- Department of Basic Medical Sciences, Neurosciences and Sensory Organs, Section of Human Anatomy and Histology, University of Bari Medical School, Bari, Italy
microRNAs (miRNAs) are small non-coding RNA molecules, evolutionary conserved. They target more than one mRNAs, thus influencing multiple molecular pathways, but also mRNAs may bind to a variety of miRNAs, either simultaneously or in a context-dependent manner. miRNAs biogenesis, including miRNA transcription, processing by Drosha and Dicer, transportation, RISC biding, and miRNA decay, are finely controlled in space and time.
miRNAs are critical regulators in various biological processes, such as differentiation, proliferation, apoptosis, and development in both health and disease. Their dysregulation is involved in tumor initiation and progression. In tumors, they can act as onco-miRNAs or oncosuppressor-miRNA participating in distinct cellular pathways, and the same miRNA can perform both activities depending on the context.
In tumor progression, the angiogenic switch is fundamental. miRNAs derived from tumor cells, endothelial cells, and cells of the surrounding microenvironment regulate tumor angiogenesis, acting as pro-angiomiR or anti-angiomiR.
In this review, we described miRNA biogenesis and function, and we update the non-classical aspects of them. The most recent role in the nucleus, as transcriptional gene regulators and the different mechanisms by which they could be dysregulated, in tumor initiation and progression, are treated. In particular, we describe the role of miRNAs in sprouting angiogenesis, vessel co-option, and vasculogenic mimicry. The role of miRNAs in lymphoma angiogenesis is also discussed despite the scarcity of data.
The information presented in this review reveals the need to do much more to discover the complete miRNA network regulating angiogenesis, not only using high-throughput computational analysis approaches but also morphological ones.
Introduction
From the discovery of the lin-4/lin-14 paradigm in Caenorhabditis elegans, microRNAs (miRNAs) have come a long way (1–4). miRNAs were found in eukaryotic organisms, viruses, and prokaryotic cells.
miRNAs of viral origin, known as v-miRNAs, in the host, can function as post-transcriptional gene regulators as well as viral genes. Their expression can prolong the longevity of infected cells, enhance their immune response evasion, and regulate the switch to lytic infection (1–4). miRNAs in prokaryotic cells is a more recent and, therefore, unexplored research field. They are comparable in size and can be export outside the cells by vesicles as eukaryotic miRNAs, but diverge considerably from these in biogenesis and presentation to the target mRNAs (5).
miRNAs are non-protein-coding regulatory genes, evolutionary conserved, partially complementary to one or more messenger RNA (mRNA) molecules (target mRNAs) (6). Mature miRNAs are short, single-stranded RNA molecules of about 21-23 nucleotides in length, which is just about right for specificity to particular target genes as well as for plasticity to evolve towards new gene targets. They regulate the expression of homologous target-gene transcripts, at transcriptional and/or post-transcriptional level, through a mechanism known as RNA interference (RNAi). They may induce both down- and up-gene expression by different mechanisms in both cytoplasm and nucleus where they have been found.
In this review, we described miRNA biogenesis, regulation, and function underline essential aspects of processes localization and timeline. Indeed, we update the non-classical aspects of miRNAs, such as their new role in the nucleus as transcriptional gene regulators. Then different mechanisms by which miRNAs could be dysregulated in tumor initiation and progression are addressed. In particular, we describe the role of miRNA in sprouting angiogenesis, vessel co-option, and vasculogenic mimicry. The role of miRNA in lymphoma angiogenesis is also discussed despite the scarcity of data.
In the end, this review demonstrates the need to do much more to discover the complete miRNA network regulating angiogenesis, not only using high-throughput analysis approaches but also morphological ones.
miRNA Biogenesis
miRNAs biogenesis starts from DNA sequences, called miRNA genes, or from clusters genes which are only transcripted as miRNA molecules or together as polycistronic transcripts, respectively (7, 8). Alternatively, the miRNAs localize within an intron or untranslated region (UTR) of a protein-coding gene (8). Two main pathways of biogenesis were identified as canonical and non-canonical ones.
Canonical miRNA Biogenesis and Functions
In the canonical biogenesis pathway, primary miRNAs (pri-miRNA) are transcribed from their genes by RNA polymerase III, and then processed into precursor miRNA (pre-miRNAs) by the Microprocessor complex, consisting of two multiprotein units (9–11). One of these is a large multiprotein unit that includes different classes of RNA-associated proteins, double-stranded RNA (dsRNA) binding proteins, ribonucleoproteins, and Ewing’s sarcoma protein family. The other one is a small multiprotein unit that includes the RNase III enzyme, named Drosha, and the RNA binding protein DiGeorge Syndrome Critical Region 8 (DGCR8) (12).
The Microprocessor recognizes specific motifs within the pri-miRNA, and cleaves it at the base of the characteristic hairpin structure, generating a 5′-monophosphate and a 3′-2-nt overhang on pre-miRNA (10, 13, 14). After, pre-miRNAs are exported, from the nucleus to the cytoplasm, by a complex of exportin5 (XPO5) and RAS-related nuclear protein-guanosine-5’-triphosphate-ase (Ran-GTPase) (11). In the cytoplasm, the terminal loop of pre-miRNA is removed by the RNase III endonuclease Dicer (13).
At this point, miRNAs appear as short RNA duplexes termed miRNA duplexes that are loaded into the RNA-induced silencing complex (RISC) (15). This machine is a multiprotein complex that includes proteins such as Argonaute RISC Catalytic Component 2 (Ago2), Aubergine (another Ago family protein) Staphylococcal Nuclease Domain-Containing Protein 1 (SND1), Astrocyte Elevated Gene-1 (AEG-1), Fragile X Mental Retardation 1 (FMR1), VIG (vasa intronic gene), R2D2 (a dsRNA binding protein with two dsRNA binding domains), and Armitage-RNA helicase1 (15–17).
Ago2 is the RISC catalytic component that leaves and/or removes one strand of the duplex (18, 19). Based on the directionality of the miRNA’s strands originated, the name of the mature miRNA will be 5p strand (miRNA-5p) if it arises from the 5′-end of the pre-miRNA hairpin, or will be 3p strand (miRNA-3p) if it originates from the 3′-end. The two mature miRNAs strands have different target-specificity (20, 21). The strand which will remain incorporated into the RISC is the active strand, and it is named the guide strand, leading strand or miR. The other strand gets degraded, and it is named the passenger strand or miR* (22). Thermodynamic features that, in turn, are related to ribonucleotide (purines vs. pyrimidines) composition of the duplex, play an essential role in miRNA-RISC assembly (23).
RISC with the loaded miRNA (miRISC) acts inhibiting multiple steps of protein synthesis and affecting mRNA target stability (24–26). In detail, the miRNA in the RISC complex drives it to an mRNA target in a complementary-nucleotides-based way. Usually, the mRNA target-site is into 3'untranslated region (3’UTR), and the complementary sequence on miRNA, called seed sequence, is located at 2-7 nucleotides. However, non-canonical seed regions have been identified, such as in the 5’UTR sequence and the coding regions of mRNAs (27).
After recognition, target mRNA is still stable, but miRISC prevents its translation by inhibition of translation elongation, or through protein degradation or ribosome drop-off. In the end, mRNA, after deadenylation and decay, is degraded (28, 29). More precisely, to date, it seems that translational repression and target mRNA degradation are spatiotemporal uncoupling events. Concerning the timing, in Drosophila, Zebrafish, and mouse embryonic stem cells (ESCs), it was demonstrated that translational repression, at an early step, always occurred before mRNA degradation (30–33). This result suggests that the block of protein translation is the first event of miRNA-mediated gene silencing (the cause), and the mRNA target degradation is a secondary consequence (the effect). Concerning space, it was demonstrated that RISC assembly, its interaction with miRNA and targeted mRNA, and translational repression are processes compartmentalized on rough endoplasmic reticulum (rER) membrane. On the contrary, the degradation of the mRNAs (deadenylation and decay) takes place on endosome/multivesicular bodies (MVBs) (34, 35).
In addition to the well-known miRNAs cytoplasmic localization and functions, the nuclear ones are noteworthy. Indeed, it has been proven that minimal miRISC complex is also present in mammalian cell nuclei, where it is 20-fold smaller (36, 37). It consists only of Ago2 and miRNA, is loaded in the cytoplasm, and is imported into the nucleus by binding to importin 8 (IPO8) (38). The combination of minimal miRISC complex with Trinucleotide Repeat Containing Adaptor 6A (TNRC6A), which possesses a nuclear localization signal (NLS) and a nuclear export signal (NES), ensure the nuclear-cytoplasmic shuttling (38) even though a more selective mechanism for miRNAs distribution could be operated by nucleotides-motif at the 3’ end of the miRNA.
In the nucleus, miRNAs contribute to the regulation of both coding and non-coding RNA transcriptome (28, 37). They can act in blocking or promoting pri-miRNA maturation regulating long non-coding RNAs (lncRNAs) levels (39–41).
Other nuclear miRNAs colocalize with ribosomal RNAs (rRNAs) in the nucleolus, where they are stored, or in the cytoplasm where they influence the abundance of rRNAs and/or regulate ribosomes interaction with accessory proteins (42–45).
Moreover, in the nucleus, miRNAs induce remodeling of chromatin structure, regulate alternative splicing, and in turn, regulate itself (46–48). miRNAs can also mediate transcriptional gene activation (TGA) or transcriptional gene silencing (TGS) (49, 50). TGA depends on the complementarity between DNA sequences of gene promoter and the 5’-seed with a part of the flanking region of miRNA. Two mechanisms that unlock gene silencing have been identified: i) in case of silencing complexes on gene promoter, such as lncRNA or promoter-associated RNA (pRNA), miRNA can act inducing the cleavage of these molecules by the recruiting of the nuclear RISC; ii) miRNA can directly recognize the nascent RNA promoter and recruit the minimal miRISC that in turn enroll the protein complex assigned to shift the chromatin structure toward a more permissive arrangement (51, 52).
About TGS, where epigenetic modifications have a more prominent role than in TGA: i) miRNA with its seed sequence may directly interact with single-stranded DNA complementary sequences in the promoter region inducing histone epigenetic modification and altering the binding of transcription factors; ii) miRISC may directly target to non-coding transcripts, sense and antisense, and serves as a molecular scaffold to recruit an inhibitory protein complex and chromatin modulators that will establish a non-permissive transcriptional status; iii) miRNA may hybridize with double-stranded DNA to form relatively stable RNA*DNA : DNA triplexes via Hoogsteen or reverse Hoogsteen interaction that induces promoter-specific transcriptional repression through the disruption of the formation of the pre-initiation complex at the promoter (28, 37).
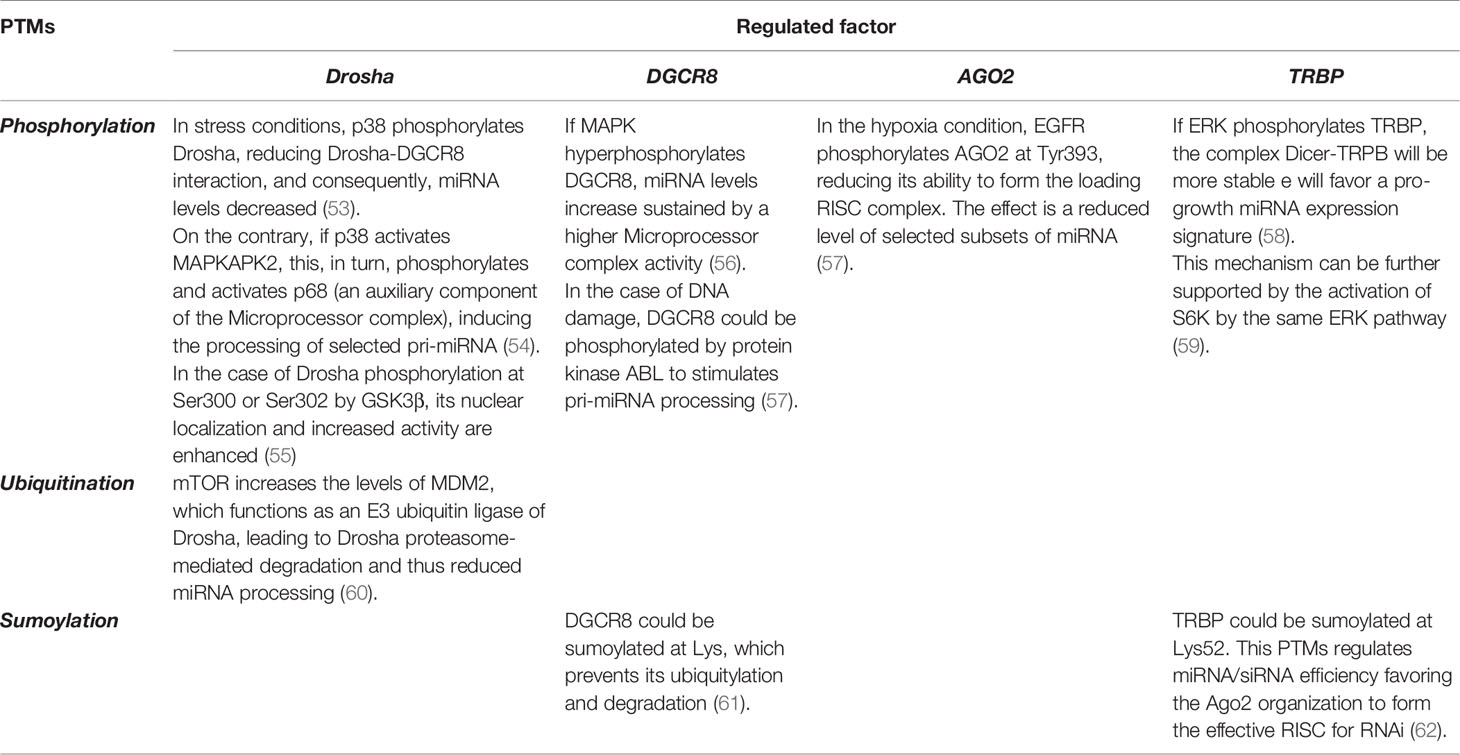
miRNAs levels and their activity can be regulated by a series of post-translational modifications (PTMs) affecting the miRNA processing machine (see Table 1 and below section PARylation post-translational modification affect miRNA activity in tumors). Moreover, co- and post-transcriptional regulation of miRNA transcripts are performed by specific RNA-biding proteins (RBPs), which affect miRNA processing and loading into RISC, and facilitate the crosstalk between various RNA pathways [reviewed in (28)].
Alterations in miRNA biogenesis can promote and support the onset of cancer. The oncogene p53, whose mutations occur in 50% of human cancers, promotes cancer interfering with Drosha in the Microprocessor complex. It inhibits miRNA processing, resulting in the accumulation of pri-miRNAs and parallel depletion of mature miRNAs (63). Mutant p53, by sequestrating the Drosha cofactors, DEAD-box RNA helicases p68 (DDX5), or p72/p82 (DDX17), promotes tumor cell migration, epithelial to mesenchymal transition (EMT), and cell survival (64, 65).
