB after myocardial infarction. Biochem. Biophys. Res. Commun. 342, 773–774. doi: 10.1016/j.bbrc.2006.02.024
Tinkum, K. L., Marpegan, L., White, L. S., Sun, J., Herzog, E. D., Piwnica-Worms, D., et al. (2011). Bioluminescence imaging captures the expression and dynamics of endogenous p21 promoter activity in living mice and intact cells. Mol. Cell Biol. 31, 3759–3772. doi: 10.1128/MCB.05243-11
Tuveson, D., and Hanahan, D. (2011). Translational medicine: cancer lessons from mice to humans. Nature 471, 316–317. doi: 10.1038/471316a
Uhrbom, L., Nerio, E., and Holland, E. C. (2004). Dissecting tumor maintenance requirements using bioluminescence imaging of cell proliferation in a mouse glioma model. Nat. Med. 10, 1257–1260. doi: 10.1038/nm1120
Vantaggiato, C., Dell’Omo, G., Ramachandran, B., Manni, I., Radaelli, E., Scanziani, E., et al. (2016). Bioluminescence imaging of estrogen receptor activity during breast cancer progression. Am. J. Nucl. Med. Mol. Imag. 6, 32–41.
Vooijs, M., Jonkers, J., Lyons, S., and Berns, A. (2002). Noninvasive imaging of spontaneous retinoblastoma pathway-dependent tumors in mice. Cancer Res. 62, 1862–1867.
Vykhovanets, E. V., MacLennan, G. T., Vykhovanets, O. V., Cherullo, E. E., Ponsky, L. E., and Gupta, S. (2012). Molecular imaging of nuclear factor-κB in bladder as a primary regulator of inflammatory response. J. Urol. 187, 330–337. doi: 10.1016/j.juro.2011.09.007
Vykhovanets, E. V., Shankar, E., Vykhovanets, O. V., Shukla, S., and Gupta, S. (2011). High-fat diet increases NF-κB signaling in the prostate of reporter mice. Prostate 71, 147–156. doi: 10.1002/pros.21230
Vykhovanets, E. V., Shukla, S., MacLennan, G. T., Resnick, M. I., Carlsen, H., Blomhoff, R., et al. (2008). Molecular imaging of NF-kappaB in prostate tissue after systemic administration of IL-1 beta. Prostate 68, 34–41. doi: 10.1002/pros.20666
Yamakoshi, K., Takahashi, A., Hirota, F., Nakayama, R., Ishimaru, N., Kubo, Y., et al. (2009). Real-time in vivo imaging of p16Ink4a reveals cross talk with p53. J. Cell Biol. 186, 393–407. doi: 10.1083/jcb.200904105
Yang, X., Jing, H., Zhao, K., Sun, R., Liu, Z., Ying, Y., et al. (2013). Functional imaging of Rel expression in inflammatory processes using bioluminescence imaging system in transgenic mice. PLoS One 8:e57632. doi: 10.1371/journal.pone.0057632
Yang, X., Sun, R., Ci, L., Wang, N., Yang, S., Shi, J., et al. (2018). Tracing the dynamic expression of the Nfκb2 gene during inflammatory processes by in vivo bioluminescence imaging in transgenic mice. Biochem. Biophys. Res. Commun. 501, 41–47. doi: 10.1016/j.bbrc.2018.04.126
Young, N. A., Bruss, M. S., Gardner, M., Willis, W. L., Mo, X., Valiente, G. R., et al. (2014). Oral administration of nano-emulsion curcumin in mice suppresses inflammatory-induced NFκB signaling and macrophage migration. PLoS One 9:e111559. doi: 10.1371/journal.pone.0111559
Zagozdzon, A. M., O’Leary, P., Callanan, J. J., Crown, J., Gallagher, W. M., and Zagozdzon, R. (2012). Generation of a new bioluminescent model for visualisation of mammary tumour development in transgenic mice. BMC Cancer 12:209. doi: 10.1186/1471-2407-12-209
Zangani, M., Carlsen, H., Kielland, A., Os, A., Hauglin, H., Blomhoff, R., et al. (2009). Tracking early autoimmune disease by bioluminescent imaging of NF-kappaB activation reveals pathology in multiple organ systems. Am. J. Pathol. 174, 1358–1367. doi: 10.2353/ajpath.2009.080700
Zhong, R., Pytynia, M., Pelizzari, C., and Spiotto, M. (2014). Bioluminescent imaging of HPV-positive oral tumor growth and its response to image-guided radiotherapy. Cancer Res. 74, 2073–2081. doi: 10.1158/0008-5472.CAN-13-2993
Keywords: cancer, BioLuminescent Imaging (BLI), macroenvironment, animal models, in vivo image system
Citation: Manni I, de Latouliere L, Gurtner A and Piaggio G (2019) Transgenic Animal Models to Visualize Cancer-Related Cellular Processes by Bioluminescence Imaging. Front. Pharmacol. 10:235. doi: 10.3389/fphar.2019.00235
Received: 20 December 2018; Accepted: 25 February 2019;
Published: 15 March 2019.
Edited by:
Lina Ghibelli, University of Rome Tor Vergata, ItalyReviewed by:
Ignacio Bejarano, University of Cantabria, SpainClemens Löwik, Erasmus University Rotterdam, Netherlands
Copyright © 2019 Manni, de Latouliere, Gurtner and Piaggio. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Isabella Manni, isabella.manni@ifo.gov.it Giulia Piaggio, giulia.piaggio@ifo.gov.it
†Co-last authors
 Isabella Manni*
Isabella Manni* Luisa de Latouliere
Luisa de Latouliere Aymone Gurtner
Aymone Gurtner Giulia Piaggio
Giulia Piaggio