Moreover, cancer is supported by altered expression levels of miRNA processing machinery components such as Drosha, DGCR8, and Dicer that are down-regulated or up-regulated in several cancers (66). For instance, in neuroblastoma and ovarian cancer, Drosha and Dicer’s reduced expression levels and overexpression of DGCR8 correlate with tumor progression and a worse clinical outcome (67–69).
Dicer mutations lead to altered expression levels or could affect the region encoding RNase III domains that cause defects in its function. In several tumors, such as pleuropulmonary blastoma, cystic nephroma, ovarian cancer, Wilms tumor, pituitary blastoma, and rhabdomyosarcoma, the pathogenesis is supported by reduced Dicer expression and/or impaired function that in turn induce an aberrant miRNA expression (70–73).
Impaired function of the Dicer cofactor TRBP also contributes to miRNA dysregulation in cancer. In the Ewing sarcoma family tumor, it was demonstrated that cancer stem cell renewal and tumor maintenance depend on TRBP expression (74).
Pre-miRNA export dysfunction may lead to tumorigenesis. By cancer tissue microarrays (TMAs) and the cancer genome atlas (TCGA) RNA seq-data, it was shown that increased level of XPO5 promotes and sustain tumor including prostate cancer, breast cancer, ovarian cancer, and bladder cancer (75–77).
Non-Canonical miRNA Biogenesis and Functions
In addition to the canonical pathway, miRNAs could be processed from unexpected non-coding RNAs by mechanisms that may depend on cell type and cellular state (78).
In the non-canonical miRNA biogenesis pathway, functionally miRNAs result from a different combination of the same proteins of the canonical pathway. Two main non-canonical miRNA biogenesis pathways are distinguished: Drosha/DGCR8-independent and Dicer-independent (79). An example of the first is miRtrons biogenesis (80). MiRtron-derived pri-miRNAs correspond to the entire intronic sequence of the mRNA encoding genes in which they are located. They are processed by splicing and not by the Microprocessor complex (81). After splicing, miRtron-derived pre-miRNAs are exported to the cytoplasm by XPO5 and are processed by Dicer to form a mature miRNA (82, 83). miRtrons could be distinguished from canonical miRNAs looking for key features such as bulges in the stem region, guanine content, hairpin free energy, and hairpin length (84).
An example of a non-canonical miRNA biogenesis Dicer-independent pathway is pre-miR-451. It is processed, initially, by Drosha to liberate the pre-miRNA, following, in the cytoplasm, it is loaded into Ago2 to catalyze the maturation of this microRNA (85). Pre-miR-451 is not processed by Dicer because its stem-loop structure is too short to be efficiently recognized and processed (86, 87). Therefore, in this mechanism, it seems that Ago2 is sufficient for RISC loading and proper guide strand selection (88). This miRNA is involved in tumorigenesis and tumor progression of several cancers. It is downregulated in epithelial cells cancer that lose their basolateral polarity, dramatically proliferate, and turn to invasive adenocarcinoma forms as demonstrated in gastric cancer, colorectal cancer, non–small cell lung carcinoma (NSCLC) (89–91)
Non-canonical miRNA could also be small nucleolar RNAs (snoRNAs) and transfer RNAs (tRNAs) (78). snoRNA-derived miRNAs are approximately 22 nucleotides in length, require Dicer activity, but are independent of Drosha/DGCR8, bind to Argonaute proteins and repress target mRNAs as miRNA do (92, 93).
tRNAs with their clover shape can generate many non-coding small RNAs, named tRNA-derived small RNAs (tsRNAs). These, based on their mechanism of biogenesis and length, are classified as tRNA-derived stress-induced RNA (tiRNA) or tRNA-derived fragment (tRF). tRFs are smaller than tiRNAs, originate from a specific sequence of mature or primary tRNAs cleaved by Dicer, or they could be processed by RNase Z or ELAC2 in the nucleus and cytoplasm, respectively (94). They can bind to the Ago proteins family in a cell-type-specific manner and can regulate mRNA stability by directly binding to it, inhibiting protein translation, or cleaving a partially complementary target site (94–96).
The role of non-canonical miRNA in the biological functions and development of cancer is poorly understood. Splicing factors, such as SRSF1 or SRSF2, alter the levels of miRNAs of miRtron origin. In the colorectal cancer cell line HCT116, it was found that increased nuclear levels of SRSF1 positively correlate to hsa-miR-1229-3p expression, while increased nuclear levels of SRSF2 positively correlate to hsa-miR-1227-3p and hsa-miR-1229-3p expression (97). The miRtron miR-6778–5p, derived from intron 5 of SHMT1 (coding cytoplasmic serine hydroxymethyltransferase), is a pivotal regulator of cancer stem cell stemness in Drosha-silenced gastric cancer cells. It promotes SHMT1 expression that sustains cancer energy intake via folate-dependent serine/glycine inter-conversion in the one-carbon mitochondrial metabolic pathway (98).
By RNA-seq-based data set, in MDA-MB-231s breast cancer cell line and more than 90% of Luminal B Her2+ human breast cancer, a small nucleolar RNA-derived RNAs, snoRNA-93, was identified as a promoter of invasion (99). In prostate cancer, the increased expression of small nucleolar RNA-derived RNAs snoRD78 was detected in a subset of patients that developed the metastatic disease (100).
PARylation Post-Translational Modification Affect miRNA Activity in Tumors
Among the PTMs affecting the miRNA processing machine, poly-ADP-ribosylation (PARylation) is critical. It is a mechanism by which poly-ADP-ribose (pADPr) macromolecular polymer is added to some proteins, acting as a post-translational modification well documented in the nucleus and the cytoplasm (101–103). The reactions are catalyzed by ADP-ribosyltransferases proteins that include poly-ADP-ribose polymerases (PARPs) (104). PARPs are involved in DNA repair, when DNA single-strand breaks are present, and induce apoptosis via exhaustion of ATP reserves (105, 106). To date, knowledge on the role of PARPs in RNA metabolism is growing (107–109). In 2011, Leung et al. demonstrated that pADPr is a crucial regulator of miRNAs PTMs in the cytoplasm and, consequently, mRNA expression levels (110). In detail, by immunoprecipitation assay and GFP fusion technology under different experimental conditions in four human cell lines, they showed that cytoplasmic stress granules were rich in mRNA binding proteins, contained six poly-ADP-ribose polymerases, two poly-ADP-ribose glycohydrolase, and Ago proteins (110, 111). Ago2, in standard and stress conditions, were PARylated by catalytically inactive PARP13 and other synthesizing PARPs (mono and poly-ADPr), but during stress, it was much more PARylated, probably for increased PARP activity and/or decreased poly-ADP-ribose glycohydrolase (PARG) activity. Ago2 increased PARylation reduces the miRNA repression activity and miRNA-directed cleavage due to disruption of electrostatic interaction between miRNA:mRNA or steric obstacles for effective silencing of miRNAs (110).
In the colorectal cancer DLD-1 cell line was found that PARylation of Ago2-associated proteins during viral infection relieves miR-17/93 family repression of the interferon-stimulated genes which contain in the 3′ UTRs the miRNA target sites. This means that cells respond to viral infection by downregulation of miRNAs pathway activity via the PARylation of Ago2 complexes (112, 113). By immunohistochemistry reactions on TMAs containing tumor and normal tissue, reduced expression of PARP13 was demonstrated in liver, colon, and bladder cancers (114). PARP13 targeted TNF-related apoptosis-inducing ligand (TRAIL) 4 transcript at the cell decay pathway, destabilizing its mRNA after transcription, via exosome, by binding to its 3 ‘UTR region, and thus increasing the sensitivity of tumor cells to TRAIL-mediated apoptosis (115). A PARP13 anticancer role was also found in Hepatitis B Virus related-liver cancer and leukemia, thanks to its antiviral activity (116, 117). PARP1 expression was found to increase in breast, uterine, lung cancer, ovarian cancer, skin cancer, and non-Hodgkin’s lymphoma (118–120). In breast cancers, including triple-negative breast cancers, PARP1 expression negatively correlates to the estrogen receptor, progesterone receptor, or epidermal growth factor receptor-2 expression (121). In breast cancer, the oncomir miR-155-5p was found mostly upregulated and associated with a more aggressive type and worse outcome (122). The combined treatment with Olaparib, an inhibitor of PARP1, with miR-155-5p ectopic overexpression was found a great combination to induce lethal effect to reduce cancer cells viability (123)
Further investigation of PARPs as a biomarker for novel cancer therapies is needed.
miRNA in Tumors
Tumors develop due to progressive DNA alterations, which will allow tumor cells to proliferate uncontrollably (124). Tumor cells acquire the ability not to respond to growth inhibitors and to resist to mechanisms of programmed cell death. Therefore, tumor cells become immortal and able to shift their metabolism to more advantageous energy mechanisms in the interest of continuous growth and proliferation. Furthermore, to grow beyond a specific size, tumors activate angiogenesis mechanisms, which will allow the tumor cells to receive more oxygen and nutrients, and to eliminate metabolism waste more efficiently. Thanks to angiogenesis, tumor cells not only proliferate but becomes able to invade the surrounding tissues giving rise to local metastases. Moreover, tumor initiation and progression are supported and favored by the inflammation processes, and by the tumor cell competence to escape the immune surveillance.
In this complex genetic disease, coding and non-coding genes are involved. miRNAs, because of their ubiquitous role in gene regulation, are critical regulators in various biological processes, such as differentiation, proliferation, apoptosis, and development in both health and disease (125).
The activity of miRNAs in tumors is regulated by the same alterations affecting protein-coding genes, such as chromosomal rearrangements, genomic amplifications or deletions or mutations, abnormal transcriptional control, dysregulation of epigenetic changes and defects in the biogenesis machinery. A typical chromosomal rearrangement is a chromosomal translocation, especially in hematological malignancies, in which it promotes tumor development and progression by the promoter exchange or by the creation of chimeric genes translated as fusion proteins. In Acute Myeloid Leukemia (AML) patients with myeloid/lymphoid leukemia gene (or mixed-lineage leukemia, MLL) rearrangement, by large-scale genome-wide microarray analysis, it was demonstrated that among 48 selected miRNAs, 47 of them are increased (126). This result suggests that MLL fusion protein promotes the transcription of its downstream targets, such as miRNA genes.
The primary miRNAs alteration in tumors is the aberrant gene expression arises from amplification or deletion of specific genomic regions that coupling with abnormal expression levels of mature and/or precursor miRNA compared with the corresponding healthy tissues. In B-cell chronic lymphocytic leukemia (B-CLL), the miR-15a/16-1 cluster gene is deleted in more than 50% of cases, resulting in decreased expression of both miRNAs (127). Single-nucleotide polymorphisms (SNPs) in miRNAs or miRNAs-targets are associated with the development of several tumors. For example, in breast cancer, polymorphisms in the 3′-UTR of the ErbB4 gene could affect miRNA binding sites resulting in post-translational dysregulation of mRNA and a predisposition to tumor development (128). In gastric cancer, SNPs in miRNA machinery genes was strictly associated with cancer susceptibility and malignant behavior (Dicer and GEMIN4), or lymphatic metastasis (GEMIN4 and Ago1) (129). In hepatocellular carcinoma, SNPs in miRNA3152 and miRNA449b were linked with the risk of tumor development (130).
Different transcription factors, such as p53, c-Myc, nuclear receptors, and RAS control miRNA expression and are often associated with tumorigenesis. For instance, p53 strengthens his tumor suppressor activity, also inducing the expression of specific miRNAs, which show the same suppressive functions. In pancreatic cancer cells, p53 directly transactivates miR34a inducing gene expression reprogramming and promoting apoptosis (131). On the contrary, there are oncogenic transcription factors that induce tumorigenesis, such as members of the RAS family. miR-143/miR-145 co-expressed in the same primary transcript, highly expressed in healthy colon, are significantly decreased in colorectal cancer in which are targeted by KRAS (132).
Dysregulated epigenetic changes of miRNA may induce tumors. The activation of pri-miRNA transcription is under epigenetic control, especially, it is ruled by promoter-associated CpG island methylation and histone modification. miR34a and other members of the same family, despite being located on different chromosomes, were found hypermethylated in both solid and hematological tumors (133, 134). The use of epigenetic drugs also demonstrates the epigenetic control of miRNA. For example, in triple-negative breast cancer cell lines it was shown that the epigenetic therapy with suberoylanilide hydroxamic acid (SAHA), a histone deacetylase (HDAC) inhibitor, in combination with epigallocatechin-3-gallate (EGCG), a DNA methyltransferase (DNMT) inhibitor, reduces the expression of oncogenic miRNA-221/222 (135).
Dysregulation of all miRNA biogenesis steps may contribute to tumor growth and spread (136). About the Microprocessor complex, both Drosha up- and down-regulation were found in different tumors such as advanced cervical squamous cell carcinoma and clear renal cell carcinoma (137, 138). Further, pre-miRNA export, Dicer, and Ago2 dysregulation are involved. Mutation of XPO5 causes the defect of pre-miRNA export, leading to its accumulation in the nucleus (139). In vitro and in an animal model of thyroid cancer, it was shown that oncogenic miR-146b-5p down-regulates miRNA biosynthesis by targeting Dicer and reducing its expression (140). Dicer deletion and/or down-expression promote tumor cell growth, migration, invasion, and EMT. The master regulator of miRNA maturation and function Ago2 was overexpressed in different tumors in which miRNAs act repressing their targets (141–143).
Additionally, nuclear miRNAs in tumor cells is documented. They are involved in the transcriptional regulation of a variety of tumor promoter/repressor genes or tumor-related genes that control tumor initiation, self-sustenance, apoptosis, metastasis, and angiogenesis (37). For example, miR-215-5p, which is nuclear-localized, is up-regulated in high-grade gliomas and promotes tumor cell proliferation, clone formation, migration, and suppresses apoptosis by directly binding to both the promoter and 3’UTR of PCDH9 gene (a member of the protocadherin family and cadherin superfamily). Therefore, PCDH9 expression will be down-regulated at the transcriptional and post-transcriptional levels in glioma (144). Other nuclear-localized miRNAs that promote transcriptional activation of oncogenes by binding to their promoters include miR-483 that binds to IGF2 promoter to increase its expression in Wilms’ tumors and miR-558 that binds to heparanase promoter to enhance its expression in neuroblastoma cells (145, 146). In silico and after mouse model validation, it was demonstrated that miR-744 promotes the transcription of Cyclin B1, favoring the recruitment of RNA polymerase II and the trimethylation of histone-3 at lysine-4 at the promoter region. If it is expressed for a short time, this miRNA promotes cell proliferation. If expressed for a long time, it causes aberrant chromosome distribution and in vivo tumor suppression (147). miR-10a acts as a transcription repressor of Hoxd4 in the nucleus of HCT116 colon cancer, THP-1 acute monocytic leukemia, MCF7, and MDA-MB-23 breast cancer cel1 lines. miR-10a induces hypermethylation and trimethylation of histone-3 in the Hoxd4 promoter, thus preventing cancer cell invasion and metastasis (148–150). miR-370, miR-1180, and miR-1236 expression levels are decreased in bladder cancer tissues. These nuclear miRNAs activate tumor suppressor gene p21 by binding to its promoter, as demonstrated in T24 and EJ bladder cancer cell lines (151). In chronic lymphocytic leukemia, the nuclear miRNA-709 controls the biogenesis of tumor suppressor miRNA miR-15a/16-1 (152). It directly binds to a 19-nt recognition element on pri-miR-15a/16-1 and prevents its processing into pre- miR-15a/16-1 and leads to maturation suppression. During apoptotic stimuli, the nuclear level of miRNA-709 decreased because it is exported in the cytoplasm while the nuclear level of miR-15a/16-1 increased, leading to enhanced cell apoptosis via anti-apoptotic gene Bcl-2 downregulation (40). The nuclear miR-223, a regulator of innate immunity, is associated with carcinogenesis. In AML, its down-expression induces an increased level of transcriptional factor E2F1, which further reduces miR-223 by binding to its promoter (153). However, the nuclear-localized miRNAs mechanisms of action are incompletely understood and have so far been largely neglected. Further investigations will better elucidate the mechanisms through which nuclear miRNAs execute their functions in different cell types and physiological/pathological conditions.
The pivotal role of miRNAs in tumors is not just the intracellular one (cytoplasm and nucleus). They were also found in extracellular fluids, as free circulating molecules or enclosed in exosomes. These extracellular molecules are intimately involved in cell-cell communication, can travel long distances to affect recipient cells, in particular, immune cells in the tumor microenvironment (154–156). Quality and quantity dysregulations of circulating miRNAs are associated with tumor origin, invasion, metastasis, angiogenesis, immune escape, metabolic switch, and chemosensitivity or chemoresistance (157, 158). Circulating free miRNAs can be released from all cell types as complexes associated with Argonaute proteins during cellular processes such as secretion, apoptosis, inflammation, and necrosis. For instance, by microarrays, in malignant tissue and plasma of tongue squamous cell carcinoma patients, it was shown that many miRNAs (miR-19a, miR-27b, miR-20a, miR-28-3p, miR-200c, miR-151-3p, miR-223, and miR-20b) were up-regulated in plasma, free and exosomes in comparison to benign tongue tissue and plasma (159).
Moreover, in breast cancer in vitro model, it was shown that secreted miRNAs from metastatic cells, in a process dependent on Neutral Sphingomyelinase 2 (nSMase2), were transported to endothelial cells by exosomes to promote angiogenesis (160).
In hematological malignancies, tumor-derived exosomes reprogram bone marrow stromal cells, promoting a tumor-supportive microenvironment by suppressing anti-tumor immunity and promoting inflammatory cell recruitment, angiogenesis, osteoclast differentiation, and drug resistance. Moreover, we have demonstrated that in multiple myeloma, cell-derived exosomes induce overexpression of miR-27b-3p and miR-214-3p in fibroblasts of the surrounding tumor microenvironment. These miRNAs increase proliferation and reduce apoptosis of fibroblasts via FBXW7 and PTEN signaling, and sustain tumor growth and progression by drug resistance (161, 162).
Summing up, miRNAs in tumors can act as oncogenes or tumor suppressors (163). However, the same miRNA can act as onco-miRNAs and oncosuppressor-miRNAs participating in distinct pathways that have different effects on cell survival, growth, and proliferation depending on the cell type and the pattern of gene expression (164). Another critical aspect of evaluating is the different expression between precursor miRNA and its active molecule. In various types of tumors, it was found altered levels of pre-miRNA but not of the active molecules (165, 166).
So ultimately, all tumors present specific signatures of miRNAs altered expression. For this reason, the tumor-specific miRNAs expression profile may represent a valid and useful biomarker for diagnosis, prognosis, follow-up, patient stratification, definition of risk groups, and for the development of new therapeutic strategy (167). However, there are many challenges to be addressed before reaching full approval for clinical use (168). It is imperative to validate targets, prevent unwanted off-target effects, and develop an efficient and specific miRNA delivery system.
miRNA in Angiogenesis—The AngiomiR
Angiogenesis is the process by which new blood vessels originate. It is a physiological process during embryonic development and reproduction (corpus luteum formation), but it has a pivotal role also in pathological processes such as wound healing, inflammation, and tumor. In this last, angiogenesis is a key process for development and progression, leading to form an efficient vasculature network that guarantees a high supply of nutrients, an efficient system to remove catabolites, and the cancer stem cell maintenance (169). Different mechanisms can contribute to the formation of the new vascular network in tumors, such as sprouting angiogenesis, intussusceptive angiogenesis, vascular co-option, vasculogenic mimicry, postnatal vasculogenesis, or by differentiation of putative cancer stem cells (CSCs) into endothelial cells (ECs) (170–172).
Angiogenesis is a multi-step process. Taking as an example the well-known sprouting angiogenesis, it begins with activation of ECs, which become able to exit their vessel of origin, invade the surrounding stroma by local enzymatic degradation of the basement membrane, proliferate and migrate in the direction of the angiogenic stimulus, and form tubes. Then the processes of blood vessel fusion, blood vessel pruning, and pericytes stabilization occur to form and complete the new capillary network.
Tumors are heterogeneous and entropic organs, in which tumor cells cohabit and codevelop with the molecular and cellular components of the surrounding microenvironment. In this plastic environment, the imbalance between pro-angiogenic and anti-angiogenic signaling guides the “angiogenic switch,” causing continuous generation of new blood vessels (173). The constant expression of pro-angiogenic factors does not allow blood vessel maturation resulting in the characteristic aberrant, leaky, disorganized, immature, thin-walled, and ill-perfused tumor vasculature (174). These aspects of tumor blood vessel cause and sustain a hypoxic microenvironment, even in highly vascularized tumors, that supports tumor growth favoring all hallmarks of the tumor, including immunosurveillance escape, metastasis, and not respond to therapies.
In addition to the well-known growth factors and vascular genes involved, angiogenesis is also regulated by epigenetic states of genes, and miRNAs expression and function (175). miRNAs act controlling the expression of angiogenesis-related factors, endothelial cell proliferation, migration, and tube formation. They are called angiomiRNAs (angiomiRs).
It is possible to distinguish four-way of miRNAs action on tumor angiogenesis: i) tumor cells derived-miRNAs may affect the activity of ECs via non-cell-autonomous mechanisms; ii) ECs derived-miRNAs may regulate the cell-autonomous behavior; iii) tumor-derived extracellular vesicles transfer miRNAs to ECs; other organs/systems derived-miRNAs might affect either tumors or ECs (Figure 1). (176). In other words, in the first way, genetic and epigenetic alterations in tumor cells affect the phenotypes of ECs through changing microenvironment or intercellular interaction. In the second way, genetic and epigenetic alterations in ECs affect the cell’s own phenotypes. In the third way, tumor cells derived-miRNAs regulate the expression of pro- or anti-angiogenic factors in a paracrine manner by extracellular vesicles (EVs) that contain tumor cells-miRNAs which will be transferred from tumor cells to ECs, where they induce the pro-or anti-angiogenic effects. The fourth ways take advantage of cellular communication across tissues. miRNAs from other organs/systems, different from tumor cells or ECs, regulate the expression of pro- or anti-angiogenic factors in a paracrine manner. In zebrafish muscle, the evolutionarily conserved myomiRs, miR-1, and miR-206 from muscle cells negatively regulate angiogenesis by directly modulating VEGFA mRNA expression in ECs by a possible binding to three sites in its 3′UTR (177). Increased expression of miR-1 and miR-206 was found in patients affected by rhabdomyosarcomas and gastric cancer compared with healthy subjects (178, 179).
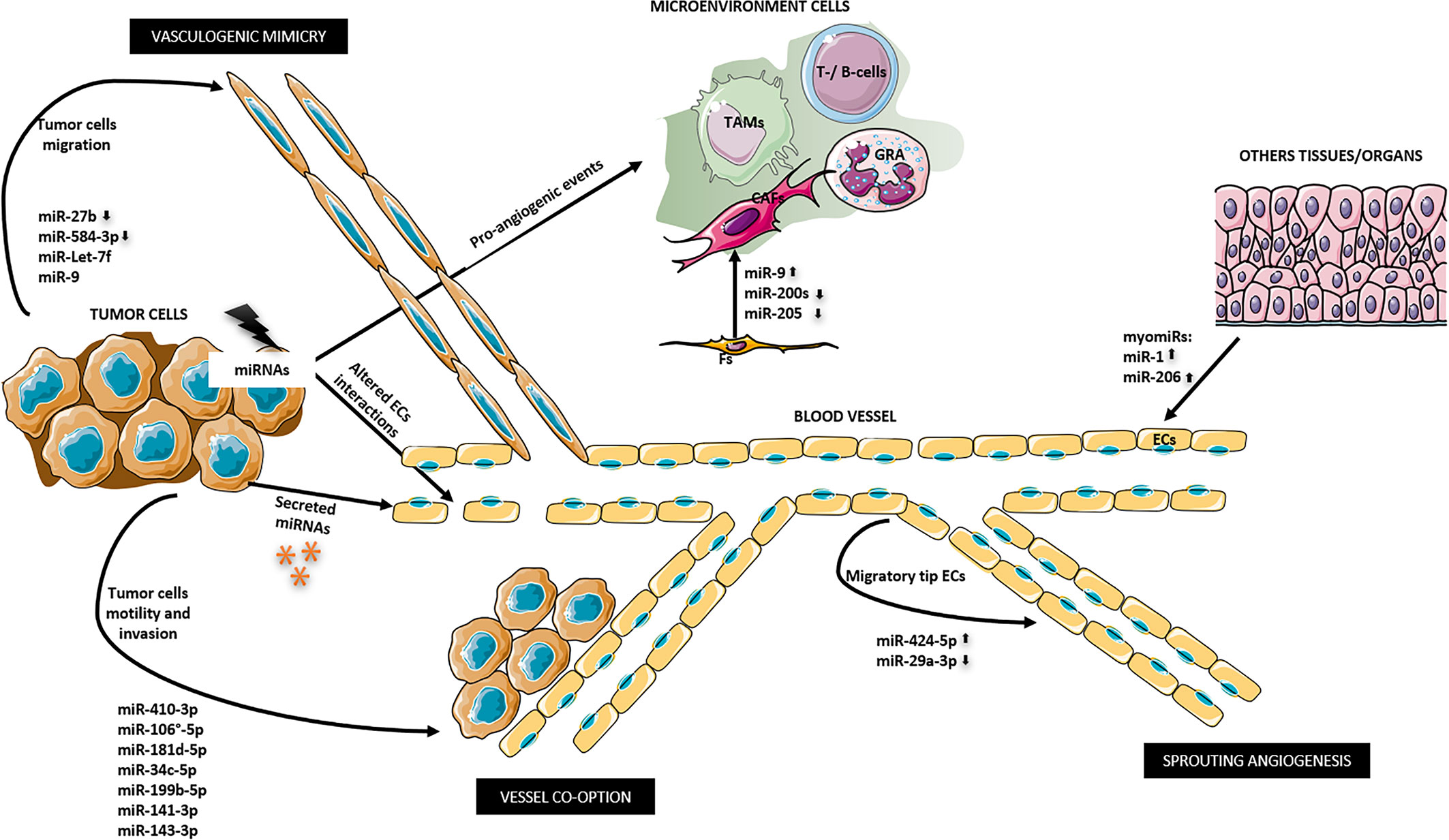
Figure 1 miRNAs ways to regulate tumor angiogenesis. 1) miRNAs derived from tumor cells indirectly may affect ECs supporting a pro-angiogenic microenvironment and altering ECs junctions. miR-9, miR-200s, and miR-205 promote the differentiation of fibroblasts into cancer associated fibroblasts (CAFs). 2) miRNA derived from ECs in autocrine manner may promote a migratory tip ECs phenotype that in turn promote sprouting angiogenesis. 3) Tumor-derived miRNAs may be secreted outside the cell and may directly regulate ECs. 4) Secreted miRNAs from other organs/tissues might affect ECs as myomiRs done. 5) Altered tumor-miRNA expression may promote tumor cell motility and invasion of the surrounding tissue that promote angiogenesis and tumor dissemination by vessel co-option. 6) Other miRNAs can induce vasculogenic mimicry. CAFs, cancer associated fibroblasts; ECs, endothelial cells; Fs, fibroblasts; GRA, granulocytes; TAMs, tumor associated macrophages; T-/B-cells.
AngiomiRs can work targeting the negative regulators of angiogenesis and so promoting neoangiogenesis (pro-angiomiRs) or by targeting positive regulators of angiogenesis and so inhibiting it (anti-angiomiRs).
Besides the altered expression levels of certain angiomiRs, it has been shown that tumor angiogenesis can depend on the dysregulation of miRNAs biogenesis enzymes, Dicer and Drosha, involved in the maturation of ECs (180–182).
Recently Tiwari et al. (168) and Leone et al. (183) reviewed the mechanisms by which miRNAs regulate new blood vessel formation involving classical and non-classical growth factors and signaling pathways of angiogenesis. Therefore, we propose a concise table (Table 2) to get a glance at the main miRNA targets and tumors involved, and below, we examine the role of miRNAs in the three most studied mechanisms by which angiogenesis can happen, sprouting angiogenesis, vessel co-option and vasculogenic mimicry.
Sprouting angiogenesis requires detailed temporal coordination of ECs migration and proliferation (171). In this mechanism, new blood vessels originate from the pre-existing one, and the two main actors are tip cells and stalk cells. The tip cells are activated ECs on the vascular front that migrate in response to angiogenic stimuli coming from the surrounding microenvironment. The stalk cells are activated ECs which are located behind the vascularization front, intensely proliferate and organize themselves to form the vessel lumen. Quiescent ECs became tip/stalk cells in response to vascular endothelial growth factor-A (VEGF-A) gradient concentration and its receptor (VEGFR) expression level in association with extensively transcriptomic alterations (226). In vitro, by a 3D-spheroid model that reproduces the initial phase of sprouting angiogenesis and by the administration of VEGF-A, it was demonstrated that quiescent ECs are activated in response to VEGF-A and differentiate into tip cells that allow the migratory process rather than cell proliferation (227). In this phenotypic switch, during initial sprouting angiogenesis, miRNA plays a pivotal role in controlling tip cell fate by activating specific gene programs (227). The computational method Gene Set Enrichment Analysis (GSEA) showed the activation of genes that regulate extracellular matrix organization, cell adhesion molecules, integrin pathway, protein translation, and collagen formation. Instead, the genes that are associated with cell-cycle progression or are involved in mitogen-associated protein kinase (MAPK) signaling are repressed (227, 228). miR-424-5p up-expression and miR-29a-3p down expression were reported as the primary miRNAs in this network where VEGF triggers tip cell specification (227). miR-424-5p expression induces the repression of target genes involved in cell proliferation, including MAPK and cell cycle-related ones (227, 229). On the contrary miR-29a-3p downregulation induces the expression of target genes involved in cell migration as extracellular matrix (ECM) -related ones (227, 230). The same authors investigated the angiogenic post-transcriptional activity of 58 sprouting-associated genes (including the two mentioned above) in human colorectal cancer, by the means of GSEA, showing the utility of miRNA network analysis for patients therapy stratification and new druggable biomarkers detection.
Vessel co-option is an alternative mechanism of angiogenesis in which cancer cells migrate towards and along the pre-existing vasculature without or before angiogenesis, or in response to anti-angiogenic therapy (231). In addition to promoting tumor growth and progression, vessel co-option mediates tumor resistance to anti-angiogenic therapy (232). In a study, in which was evaluated the role of vessel co-option concerning sorafenib [a tyrosine kinase inhibitor (TKI)] resistance in hepatocellular carcinoma, by miRNA sequencing and qRT-PCR, four signaling pathways that are up-regulated were found (233). These pathways, involved in cellular motility and invasion, were including axonal guidance, EMT, STAT3, and Wnt/βcatenin signaling. Evaluating the miRNA cluster involved in EMT, three transcripts coding for Vimentin, and Zinc Finger E-Box Binding Homeobox-1/-2 (ZEB1 and ZEB2), were found significantly up-regulated in resistant tumor vs. control (233). These results highlight the importance of targeting different types of angiogenesis mechanisms because the tumor can activate alternative ways that non respond to classic anti-VEGF or TKI therapies.
Vasculogenic mimicry is a tumor neovascularization mechanism different from the endothelium-dependent ones. This mechanism is often present in highly aggressive and genetically dysregulated tumors in which tumor cells mimic endothelial cells. In ovarian cancer were found that low miR-27b expression levels are closely correlated with high vascular endothelial cadherin (VE-cadherin) expression and the high vasculogenic mimicry capability of tumor cells (234). VE-cadherin has been recognized as a master gene of tumor vasculogenic mimicry (235, 236). MiR-27b targets the 3′ UTR of VE-cadherin mRNA to suppress ovarian cancer cell migration and invasion as well as vasculogenic mimicry, as also demonstrated in hepatocellular carcinoma (234, 237).
Another miRNA related to vasculogenic mimicry is miR-584-3p. This miRNA function as a potent tumor suppressor by inhibiting vasculogenic mimicry of malignant glioma targeting Rho Associated Coiled-Coil Containing Protein Kinase 1(ROCK1) that disturb hypoxia-induced stress fiber formation and migration of glioma cells (238). In glioma, vasculogenic mimicry could also be suppressed by miR-Let-7f disturbing periostin-induced migration of tumor cells and by miR-9 disturbing Stathmin (a cytosolic phosphoprotein which regulates microtubule dynamics during cell-cycle progression) expression in vascular endothelial cells and glioma cells (239–241).
No specific studies were found concerning miRNA and tumor angiogenesis that occurs by intussusception or vasculogenesis.
In addition to the proliferation and migration of ECs and tumor cells to promote angiogenesis, miRNAs can control new blood vessel formation by regulating the tumor microenvironment. This means that miRNAs can regulate all the cellular and non-cellular components of the surrounding tissue. A recent review of Pan et al. elucidates the role of miRNA in regulating cancer-associated fibroblasts (CAFs), tumor-associated bone marrow mesenchymal cells, tumor-associated macrophages (TAMs), T-cells, and ECs (242). For instance, in breast cancer, it was shown that several miRNAs are implicated in the switch from normal fibroblast to CAFs, including miRNA-9 (up-regulated), miR-200s, and miR-205 (down-regulated). The reduced expression of miR-205 converts the normal fibroblasts into CAFs by promoting YAP1 expression, which is also involved in angiogenesis (243).
Summing up, miRNAs provide new insights into understanding the molecular mechanisms concerning angiogenesis. Their dysregulation may be responsible for the failure of current anti-angiogenic therapy.
In addition to targeting the different types of angiogenesis mechanisms, the new therapeutic approach should also take into account the vascular morphology. As said above, the tumor blood vessels are not mature, so vascular normalization is necessary. Vascular normalization can be accomplished by attenuation of hyperpermeability, increasing vascular pericyte coverage, inducing a more normal basement membrane, resulting in improved tumor perfusion, reduced hypoxia, and therapeutic outcomes (244).
Therapeutic strategies involve the use of miRNA as antagonists or mimics. Antagonists act inhibiting endogenous miRNAs with a gain-of-function in disease. Mimics act simulating miRNAs with a beneficial effect, but that are lost in the disease. miRNAs antagonists or mimics may be employed in transient normalization of tumor vasculature to make it more competent for oxygen and drug delivery (245).
miRNA in Lymphoma
Lymphoma is a group of tumors that begins in the cells of the immune system, lymphocytes-T and -B, that grow out of control. These cells reside in the lymph nodes, spleen, thymus, bone marrow, and other parts of the body. All these parts of the lymphatic system can be affected by lymphoma.
There are two main types of lymphoma, Non-Hodgkin Lymphoma (NHL) and Hodgkin Lymphoma (HL) (246, 247). The main discriminant between the two types of lymphoma is the presence of a specific type of abnormal cells called Reed-Sternberg cells. These cells are also known as lacunar histiocytes and are giant cells found by light microscopy in biopsies from individuals affected by HL. In NHL, these cells are not present. Moreover, other features distinguish the two major groups of lymphoma are: NHL is more common than Hodgkin lymphoma; the median age for diagnosis of NHL is 55 whereas for HL is 39; NHL may start in lymph nodes anywhere in the body, whereas HL usually starts in the upper part of the body (neck, chest or armpits); NHL is typically not diagnosed before it has reached a more advanced stage, whereas, HL is often diagnosed at an early stage and consequently it is one of the most treatable cancers.
Despite these differences, both NHL and HL have similar symptoms, like enlarged lymph nodes, fatigue, weight loss, and fever. In both, different subtypes are distinguished (246, 247). NHL could affect adults, children, and non-lymphatic tissue as skin. Primarily, they are classified as B- or T-cell lymphoma based on the white blood cell category affected, and secondarily as indolent or aggressive based on how tumor cells grow and spread (slow or quickly, respectively) (248). HL subtypes are Nodular sclerosis Hodgkin lymphoma (NSCHL), Mixed cellularity Hodgkin lymphoma (MCCHL), Lymphocyte-rich Hodgkin lymphoma, and Lymphocyte-depleted Hodgkin lymphoma (248).
Lymphocytes, like all the other classes of blood cells, are derived from bone marrow resident hematopoietic stem cells (HSCs), through progressive loss of pluripotency caused by a response to lineage determining signals. Intrinsic and extrinsic molecules, including miRNAs, regulate cell fate and lineage commitment in stem and progenitor cells (249). miRNAs dysregulation has been found in both solid and hematologic tumors by various genome-wide techniques, including oligonucleotide miRNA microarray analysis, bead-based flow-cytometric technique, quantitative real-time PCR for precursor/active miRNAs, miRAGE or RAKE assay (250–253). Metzler et al. provided the first evidence of miRNA involvement in human lymphoma. In children with Burkitt lymphoma, He and Coworkers found that miR‐155 was 100-fold increased compared to healthy blood and pediatric leukemia patients (254).
Different studies have demonstrated the essential role of Dicer1 for stem cell persistence and differentiation in vivo. In HSCs, Dicer1 deletion compromises the hematopoietic stem/progenitor cell (HSPC) function in a manner consistent with stem cell death (255). In the same work, a specific miRNA, miR-125a, was found to control the HSCs population size by regulating HSPCs apoptosis. In fact, miR-125a protects HSPCs from apoptosis and promotes the extensive expansion of the HSCs (255). Moreover, the ablation of Dicer1 in B- cell progenitors blocks the transition from pro- to pre-B cells because of mir-17~92 targeting the pro-apoptotic molecule Bim (256). Other miRNAs of the molecular circuitry that controls hematopoiesis are miR-223, miR-181, and miR-142. These miRNAs are specifically expressed in hematopoietic cells, and their expression is dynamically regulated during early hematopoiesis and lineage commitment (257). In detail, the Authors have shown that miR-181 is preferentially expressed in the B-lymphocytes of bone marrow, and its expression in HSPCs led to an increased fraction of B-lineage.
miRNAs are also profiled in several distinct stages of T-lymphocyte development. miR-181a represses, biding their 3′ UTR elements, the expression of Bcl-2, CD69, and the T-cell receptor genes, all of which are coordinately involved in positive selection of CD4+/CD8+ cells during thymocyte maturation (258, 259).
miR-155 regulates both B- and T-lymphocytes functions. Its deletion causes immunodeficiency because of B-lymphocytes produce lower levels of immunoglobulins, tumor necrosis factor (TNF) and lymphotoxin, and because of T-lymphocytes produce lower levels of interferon-gamma (IFNγ) and interleukin-12 (IL-12) promoting differentiation to Th2 phenotype (260, 261). For miR-155 is currently underway the first-in-human phase 1 clinical trial (Identifier NCT02580552) and phase 2 clinical trial (Identifier NCT03713320). The biopharmaceutical company, MiRagen Therapeutics is developing a locked nucleic acid-modified oligonucleotide inhibitor of miR-155, called MRG-106, and also known as Cobomarsen. The preliminary results have shown that miR-155, which is usually repressed in lymphoma, might be de-repressed by Cobomarsen in patients with mycosis fungoides, the most common form of cutaneous T-cell lymphoma. miR-155 overexpression will deactivate/activates signaling pathways, including JAK/STAT, MAPK/ERK, and PI3K/AKT, associated with tumor cell proliferation, survival, and apoptosis (262).
Angiogenesis in hematological malignancies, as in solid tumors, correlates with tumor growth and development (263). As discussed in the previous section, miRNAs regulate various aspects of angiogenesis, including proliferation, migration, and morphogenesis of endothelial cells. Therefore, the knowledge of all these mechanisms will permit to improve the management of malignancies, and therefore the patients’ prognosis by targeting specific molecules and processes involved in tumor angiogenesis. Despite the several data on miRNome characterization in lymphoma, the data about the specific regulation of angiogenesis by miRNAs are scarce (264–266).
miR-135b up-regulation was demonstrated in anaplastic large-cell lymphoma (ALCL) cell lines and clinical samples positive to the oncogenic translocation nucleophosmin-anaplastic lymphoma kinase (NPM-ALK) (267). NPM-ALK promotes the expression of miR-135b and its host gene LEMD1 through STAT3 activation. By the decoy RNA system to achieve long-term miR-135b suppression and by GSEA, it was shown that miR-135b targets FOXO1, TGFBR1, SIRT1, cyclin G2, CREG1, Bcl11b, and STAT6 and confer chemoresistance to ALCL cells. Moreover, NPM-ALK/STAT3–miR-135b axis polarizes the identity of ALCL cells to a Th17 immunophenotype (pro-inflammatory cells) downregulating GATA3 and STAT6. This pro-inflammatory immunophenotype shift was also demonstrated in a xenograft model of ALCL in which the tumor growth and angiogenesis support and are supported by the pro-inflammatory microenvironment (267, 268).
In the same tumors, ALK-positive ALCL, the angiogenesis is still sustained by hypoxia-miR-16 downregulation. miR-16 downregulates VEGF expression biding the 3’UTR- of its mRNA directly. In onco-ALK mouse MEF cell lines and transgenic models, it was shown that ALK and hypoxia-inducible factor 1 alfa (HIF-1α) co-expression induce miR-16 downregulation, which therefore does not allow VEGF downregulation by post-transcriptional regulation (269).
The polycistronic miR-17-92 cluster, encoding six miRNAs (miR-17, miR-18a, miR-19a, miR-19b, miR-20a, and miR-92), is involved in immune cell development, have an oncogenic role in lymphoma, and a controversial role in angiogenesis (270, 271). In p53-null colonocytes, co-expressing K-RAS and c-Myc, was shown an increased neoangiogenesis correlated with the downregulation of anti-angiogenic thrombospondin-1 (Tsp1) via miR-18, and of the connective tissue growth factor via miR-18 and miR-19 (272). On the contrary, other authors demonstrated an anti-angiogenic role of the same cluster showing that the overexpression of miR-20a inhibits sprouting in vitro, whereas its inhibition increases ECs sprout formation in perfused vessels Matrigel plugs, but not following systemic inhibition (273). The dispute can be overcome, considering that tumor angiogenesis regulation is context-dependent.
A schematic diagram of the significant lymphoma-miRNAs above mentioned is represented in Figure 2.
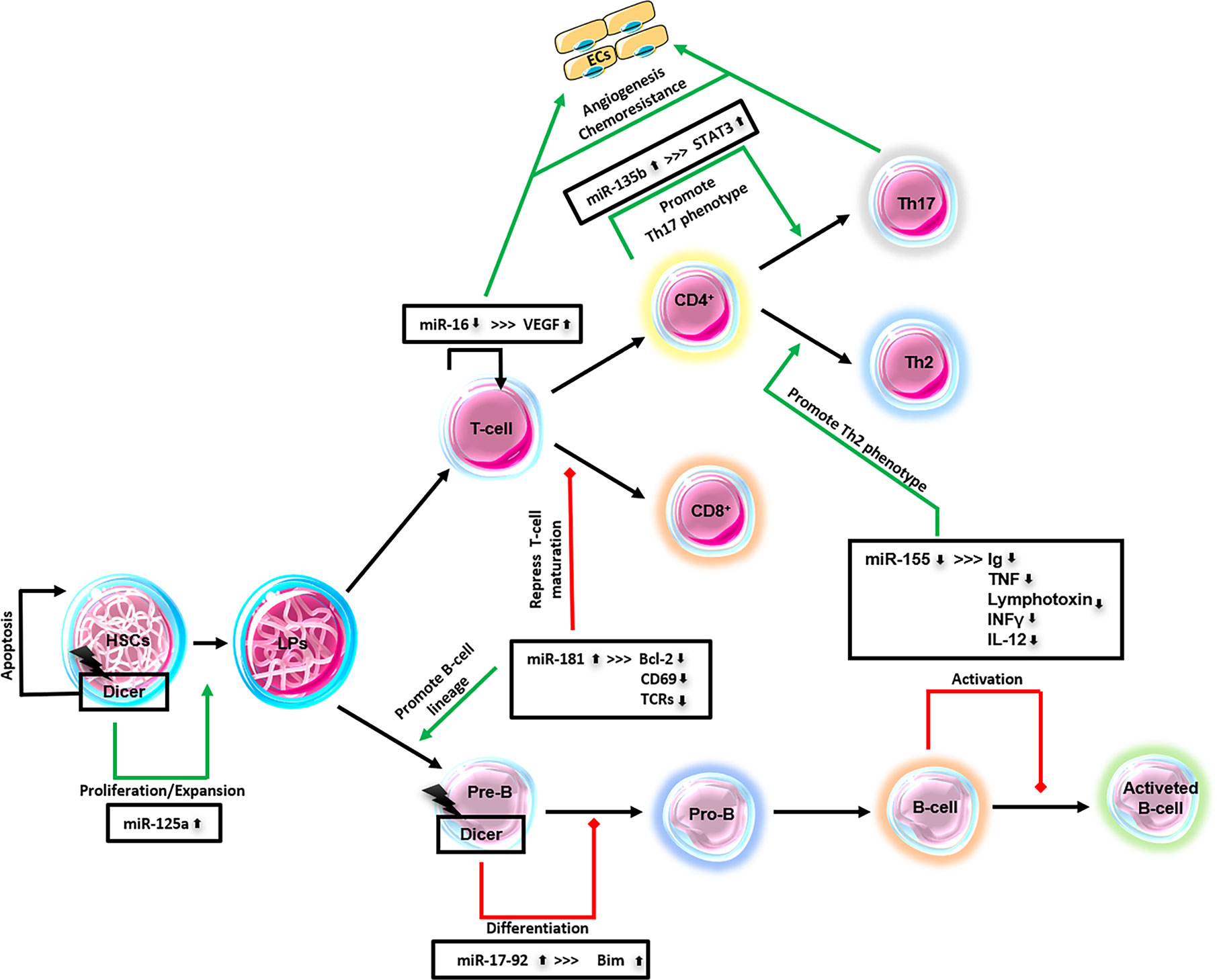
Figure 2 Major lymphoma-derived miRNAs. This schematic diagram represents miRNAs that have been identified in lymphomas and their relationship to lymphopoiesis. ECs, endothelial cells; HSCs, hematopoietic stem cells; LPs, lymphocytes precursors; Pre-B, pre-B-cells; Pro-B, pro-B-cells; CD4+, T-cells; CD8+, T-cells; Th2, T-helper cells; Th17, T-helper cells; T-/B-cells.
Challenges and Future Directions
Large-scale gene expression analysis evidence that the expression of individual miRNA or miRNA signatures will become routine biomarkers for cancer diagnosis and prognosis, and personalized therapy, thanks to their accessibility, high specificity, and sensitivity.
miRNAs induce epigenetic alterations that are reversible and targeted multiple regulators of related pathways (274–277). Tumor angiogenesis can be ruled by derepressing/miming tumor suppressor genes with angiostatic properties that are instead epigenetically silenced in a tumor, endothelial, and/or microenvironment cells, or by repressing oncogenes codifying for pro-angiogenic molecules (278, 279). Some argue that miRNA inhibitors are prevalently used for therapies because endogenous miRNA inhibition is less hazardous than overexpressing. On the contrary, others sustained that miRNA therapy to replace endogenous miRNAs repressed in cancer cells, using miRNA mimics, is less dangerous because they will not damage normal cells since they already express it.
miRNAs for therapy are usually chemically modified to stabilize the oligonucleotide once injected by intravenous, intraperitoneal, or intramuscular ways. miRNAs could be embedded in a liposome or viral backbone, or associated with targeted nanoparticles to improve affected cells’ yield.
Anti-angiogenic therapies usually directly targeted ECs, because ECs are in direct contact with blood carrying drugs, a small amount of ECs regulate many tumor cells, ECs are genetically more stable than tumor cells and so therapeutic effects are more predictable, and finally, only the ECs in the tumor site are activated and could be specifically targeted. Tumor ECs “angiogenic stage-related profiles” was extensively characterized by in vitro ECs culture under different cytokines and growth factors stimuli, using tube formation models, and ECs isolated from in vivo sources (280–282).
miRNA combination therapies with anti-angiogenic strategies, chemotherapy, radiotherapy, photodynamic therapy, and immunotherapy are in-depth reviewed elsewhere (278, 283). Noteworthy are two approved clinical trials about miR-16 and miR-34a. miR-16-based microRNA mimic is employed in a clinical trial to treat malignant pleural mesothelioma and non–small-cell lung cancer (Identifier NCT02369198). miR-34a mimic, in combination with the antiangiogenic drug doxorubicin, is in phase I clinical trial to treat patients with advanced solid and hematological tumors (Identifier NCT01829971).
Given the scarcity of miRNAs in clinical studies, their clinical application is not easy. The significant challenges are to prevent unwanted off-target effects, to develop an efficient and specific miRNA delivery system, to optimize effective dose (167). Well-defined pre-clinical studies, using cell lines and animal models and a broad set of human samples, are needed to solve these challenges. For example, coculture and 3D-culture for ECs with other microenvironment cells to study angiogenesis represents a more physiological condition. In order to improve clinical trials, it will be necessary to follow miRNA-treated patients for a long time to prove that miRNAs are unique biomarkers of angiogenesis‐related cancers. Moreover, miRNAs signatures from serum, plasma, or urinary should be further investigated as less invasive procedures. Recently, circulating exosomal miRNAs, thanks to reasonable stability, higher specificity, and sensitivity, appear potential biomarkers for clinical applications.
Standardized miRNAs application protocols in clinical practice are missing, maybe due to the lack of extensive studies on miRNAs with limited data comparability. More attention must be paid to selecting the starting material, sample collection, detection platform, and statistical analysis (284). For instance, if we wanted to quantify the circulating miRNAs, we should be attentive to the source that could be the whole blood, serum, plasma, exosomes, or microvesicles. When whole blood is used, miRNAs from blood cells will influence the total amount of the only circulating ones (273, 274). For sample collection, extreme pH changes or repeated freezing and thawing cycles should be avoided. The best commercial kit available for miRNAs extraction should choose to produce the highest yield of total RNA extraction (275). Moreover, it must be considered that in the samples of cancer patients, there will be a greater quantity of miRNA, and therefore to compare the miRNA expression data with healthy ones, it is more appropriate to use the same volume of starting material (biological fluid, cells, tissue) instead to use the same amount of total RNA.
Normalization of miRNAs quantification is another challenge. The small nuclear U6, or small nucleolar RNAs SNORD44, or some validated miRNAs, as miR-16, were usually used for normalization, but it was demonstrated that these common reference genes for miRNA research are poor normalizers because differentially regulated (285, 286). Therefore, it is necessary to verify the expression stability of putative normalizers in each experiment.
Apart from these technical problems, the difficulties in translate microRNA biomarkers from bench to clinic are associated with the inconsistency of miRNAs as unique biomarkers since: several of them are detectable in patients with different tumor types; other studies have shown different results for the same miRNA in the same tumors; sometimes is difficult to discriminate among closely related miRNAs (286, 287).
In order to improve and implement the clinical application of miRNAs, it is necessary to start with a careful evaluation of the preliminary study approaches. miRNAs that are already known and that are present in the miRNA database (miRbase) can be detected and quantified by polymerase chain reaction (PCR)-based or hybridization-based methods. Quantitative reverse transcription-polymerase chain reaction, qRT-PCR, is the gold-standard method in miRNA profiling in human (288). TaqMan TLDA microfluidic cards and miRCURY LNA qPCR detection platforms are the most used (276). Hybridization techniques are also used. miRNA microarrays analyze thousands of miRNAs in one assay but have less sensitivity than PCR-based methods (289).
On the other hand, to know and quantify new miRNAs, miRNA-seq is applied (290). The computational and prediction methods developed to detect tumor-related miRNAs are not satisfactory enough (291). However, despite the improvement in miRNA expression profiling technologies and big data in public databases, to date, these public portals with miRNA expression data from patients around the world are underemployed.
Conclusions
The human genome encodes approximately 2,600 mature miRNAs and more than 200,000 mRNAs (279). miRNAs may target more than one mRNAs, thus influencing multiple molecular pathways, but also mRNAs may bind to a variety of miRNAs, either simultaneously or in a context-dependent manner (292, 293).
Computational analysis suggests that around 30% of protein-coding genes in humans are regulated by miRNAs (177). Each step of miRNA biogenesis, including miRNA transcription, processing by Drosha and Dicer, transportation, RISC binding, and miRNA decay, is finely controlled. Therefore, minimal dysregulation in miRNA biogenesis and their control of gene expression can lead to tumorigenesis.
Here we have provided updates in miRNAs biogenesis, function, and role in health and tumor and especially in angiogenesis. The identification of miRNAs as onco-miRNAs or oncosuppressor-miRNA with a critical role in angiogenesis has raised new hopes for the treatment of tumors. They may be candidates even for target-specific therapies as antagonists or mimics (294). However, their diffusion as drugs is still far from the full clinical application because there are several criticalities to be carefully considered as tissue-specific delivery and cellular uptake sufficient to produce the therapeutic effect.
The relationship between angiogenesis and progression of several hematological malignancies is well established, and anti-angiogenic drugs appear a promising therapeutic strategy (295). Endostatin, immunomodulatory drugs (e.g., Thalidomide), neutralizing antibodies (e.g., Bevacizumab), and histone deacetylase inhibitors (e.g., Vorinostatat) are used in the treatment of lymphoma, but they are not enough to re-establish “normal” angiogenesis. miRNAs could cover part of the leaks. Among the hematological tumors, we decide to evaluate the literature concerning the role of miRNAs in the regulation of angiogenesis in lymphoma, and we observed that although the studies are scarce, there are two clinical trials for miR-155 in progress.
In addition to the critical issues concerning the pharmacokinetics and pharmacodynamics of miRNA-drugs, it should be stressed that there is still much to discover in the field of miRNAs. The sequencing of miRNome and the mapping of miRNA–mRNA interactions is far from being complete due to the recognized challenge of computational prediction of mRNA–microRNA interactions. In most studies, methods of high-throughput analysis based on Next Generation Sequencing technology (NGS) and computational analysis are applied. However, these experimental approaches to define the role of small non-coding RNAs in the biological and pathological processes are not enough. The morphology-based approach, such as dual in situ Dual ISH-IHC Technology, should walk together with the most modern computational and molecular technologies for a more precise characterization of the biological events in a more realistic molecular context (296–298).
Author Contributions
This work was conceived and planned by TA and DR. The original draft preparation and writing were made by TA, RT, and MG. DR reviewed and edited the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by Associazione “Il Sorriso di Antonio”, Corato, Italy.
Abbreviations
AML, acute myeloid leukemia; Ago, argonaute family of proteins; AEG-1, astrocyte elevated gene-1; ALCL, anaplastic large-cell lymphoma; AML, acute myeloid leukemia; ANN, axillary lymph node−negative; B-CLC, B-cell chronic lymphatic leukemia; CAFs, cancer-associated fibroblasts; CSCs, cancer stem cells; DGCR8, DiGeorge syndrome critical region 8; DNMT, DNA methyltransferase; dsRNA, double-stranded RNA; ECM, extracellular matrix; EGCG, epigallocatechin-3-gallate; ECs, endothelial cells; EMT, epithelial-mesenchymal transition; ESCs, embryonic stem cells; FBXW7, F-box/WD repeat-containing protein 7; FMR1, fragile X mental retardation 1; GBM, glioblastoma multiforme; GEMIN4, gem nuclear organelle associated protein 4; GSEA, Gene Set Enrichment Analysis; HCC, hepatocellular carcinoma; HDAC, histone deacetylase; HIF-1α, hypoxia-inducible factor 1 alfa; HL, Hodgkin lymphoma; HSC, hematopoietic stem cells; HSPC, hematopoietic stem/progenitor cell; IFNγ, interferon-gamma; IL-12, interleukin-12; IPO8, importin 8; lncRNA, long non-coding RNA; MAPK, mitogen-activated protein kinase; MCCHL, mixed cellularity Hodgkin lymphoma; miRISC, miRNA-RISC; miRNA, microRNAs; MLL, myeloid/lymphoid leukemia or mixed-lineage leukemia; mRNA, messenger RNA; MVBs, multivesicular bodies; MVD, microvascular density; NES, nuclear export signal; NHL, non-Hodgkin lymphoma; NLS, nuclear localization signal; NPM-ALK, nucleophosmin-anaplastic lymphoma kinase; NSCHL, nodular sclerosis Hodgkin lymphoma; NSCLC, non-small cell lung cancer; nSMase2, neutral sphingomyelinase 2; OSCC, oral squamous cell carcinoma; pADPr, poly-ADP-ribose; PARG, poly-ADP-ribose glycohydrolase; PARylation, poly-ADP-ribosylation; PARPs, poly-ADP-ribose polymerases; PCDH9, Protocadherin 9; pre-miRNA, precursor miRNA; pri-miRNA, primary miRNA; pRNA, promoter-associated RNA; PTEN, phosphatase and tensin homolog; PTMs, post-translational modifications; Ran-GTPase, RAS-related nuclear protein-guanosine-5’-triphosphate-ase; RBPs, RNA- binding proteins; rER, rough endoplasmic reticulum; RISC, RNA-induced silencing complex; RNAi, RNA interference; ROCK1, Rho-associated coiled-coil containing protein kinase 1; R2D2, dsRNA binding protein with two dsRNA binding domains; SAHA, suberoylanilide hydroxamic acid; shRNA, short hairpin RNA; SND1, staphylococcal nuclease domain-containing protein 1; snoRNAs, small nucleolar RNAs; SNPs, single-nucleotide polymorphisms; SQCLC, squamous cell lung cancer; TAMs, tumor-associated macrophages; TGA, transcriptional gene activation; TGS, transcriptional gene silencing; TCGA, the cancer genome atlas; TKI, tyrosine kinase inhibitor; TMAs, tissue microarrays; TNF, tumor necrosis factor; TNRC6A, trinucleotide repeat-containing adaptor 6a; tRFs, tRNA derived RNA fragments; tRNAs, transfer RNAs; tsRNAs, tRNA-derived small RNAs; UTR, untranslated region; VE-cadherin, vascular endothelial cadherin; VEGF, vascular endothelial growth factor; VEGFR, vascular endothelial growth factor receptor; VIG, vasa intronic gene; XPO5, exportin5; ZEB1/2, zinc finger e-box binding homeobox 1/2.
References
1. Kincaid RP, Sullivan CS. Virus-Encoded microRNAs: An Overview and a Look to the Future. PLoS Pathog (2012) 8:e1003018. doi: 10.1371/journal.ppat.1003018
2. Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell (1993) 75:843–54. doi: 10.1016/0092-8674(93)90529-Y
3. Ruvkun G, Wightman B, Ha I. The 20 years it took to recognize the importance of tiny RNAs. Cell (2004) 116:S93–6, 2 p following S6. doi: 10.1016/s0092-8674(04)00034-0
4. Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell (1993) 75:855–62. doi: 10.1016/0092-8674(93)90530-4
5. Bloch S, Wegrzyn A, Wegrzyn G, Nejman-Falenczyk B. Small and Smaller-sRNAs and MicroRNAs in the Regulation of Toxin Gene Expression in Prokaryotic Cells: A Mini-Review. Toxins (Basel) (2017) 9:181. doi: 10.3390/toxins9060181
6. Farazi TA, Juranek SA, Tuschl T. The growing catalog of small RNAs and their association with distinct Argonaute/Piwi family members. Dev (Cambridge England) (2008) 135:1201–14. doi: 10.1242/dev.005629
7. Olena AF, Patton JG. Genomic organization of microRNAs. J Cell Physiol (2010) 222:540–5. doi: 10.1002/jcp.21993
8. Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res (2004) 14:1902–10. doi: 10.1101/gr.2722704
9. Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol (2014) 15:509–24. doi: 10.1038/nrm3838
10. Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev (2004) 18:3016–27. doi: 10.1101/gad.1262504
11. Okada C, Yamashita E, Lee SJ, Shibata S, Katahira J, Nakagawa A, et al. A high-Resolution structure of the pre-microrna nuclear export machinery. Science (2009) 326:1275–9. doi: 10.1126/science.1178705
12. Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, et al. The Microprocessor complex mediates the genesis of microRNAs. Nature (2004) 432:235–40. doi: 10.1038/nature03120
13. Cullen BR. Transcription and processing of human microRNA precursors. Mol Cell (2004) 16:861–5. doi: 10.1016/j.molcel.2004.12.002
14. Liu X, Fortin K, Mourelatos Z. MicroRNAs: biogenesis and molecular functions. Brain Pathol (2008) 18:113–21. doi: 10.1111/j.1750-3639.2007.00121.x
15. Noland CL, Doudna JA. Multiple sensors ensure guide strand selection in human RNAi pathways. RNA (2013) 19:639–48. doi: 10.1261/rna.037424.112
16. Santhekadur PK, Kumar DP. RISC assembly and post-transcriptional gene regulation in Hepatocellular Carcinoma. Genes Dis (2020) 7:199–204. doi: 10.1016/j.gendis.2019.09.009
17. van den Berg A, Mols J, Han J. RISC-target interaction: cleavage and translational suppression. Biochim Biophys Acta (2008) 1779:668–77. doi: 10.1016/j.bbagrm.2008.07.005
18. Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science (2004) 305:1437–41. doi: 10.1126/science.1102513
19. Rivas FV, Tolia NH, Song JJ, Aragon JP, Liu J, Hannon GJ, et al. Purified Argonaute2 and an siRNA form recombinant human RISC. Nat Struct Mol Biol (2005) 12:340–9. doi: 10.1038/nsmb918
20. Li SC, Liao YL, Ho MR, Tsai KW, Lai CH, Lin WC. miRNA arm selection and isomiR distribution in gastric cancer. BMC Genomics (2012) 13 Suppl 1:S13. doi: 10.1186/1471-2164-13-S1-S13
21. Siomi H, Siomi MC. On the road to reading the RNA-interference code. Nature (2009) 457:396–404. doi: 10.1038/nature07754
22. Meijer HA, Smith EM, Bushell M. Regulation of miRNA strand selection: follow the leader? Biochem Soc Trans (2014) 42:1135–40. doi: 10.1042/BST20140142
23. Miller S, Jones LE, Giovannitti K, Piper D, Serra MJ. Thermodynamic analysis of 5′ and 3′ single- and 3′ double-nucleotide overhangs neighboring wobble terminal base pairs. Nucleic Acids Res (2008) 6:5652–9. doi: 10.1093/nar/gkn525
24. Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet (2008) 9:102–14. doi: 10.1038/nrg2290
25. Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature (2010) 466:835–40. doi: 10.1038/nature09267
26. Nilsen TW. Mechanisms of microRNA-mediated gene regulation in animal cells. Trends Genet (2007) 23:243–9. doi: 10.1016/j.tig.2007.02.011
27. Seok H, Ham J, Jang ES, Chi SW. MicroRNA Target Recognition: Insights from Transcriptome-Wide Non-Canonical Interactions. Mol Cells (2016) 39:375–81. doi: 10.14348/molcells.2016.0013
28. Catalanotto C, Cogoni C, Zardo G. MicroRNA in Control of Gene Expression: An Overview of Nuclear Functions. Int J Mol Sci (2016) 17:1712. doi: 10.3390/ijms17101712
29. Jonas S, Izaurralde E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat Rev Genet (2015) 16:421–33. doi: 10.1038/nrg3965
30. Bazzini AA, Lee MT, Giraldez AJ. Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in Zebrafish. Science (2012) 336:233–7. doi: 10.1126/science.1215704
31. Djuranovic S, Nahvi A, Green R. miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science (2012) 336:237–40. doi: 10.1126/science.1215691
32. Freimer JW, Hu TJ, Blelloch R. Decoupling the impact of MicroRNAs on translational repression versus RNA degradation in embryonic stem cells. eLife (2018) 7:e38014. doi: 10.7554/eLife.38014
33. Hu W, Coller J. What comes first: Translational repression or mRNA degradation? the deepening mystery of microRNA function. Cell Res (2012) 22:1322–4. doi: 10.1038/cr.2012.80
34. Bose M, Barman B, Goswami A, Bhattacharyya SN. Spatiotemporal Uncoupling of MicroRNA-Mediated Translational Repression and Target RNA Degradation Controls MicroRNP Recycling in Mammalian Cells. Mol Cell Biol (2017) 37:e00464–16. doi: 10.1128/mcb.00464-16
35. Stalder L, Heusermann W, Sokol L, Trojer D, Wirz J, Hean J, et al. The rough endoplasmatic reticulum is a central nucleation site of siRNA-mediated RNA silencing. EMBO J (2013) 32:1115–27. doi: 10.1038/emboj.2013.52
36. Gagnon KT, Li L, Chu Y, Janowski BA, Corey DR. RNAi factors are present and active in human cell nuclei. Cell Rep (2014) 6:211–21. doi: 10.1016/j.celrep.2013.12.013
37. Liu H, Lei C, He Q, Pan Z, Xiao D, Tao Y. Nuclear functions of mammalian MicroRNAs in gene regulation, immunity and cancer. Mol Cancer (2018) 17:64. doi: 10.1186/s12943-018-0765-5
38. Wei Y, Li L, Wang D, Zhang CY, Zen K. Importin 8 regulates the transport of mature microRNAs into the cell nucleus. J Biol Chem (2014) 289:10270–5. doi: 10.1074/jbc.C113.541417
39. López-Urrutia E, Bustamante Montes LP, Ladrón de Guevara Cervantes D, Pérez-Plasencia C, Campos-Parra AD. Crosstalk Between Long Non-coding RNAs, Micro-RNAs and mRNAs: Deciphering Molecular Mechanisms of Master Regulators in Cancer. Front Oncol (2019) 9:669. doi: 10.3389/fonc.2019.00669
40. Tang R, Li L, Zhu D, Hou D, Cao T, Gu H, et al. Mouse miRNA-709 directly regulates miRNA-15a/16-1 biogenesis at the posttranscriptional level in the nucleus: Evidence for a microRNA hierarchy system. Cell Res (2012) 22:504–15. doi: 10.1038/cr.2011.137
41. Zisoulis DG, Kai ZS, Chang RK, Pasquinelli AE. Autoregulation of microRNA biogenesis by let-7 and Argonaute. Nature (2012) 486:541–4. doi: 10.1038/nature11134
42. Atwood BL, Woolnough JL, Lefevre GM, Ribeiro MSJ, Felsenfeld G, Giles KE. Human argonaute 2 is tethered to ribosomal RNA through MicroRNA interactions. J Biol Chem (2016) 291:17919–28. doi: 10.1074/jbc.M116.725051
43. Li ZF, Liang YM, Lau PN, Shen W, Wang DK, Cheung WT, et al. Dynamic Localisation of Mature MicroRNAs in Human Nucleoli is Influenced by Exogenous Genetic Materials. PLoS One (2013) 8:e70869. doi: 10.1371/journal.pone.0070869
44. Politz JCR, Zhang F, Pederson T. MicroRNA-206 colocalizes with ribosome-rich regions in both the nucleolus and cytoplasm of rat myogenic cells. Proc Natl Acad Sci USA (2006) 103:18957–62. doi: 10.1073/pnas.0609466103
45. Reyes-Gutierrez P, Ritland Politz JC, Pederson T. A mRNA and cognate MicroRNAs localize in the nucleolus. Nucleus (2014) 5:636–42. doi: 10.4161/19491034.2014.990864
46. Kalsotra A, Wang K, Li PF, Cooper TA. MicroRNAs coordinate an alternative splicing network during mouse postnatal heart development. Genes Dev (2010) 24:653–8. doi: 10.1101/gad.1894310
47. Li LC. Chromatin remodeling by the small rna machinery in mammalian cells. Epigenetics (2014) 9:45–52. doi: 10.4161/epi.26830
48. Melamed Ze, Levy A, Ashwal-Fluss R, Lev-Maor G, Mekahel K, Atias N, et al. Alternative Splicing Regulates Biogenesis of miRNAs Located across Exon-Intron Junctions. Mol Cell (2013) 50:869–81. doi: 10.1016/j.molcel.2013.05.007
49. Kim DH, Sætrom P, Snøve O, Rossi JJ. MicroRNA-directed transcriptional gene silencing in mammalian cells. Proc Natl Acad Sci USA (2008) 105:16230–5. doi: 10.1073/pnas.0808830105
50. Xiao M, Li J, Li W, Wang Y, Wu F, Xi Y, et al. MicroRNAs activate gene transcription epigenetically as an enhancer trigger. RNA Biol (2017) 14:1326–34. doi: 10.1080/15476286.2015.1112487
51. Matsui M, Chu Y, Zhang H, Gagnon KT, Shaikh S, Kuchimanchi S, et al. Promoter RNA Links Transcriptional Regulation of Inflammatory Pathway Genes - PubMed. Nucleic Acids Res (2013) 41:10086–109. doi: 10.1093/nar/gkt777
52. Modarresi F, Faghihi MA, Lopez-Toledano MA, Fatemi RP, Magistri M, Brothers SP, et al. Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional upregulation. Nat Biotechnol (2012) 30:453–9. doi: 10.1038/nbt.2158
53. Yang Q, Li W, She H, Dou J, Duong DM, Du Y, et al. Stress induces p38 MAPK-mediated phosphorylation and inhibition of drosha-dependent cell survival. Mol Cell (2015) 57:721–34. doi: 10.1016/j.molcel.2015.01.004
54. Hong S, Noh H, Chen H, Padia R, Pan ZK, Su SB, et al. Signaling by p38 MAPK stimulates nuclear localization of the microprocessor component p68 for processing of selected primary MicroRNAs. Sci Signaling (2013) 6:ra16. doi: 10.1126/scisignal.2003706
55. Fletcher CE, Godfrey JD, Shibakawa A, Bushell M, Bevan CL. A novel role for GSK3β as a modulator of Drosha microprocessor activity and MicroRNA biogenesis. Nucleic Acids Res (2017) 45:2809–28. doi: 10.1093/nar/gkw938
56. Herbert KM, Pimienta G, DeGregorio SJ, Alexandrov A, Steitz JA. Phosphorylation of DGCR8 Increases Its Intracellular Stability and Induces a Progrowth miRNA Profile. Cell Rep (2013) 5:1070–81. doi: 10.1016/j.celrep.2013.10.017
57. Tu CC, Zhong Y, Nguyen L, Tsai A, Sridevi P, Tarn WY, et al. The kinase ABL phosphorylates the microprocessor subunit DGCR8 to stimulate primary microRNA processing in response to DNA damage. Sci Signaling (2015) 8:ra64. doi: 10.1126/scisignal.aaa4468
58. Paroo Z, Ye X, Chen S, Liu Q. Phosphorylation of the Human MicroRNA-Generating Complex Mediates MAPK/Erk Signaling. Cell (2009) 139:112–22. doi: 10.1016/j.cell.2009.06.044
59. Warner MJ, Bridge KS, Hewitson JP, Hodgkinson MR, Heyam A, Massa BC, et al. S6K2-mediated Regulation of TRBP as a Determinant of miRNA Expression in Human Primary Lymphatic Endothelial Cells. Nucleic Acids Res (2016) 44:9942–55. doi: 10.1093/nar/gkw631
60. Ye P, Liu Y, Chen C, Tang F, Wu Q, Wang X, et al. An mTORC1-Mdm2-Drosha axis for miRNA biogenesis in response to glucose- and amino acid-deprivation. Mol Cell (2015) 57:708–20. doi: 10.1016/j.molcel.2014.12.034
61. Zhu C, Chen C, Huang J, Zhang H, Zhao X, Deng R, et al. SUMOylation at K707 of DGCR8 Controls Direct Function of Primary microRNA - PubMed. Nucleic Acids Res (2015) 43:7945–460. doi: 10.1093/nar/gkv741
62. Chen C, Zhu C, Huang J, Zhao X, Deng R, Zhang H, et al. SUMOylation of TARBP2 regulates miRNA/siRNA efficiency. Nat Commun (2015) 6:8899. doi: 10.1038/ncomms9899
63. Gurtner A, Falcone E, Garibaldi F, Piaggio G. Dysregulation of microRNA biogenesis in cancer: the impact of mutant p53 on Drosha complex activity. J Exp Clin Canc Res (2016) 35:45. doi: 10.1186/s13046-016-0319-x
64. Garibaldi F, Falcone E, Trisciuoglio D, Colombo T, Lisek K, Walerych D, et al. Mutant p53 inhibits miRNA biogenesis by interfering with the microprocessor complex. Oncogene (2016) 35:3760–70. doi: 10.1038/onc.2016.51
65. Jiang FZ, He YY, Wang HH, Zhang HL, Zhang J, Yan XF, et al. Mutant p53 induces EZH2 expression and promotes epithelial-mesenchymal transition by disrupting p68-Drosha complex assembly and attenuating miR-26a processing. Oncotarget (2015) 6:44660–74. doi: 10.18632/oncotarget.6350
66. Lin S, Gregory RI. MicroRNA biogenesis pathways in cancer. Nat Rev Cancer (2015) 15:321–33. doi: 10.1038/nrc3932
67. Lin RJ, Lin YC, Chen J, Kuo HH, Chen YY, Diccianni MB, et al. microRNA signature and expression of Dicer and Drosha can predict prognosis and delineate risk groups in neuroblastoma. Cancer Res (2010) 70:7841–50. doi: 10.1158/0008-5472.CAN-10-0970
68. Merritt WM, Lin YG, Han LY, Kamat AA, Spannuth WA, Schmandt R, et al. Dicer, Drosha, and outcomes in patients with ovarian cancer. N Engl J Med (2008) 359:2641–50. doi: 10.1056/NEJMoa0803785
69. Guo Y, Tian P, Yang C, Liang Z, Li M, Sims M, et al. Silencing the double-stranded RNA binding protein DGCR8 inhibits ovarian cancer cell proliferation, migration, and invasion. Pharm Res (2015) 32:769–78. doi: 10.1007/s11095-013-1219-9
70. Robertson JC, Jorcyk CL, Oxford JT. DICER1 Syndrome: DICER1 Mutations in Rare Cancers. Cancers (Basel) (2018) 10:143. doi: 10.3390/cancers10050143
71. Doros L, Yang J, Dehner L, Rossi CT, Skiver K, Jarzembowski JA, et al. DICER1 mutations in embryonal rhabdomyosarcomas from children with and without familial PPB-tumor predisposition syndrome. Pediatr Blood Cancer (2012) 59:558–60. doi: 10.1002/pbc.24020
72. Schultz KA, Yang J, Doros L, Williams GM, Harris A, Stewart DR, et al. DICER1-pleuropulmonary blastoma familial tumor predisposition syndrome: a unique constellation of neoplastic conditions. Pathol Case Rev (2014) 19:90–100. doi: 10.1097/PCR.0000000000000027
73. Schultz KA, Harris A, Messinger Y, Sencer S, Baldinger S, Dehner LP, et al. Ovarian tumors related to intronic mutations in DICER1: a report from the international ovarian and testicular stromal tumor registry. Fam Cancer (2016) 15:105–10. doi: 10.1007/s10689-015-9831-y
74. De Vito C, Riggi N, Cornaz S, Suva ML, Baumer K, Provero P, et al. A TARBP2-dependent miRNA expression profile underlies cancer stem cell properties and provides candidate therapeutic reagents in Ewing sarcoma. Cancer Cell (2012) 21:807–21. doi: 10.1016/j.ccr.2012.04.023
75. Khan M, Khan Z, Uddin Y, Mustafa S, Shaukat I, Pan J, et al. Evaluating the Oncogenic and Tumor Suppressor Role of XPO5 in Different Tissue Tumor Types. Asian Pac J Cancer Prev (2018) 19:1119–25. doi: 10.22034/APJCP.2018.19.4.1119
76. Leaderer D, Hoffman AE, Zheng T, Fu A, Weidhaas J, Paranjape T, et al. Genetic and epigenetic association studies suggest a role of microRNA biogenesis gene exportin-5 (XPO5) in breast tumorigenesis. Int J Mol Epidemiol Genet (2011) 2:9–18.
77. Melo SA, Moutinho C, Ropero S, Calin GA, Rossi S, Spizzo R, et al. A genetic defect in exportin-5 traps precursor microRNAs in the nucleus of cancer cells. Cancer Cell (2010) 18:303–15. doi: 10.1016/j.ccr.2010.09.007
78. Stavast CJ, Erkeland SJ. The Non-Canonical Aspects of MicroRNAs: Many Roads to Gene Regulation. Cells (2019) 8:1465–. doi: 10.3390/cells8111465
79. Felekkis K, Touvana E, Stefanou C, Deltas C. microRNAs: a newly described class of encoded molecules that play a role in health and disease. Hippokratia (2010) 14:236–40.
80. Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC. Mammalian Mirtron Genes. Mol Cell (2007) 28:328–36. doi: 10.1016/j.molcel.2007.09.028
81. Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature (2007) 448:83–6. doi: 10.1038/nature05983
83. Havens MA, Reich AA, Duelli DM, Hastings ML. Biogenesis of Mammalian microRNAs by a Non-Canonical Processing Pathway. Nucleic Acids Res (2012) 40:4626–40. doi: 10.1093/nar/gks026
84. Rorbach G, Unold O, Konopka BM. Distinguishing mirtrons from canonical miRNAs with data exploration and machine learning methods. Sci Rep (2018) 8:7560. doi: 10.1038/s41598-018-25578-3
85. Cheloufi S, Dos Santos CO, Chong MMW, Hannon GJ. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature (2010) 465:584–9. doi: 10.1038/nature09092
86. Herrera-Carrillo E, Harwig A, Liu YP, Berkhout B. Probing the shRNA characteristics that hinder Dicer recognition and consequently allow Ago-mediated processing and AgoshRNA activity. RNA (2014) 20:1410–8. doi: 10.1261/rna.043950.113
87. Herrera-Carrillo EE, Berkhout B. Dicer-independent processing of small RNA duplexes: mechanistic insights and applications. Nucleic Acids Res (2017) 45:10369–79. doi: 10.1093/nar/gkx779
88. Harwig A, Kruize Z, Yang Z, Restle T, Berkhout B. Analysis of AgoshRNA maturation and loading into Ago2. PLoS One (2017) 12:e018326. doi: 10.1371/journal.pone.0183269
89. Bandres E, Bitarte N, Arias F, Agorreta J, Fortes P, Agirre X, et al. microRNA-451 regulates macrophage migration inhibitory factor production and proliferation of gastrointestinal cancer cells. Clin Cancer Res (2009) 15:2281–90. doi: 10.1158/1078-0432.CCR-08-1818
90. Tsuchiya S, Oku M, Imanaka Y, Kunimoto R, Okuno Y, Terasawa K, et al. MicroRNA-338-3p and microRNA-451 contribute to the formation of basolateral polarity in epithelial cells. Nucleic Acids Res (2009) 37:3821–7. doi: 10.1093/nar/gkp255
91. Wang R, Wang ZX, Yang JS, Pan X, De W, Chen LB. MicroRNA-451 functions as a tumor suppressor in human non-small cell lung cancer by targeting ras-related protein 14 (RAB14). Oncogene (2011) 30:2644–58. doi: 10.1038/onc.2010.642
92. Dupuis-Sandoval F, Poirier M, Scott MS. The emerging landscape of small nucleolar RNAs in cell biology. Wiley Interdiscip Rev RNA (2015) 6:381–97. doi: 10.1002/wrna.1284
93. Scott MS, Ono M. From snoRNA to miRNA: Dual function regulatory non-coding RNAs. Biochimie (2011) 93:1987–92. doi: 10.1016/j.biochi.2011.05.026
94. Li S, Xu Z, Sheng J. tRNA-derived small RNA: A novel regulatory small non-coding RNA. MDPI AG (2018) 9:246. doi: 10.3390/genes9050246
95. Haussecker D, Huang Y, Lau A, Parameswaran P, Fire AZ, Kay MA. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA (2010) 16:673–95. doi: 10.1261/rna.2000810
96. Kumar P, Anaya J, Mudunuri SB, Dutta A. Meta-analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets. BMC Med (2014) 12:78. doi: 10.1186/s12915-014-0078-0
97. Butkyte S, Ciupas L, Jakubauskiene E, Vilys L, Mocevicius P, Kanopka A, et al. Splicing-dependent expression of microRNAs of mirtron origin in human digestive and excretory system cancer cells. Clin Epigenet (2016) 8:33. doi: 10.1186/s13148-016-0200-y
98. Zhao M, Hou Y, Du YE, Yang L, Qin Y, Peng M, et al. Drosha-independent miR-6778-5p strengthens gastric cancer stem cell stemness via regulation of cytosolic one-carbon folate metabolism. Cancer Lett (2020) 478:8–21. doi: 10.1016/j.canlet.2020.02.040
99. Patterson DG, Roberts JT, King VM, Houserova D, Barnhill EC, Crucello A, et al. Human snoRNA-93 is processed into a microRNA-like RNA that promotes breast cancer cell invasion. NPJ Breast Cancer (2017) 3:25. doi: 10.1038/s41523-017-0032-8
100. Martens-Uzunova ES, Hoogstrate Y, Kalsbeek A, Pigmans B, Vredenbregt-van den Berg M, Dits N, et al. C/D-box snoRNA-derived RNA production is associated with malignant transformation and metastatic progression in prostate cancer. Oncotarget (2015) 6:17430–44. doi: 10.18632/oncotarget.4172
101. Chang P, Jacobson MK, Mitchison TJ. Poly(ADP-ribose) is required for spindle assembly and structure. Nature (2004) 432:645–9. doi: 10.1038/nature03061
102. Krishnakumar R, Kraus WL. The PARP side of the nucleus: molecular actions, physiological outcomes, and clinical targets. Mol Cell (2010) 39:8–24. doi: 10.1016/j.molcel.2010.06.017
103. Schreiber V, Dantzer F, Ame JC, de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat Rev Mol Cell Biol (2006) 7:517–28. doi: 10.1038/nrm1963
104. Liu C, Yu X. ADP-ribosyltransferases and poly ADP-ribosylation. Curr Protein Pept Sci (2015) 16:491–501. doi: 10.2174/1389203716666150504122435
105. Piskunova TS, Yurova MN, Ovsyannikov AI, Semenchenko AV, Zabezhinski MA, Popovich IG, et al. Deficiency in Poly(ADP-ribose) Polymerase-1 (PARP-1) Accelerates Aging and Spontaneous Carcinogenesis in Mice. Curr Gerontol Geriatr Res (2008) 2008:754190. doi: 10.1155/2008/754190
106. Yu SW, Andrabi SA, Wang H, Kim NS, Poirier GG, Dawson TM, et al. Apoptosis-inducing factor mediates poly(ADP-ribose) (PAR) polymer-induced cell death. Proc Natl Acad Sci USA (2006) 103:18314–9. doi: 10.1073/pnas.0606528103
107. Ke Y, Zhang J, Lv X, Zeng X, Ba X. Novel insights into PARPs in gene expression: regulation of RNA metabolism. Cell Mol Life Sci (2019) 76:3283–99. doi: 10.1007/s00018-019-03120-6
108. Malanga M, Czubaty A, Girstun A, Staron K, Althaus FR. Poly(ADP-ribose) binds to the splicing factor ASF/SF2 and regulates its phosphorylation by DNA topoisomerase I. J Biol Chem (2008) 283:19991–8. doi: 10.1074/jbc.M709495200
109. Ryu KW, Kim DS, Kraus WL. New facets in the regulation of gene expression by ADP-ribosylation and poly(ADP-ribose) polymerases. Chem Rev (2015) 115:2453–81. doi: 10.1021/cr5004248
110. Leung AK, Vyas S, Rood JE, Bhutkar A, Sharp PA, Chang P. Poly(ADP-ribose) regulates stress responses and microRNA activity in the cytoplasm. Mol Cell (2011) 42:489–99. doi: 10.1016/j.molcel.2011.04.015
111. Vyas S, Chesarone-Cataldo M, Todorova T, Huang YH, Chang P. A systematic analysis of the PARP protein family identifies new functions critical for cell physiology. Nat Commun (2013) 4:2240. doi: 10.1038/ncomms3240
112. Jee D, Lai EC. Alteration of miRNA activity via context-specific modifications of Argonaute proteins. Trends Cell Biol (2014) 24:546–53. doi: 10.1016/j.tcb.2014.04.008
113. Seo GJ, Kincaid RP, Phanaksri T, Burke JM, Pare JM, Cox JE, et al. Reciprocal inhibition between intracellular antiviral signaling and the RNAi machinery in mammalian cells. Cell Host Microbe (2013) 14:435–45. doi: 10.1016/j.chom.2013.09.002
114. Lin Y, Zhang H, Liang J, Li K, Zhu W, Fu L, et al. Identification and characterization of alphavirus M1 as a selective oncolytic virus targeting ZAP-defective human cancers. Proc Natl Acad Sci USA (2014) 111:E4504–12. doi: 10.1073/pnas.1408759111
115. Todorova T, Bock FJ, Chang P. PARP13 regulates cellular mRNA post-transcriptionally and functions as a pro-apoptotic factor by destabilizing TRAILR4 transcript. Nat Commun (2014) 5:5362. doi: 10.1038/ncomms6362
116. Gao G, Guo X, Goff SP. Inhibition of retroviral RNA production by ZAP, a CCCH-type zinc finger protein. Science (2002) 297:1703–6. doi: 10.1126/science.1074276
117. Mao R, Nie H, Cai D, Zhang J, Liu H, Yan R, et al. Inhibition of hepatitis B virus replication by the host zinc finger antiviral protein. PLoS Pathog (2013) 9:e1003494. doi: 10.1371/journal.ppat.1003494
118. Nomura F, Yaguchi M, Togawa A, Miyazaki M, Isobe K, Miyake M, et al. Enhancement of poly-adenosine diphosphate-ribosylation in human hepatocellular carcinoma. J Gastroenterol Hepatol (2000) 15:529–35. doi: 10.1046/j.1440-1746.2000.02193.x
119. Singh N. Enhanced poly ADP-ribosylation in human leukemia lymphocytes and ovarian cancers. Cancer Lett (1991) 58:131–5. doi: 10.1016/0304-3835(91)90035-g
120. Yalcintepe L, Turker-Sener L, Sener A, Yetkin G, Tiryaki D, Bermek E. Changes in NAD/ADP-ribose metabolism in rectal cancer. Braz J Med Biol Res (2005) 38:361–5. doi: 10.1590/s0100-879x2005000300006
121. Ossovskaya V, Koo IC, Kaldjian EP, Alvares C, Sherman BM. Upregulation of Poly (ADP-Ribose) Polymerase-1 (PARP1) in Triple-Negative Breast Cancer and Other Primary Human Tumor Types. Genes Cancer (2010) 1:812–21. doi: 10.1177/1947601910383418
122. Hui AB, Shi W, Boutros PC, Miller N, Pintilie M, Fyles T, et al. Robust global micro-RNA profiling with formalin-fixed paraffin-embedded breast cancer tissues. Lab Invest (2009) 89:597–606. doi: 10.1038/labinvest.2009.12
123. Pasculli B, Barbano R, Fontana A, Biagini T, Di Viesti MP, Rendina M, et al. Hsa-miR-155-5p Up-Regulation in Breast Cancer and Its Relevance for Treatment With Poly[ADP-Ribose] Polymerase 1 (PARP-1) Inhibitors. Front Oncol (2020) 10:1415. doi: 10.3389/fonc.2020.01415
124. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell (2011) 144:646–74. doi: 10.1016/j.cell.2011.02.013
125. Ha T-Y. MicroRNAs in Human Diseases: From Autoimmune Diseases to Skin, Psychiatric and Neurodegenerative Diseases. Immune Network (2011) 11:227–. doi: 10.4110/in.2011.11.5.227
126. Li Z, Lu J, Sun M, Mi S, Zhang H, Luo RT, et al. Distinct microRNA expression profiles in acute myeloid leukemia with common translocations. Proc Natl Acad Sci USA (2008) 105:15535–40. doi: 10.1073/pnas.0808266105
127. Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA (2002) 99:15524–9. doi: 10.1073/pnas.242606799
128. Bagheri F, Mesrian Tanha H, Mojtabavi Naeini M, Ghaedi K, Azadeh M. Tumor-promoting function of single nucleotide polymorphism rs1836724 (C3388T) alters multiple potential legitimate microRNA binding sites at the 3’-untranslated region of ErbB4 in breast cancer. Mol Med Rep (2016) 13:4494–8. doi: 10.3892/mmr.2016.5078
129. Song X, Zhong H, Wu Q, Wang M, Zhou J, Zhou Y, et al. Association between SNPs in microRNA machinery genes and gastric cancer susceptibility, invasion, and metastasis in Chinese Han population. Oncotarget (2017) 8:86435–46. doi: 10.18632/oncotarget.21199
130. Li W, Ma Y, Zeng D, Zhang J, Wang R, Hu J, et al. [Association between microRNA single nucleotide polymorphisms and the risk of hepatocellular carcinoma]. Rev Med Chil (2016) 144:508–15. doi: 10.4067/S0034-98872016000400013
131. Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell (2007) 26:745–52. doi: 10.1016/j.molcel.2007.05.010
132. Pagliuca A, Valvo C, Fabrizi E, di Martino S, Biffoni M, Runci D, et al. Analysis of the combined action of miR-143 and miR-145 on oncogenic pathways in colorectal cancer cells reveals a coordinate program of gene repression. Oncogene (2013) 32:4806–13. doi: 10.1038/onc.2012.495
133. Schmid G, Notaro S, Reimer D, Abdel-Azim S, Duggan-Peer M, Holly J, et al. Expression and promotor hypermethylation of miR-34a in the various histological subtypes of ovarian cancer. BMC Cancer (2016) 16:102. doi: 10.1186/s12885-016-2135-2
134. Wong MY, Yu Y, Walsh WR, Yang JL. microRNA-34 family and treatment of cancers with mutant or wild-type p53 (Review). Int J Oncol (2011) 38:1189–95. doi: 10.3892/ijo.2011.970
135. Lewis KA, Jordan HR, Tollefsbol TO. Effects of SAHA and EGCG on Growth Potentiation of Triple-Negative Breast Cancer Cells. Cancers (Basel) (2018) 11:23. doi: 10.3390/cancers11010023
136. Ali Syeda Z, Langden SSS, Munkhzul C, Lee M, Song SJ. Regulatory Mechanism of MicroRNA Expression in Cancer. Int J Mol Sci (2020) 21:1723. doi: 10.3390/ijms21051723
137. Lee SS, Min H, Ha JY, Kim BH, Choi MS, Kim S. Dysregulation of the miRNA biogenesis components DICER1, DROSHA, DGCR8 and AGO2 in clear cell renal cell carcinoma in both a Korean cohort and the cancer genome atlas kidney clear cell carcinoma cohort. Oncol Lett (2019) 18:4337–45. doi: 10.3892/ol.2019.10759
138. Muralidhar B, Winder D, Murray M, Palmer R, Barbosa-Morais N, Saini H, et al. Functional evidence that Drosha overexpression in cervical squamous cell carcinoma affects cell phenotype and microRNA profiles. J Pathol (2011) 224:496–507. doi: 10.1002/path.2898
139. Wu K, He J, Pu W, Peng Y. The Role of Exportin-5 in MicroRNA Biogenesis and Cancer. Genomics Proteomics Bioinf (2018) 16:120–6. doi: 10.1016/j.gpb.2017.09.004
140. Ramirez-Moya J, Wert-Lamas L, Riesco-Eizaguirre G, Santisteban P. Impaired microRNA processing by DICER1 downregulation endows thyroid cancer with increased aggressiveness. Oncogene (2019) 38:5486–99. doi: 10.1038/s41388-019-0804-8
141. Casey MC, Prakash A, Holian E, McGuire A, Kalinina O, Shalaby A, et al. Quantifying Argonaute 2 (Ago2) expression to stratify breast cancer. BMC Cancer (2019) 19:712. doi: 10.1186/s12885-019-5884-x
142. Zhang J, Jin H, Liu H, Lv S, Wang B, Wang R, et al. MiRNA-99a directly regulates AGO2 through translational repression in hepatocellular carcinoma. Oncogenesis (2014) 3:e97. doi: 10.1038/oncsis.2014.11
143. Zhang Y, Wang B, Chen X, Li W, Dong P. AGO2 involves the malignant phenotypes and FAK/PI3K/AKT signaling pathway in hypopharyngeal-derived FaDu cells. Oncotarget (2017) 8:54735–46. doi: 10.18632/oncotarget.18047
144. Wang C, Chen Q, Li S, Li S, Zhao Z, Gao H, et al. Dual inhibition of PCDH9 expression by miR-215-5p up-regulation in gliomas. Oncotarget (2017) 8:10287–97. doi: 10.18632/oncotarget.14396
145. Liu M, Roth A, Yu M, Morris R, Bersani F, Rivera MN, et al. The IGF2 intronic miR-483 selectively enhances transcription from IGF2 fetal promoters and enhances tumorigenesis. Genes Dev (2013) 27:2543–8. doi: 10.1101/gad.224170.113
146. Qu H, Zheng L, Pu J, Mei H, Xiang X, Zhao X, et al. miRNA-558 promotes tumorigenesis and aggressiveness of neuroblastoma cells through activating the transcription of heparanase. Hum Mol Genet (2015) 24:2539–51. doi: 10.1093/hmg/ddv018
147. Huang V, Place RF, Portnoy V, Wang J, Qi Z, Jia Z, et al. Upregulation of Cyclin B1 by miRNA and its implications in cancer. Nucleic Acids Res (2012) 40:1695–707. doi: 10.1093/nar/gkr934
148. Park CW, Zeng Y, Zhang X, Subramanian S, Steer CJ. Mature microRNAs identified in highly purified nuclei from HCT116 colon cancer cells. RNA Biol (2010) 7:606–14. doi: 10.4161/rna.7.5.13215
149. Taft RJ, Simons C, Nahkuri S, Oey H, Korbie DJ, Mercer TR, et al. Nuclear-localized tiny RNAs are associated with transcription initiation and splice sites in metazoans. Nat Struct Mol Biol (2010) 17:1030–4. doi: 10.1038/nsmb.1841
150. Tan Y, Zhang B, Wu T, Skogerbo G, Zhu X, Guo X, et al. Transcriptional inhibiton of Hoxd4 expression by miRNA-10a in human breast cancer cells. BMC Mol Biol (2009) 10:12. doi: 10.1186/1471-2199-10-12
151. Wang C, Chen Z, Ge Q, Hu J, Li F, Hu J, et al. Up-regulation of p21(WAF1/CIP1) by miRNAs and its implications in bladder cancer cells. FEBS Lett (2014) 588:4654–64. doi: 10.1016/j.febslet.2014.10.037
152. Aqeilan RI, Calin GA, Croce CM. miR-15a and miR-16-1 in cancer: discovery, function and future perspectives. Cell Death Differ (2010) 17:215–20. doi: 10.1038/cdd.2009.69
153. Taibi F, Metzinger-Le Meuth V, Massy ZA, Metzinger L. miR-223: An inflammatory oncomiR enters the cardiovascular field. Biochim Biophys Acta (2014) 1842:1001–9. doi: 10.1016/j.bbadis.2014.03.005
154. Gong J, Jaiswal R, Mathys JM, Combes V, Grau GE, Bebawy M. Microparticles and their emerging role in cancer multidrug resistance. Cancer Treat Rev (2012) 38:226–34. doi: 10.1016/j.ctrv.2011.06.005
155. Lasser C. Exosomal RNA as biomarkers and the therapeutic potential of exosome vectors. Expert Opin Biol Ther (2012) 12 Suppl 1:S189–97. doi: 10.1517/14712598.2012.680018
156. Dittmar RL SS. Chapter 4 - MicroRNAs in Exosomes in Cancer. Cancer and Noncoding RNAs. Elsevier (2018) p. 59–78. doi: 10.1016/B978-0-12-811022-5.00004-8
157. Cui M, Wang H, Yao X, Zhang D, Xie Y, Cui R, et al. Circulating MicroRNAs in Cancer: Potential and Challenge. Front Genet (2019) 10:626. doi: 10.3389/fgene.2019.00626
158. Wang H, Peng R, Wang J, Qin Z, Xue L. Circulating microRNAs as potential cancer biomarkers: the advantage and disadvantage. Clin Epigenet (2018) 10:59. doi: 10.1186/s13148-018-0492-1
159. Rabinowits G, Bowden M, Flores LM, Verselis S, Vergara V, Jo VY, et al. Comparative Analysis of MicroRNA Expression among Benign and Malignant Tongue Tissue and Plasma of Patients with Tongue Cancer. Front Oncol (2017) 7:191. doi: 10.3389/fonc.2017.00191
160. Kosaka N, Iguchi H, Hagiwara K, Yoshioka Y, Takeshita F, Ochiya T. Neutral sphingomyelinase 2 (nSMase2)-dependent exosomal transfer of angiogenic microRNAs regulate cancer cell metastasis. J Biol Chem (2013) 288:10849–59. doi: 10.1074/jbc.M112.446831
161. Di Marzo L, Desantis V, Solimando AG, Ruggieri S, Annese T, Nico B, et al. Microenvironment drug resistance in multiple myeloma: emerging new players. Oncotarget (2016) 7:60698–711. doi: 10.18632/oncotarget.10849
162. Frassanito MA, Desantis V, Di Marzo L, Craparotta I, Beltrame L, Marchini S, et al. Bone marrow fibroblasts overexpress miR-27b and miR-214 in step with multiple myeloma progression, dependent on tumour cell-derived exosomes. J Pathol (2019) 247:241–53. doi: 10.1002/path.5187
163. Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev Biol (2007) 302:1–12. doi: 10.1016/j.ydbio.2006.08.028
164. Reddy KB. MicroRNA (miRNA) in cancer. Cancer Cell Int (2015) 15:38. doi: 10.1186/s12935-015-0185-1
165. Eun JL, Baek M, Gusev Y, Brackett DJ, Nuovo GJ, Schmittgen TD. Systematic evaluation of microRNA processing patterns in tissues, cell lines, and tumors. RNA (2008) 14:35–42. doi: 10.1261/rna.804508
166. Thomson JM, Newman M, Parker JS, Morin-Kensicki EM, Wright T, Hammond SM. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev (2006) 20:2202–7. doi: 10.1101/gad.1444406
167. Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer (2006) 6:857–66. doi: 10.1038/nrc1997
168. Tiwari A, Mukherjee B, Dixit M. MicroRNA Key to Angiogenesis Regulation: MiRNA Biology and Therapy. Curr Cancer Drug Targets (2018) 18:266–77. doi: 10.2174/1568009617666170630142725
169. Folkman J. Tumor angiogenesis: therapeutic implications. N Engl J Med (1971) 285:1182–6. doi: 10.1056/NEJM197111182852108
170. Annese T, Tamma R, Ruggieri S, Ribatti D. Erythropoietin in tumor angiogenesis. Exp Cell Res (2019) 374:266–73. doi: 10.1016/j.yexcr.2018.12.013
171. Carmeliet P, Jain RK. Molecular mechanisms and clinical applications of angiogenesis. Nature (2011) 473:298–307. doi: 10.1038/nature10144
172. Ronca R, Benkheil M, Mitola S, Struyf S, Liekens S. Tumor angiogenesis revisited: Regulators and clinical implications. Med Res Rev (2017) 37:1231–74. doi: 10.1002/med.21452
173. Ribatti D. Inflammation and Angiogenesis. VIII ed. Springer Nature Switzerland AG: Springer International Publishing (2017). 111 p. doi: 10.1007/978-3-319-68448-2
174. Schaaf MB, Garg AD, Agostinis P. Defining the role of the tumor vasculature in antitumor immunity and immunotherapy. Cell Death Dis (2018) 9:115. doi: 10.1038/s41419-017-0061-0
175. Ribatti D, Tamma R. Epigenetic control of tumor angiogenesis. Microcirculation (2020) 27:e12602. doi: 10.1111/micc.12602
176. Wang Y, Wang L, Chen C, Chu X. New insights into the regulatory role of microRNA in tumor angiogenesis and clinical implications. Mol Cancer (2018) 17:22. doi: 10.1186/s12943-018-0766-4
177. Stahlhut C, Suarez Y, Lu J, Mishima Y, Giraldez AJ. miR-1 and miR-206 regulate angiogenesis by modulating VegfA expression in zebrafish. Development (2012) 139:4356–64. doi: 10.1242/dev.083774
178. Liu R, Zhang C, Hu Z, Li G, Wang C, Yang C, et al. A five-microRNA signature identified from genome-wide serum microRNA expression profiling serves as a fingerprint for gastric cancer diagnosis. Eur J Cancer (2011) 47:784–91. doi: 10.1016/j.ejca.2010.10.025
179. Miyachi M, Tsuchiya K, Yoshida H, Yagyu S, Kikuchi K, Misawa A, et al. Circulating muscle-specific microRNA, miR-206, as a potential diagnostic marker for rhabdomyosarcoma. Biochem Biophys Res Commun (2010) 400:89–93. doi: 10.1016/j.bbrc.2010.08.015
180. Kuehbacher A, Urbich C, Zeiher AM, Dimmeler S. Role of Dicer and Drosha for endothelial microRNA expression and angiogenesis. Circ Res (2007) 101:59–68. doi: 10.1161/CIRCRESAHA.107.153916
181. Suarez Y, Fernandez-Hernando C, Yu J, Gerber SA, Harrison KD, Pober JS, et al. Dicer-dependent endothelial microRNAs are necessary for postnatal angiogenesis. Proc Natl Acad Sci USA (2008) 105:14082–7. doi: 10.1073/pnas.0804597105
182. Yang WJ, Yang DD, Na S, Sandusky GE, Zhang Q, Zhao G. Dicer is required for embryonic angiogenesis during mouse development. J Biol Chem (2005) 280:9330–5. doi: 10.1074/jbc.M413394200
183. Leone P, Buonavoglia A, Fasano R, Solimando AG, De Re V, Cicco S, et al. Insights into the Regulation of Tumor Angiogenesis by Micro-RNAs. J Clin Med (2019) 8:2030. doi: 10.3390/jcm8122030
184. Wang Z, Xu L, Hu Y, Huang Y, Zhang Y, Zheng X, et al. miRNA let-7b modulates macrophage polarization and enhances tumor-associated macrophages to promote angiogenesis and mobility in prostate cancer. Sci Rep (2016) 6:25602. doi: 10.1038/srep25602
185. Jusufovic E, Rijavec M, Keser D, Korosec P, Sodja E, Iljazovic E, et al. let-7b and miR-126 are down-regulated in tumor tissue and correlate with microvessel density and survival outcomes in non–small–cell lung cancer. PLoS One (2012) 7:e45577. doi: 10.1371/journal.pone.0045577
186. Tao WY, Liang XS, Liu Y, Wang CY, Pang D. Decrease of let-7f in low-dose metronomic Paclitaxel chemotherapy contributed to upregulation of thrombospondin-1 in breast cancer. Int J Biol Sci (2015) 11:48–58. doi: 10.7150/ijbs.9969
187. Chen X, Yang F, Zhang T, Wang W, Xi W, Li Y, et al. MiR-9 promotes tumorigenesis and angiogenesis and is activated by MYC and OCT4 in human glioma. J Exp Clin Cancer Res (2019) 38:99. doi: 10.1186/s13046-019-1078-2
188. Wei YQ, Jiao XL, Zhang SY, Xu Y, Li S, Kong BH. MiR-9-5p could promote angiogenesis and radiosensitivity in cervical cancer by targeting SOCS5. Eur Rev Med Pharmacol Sci (2019) 23:7314–26. doi: 10.26355/eurrev_201909_18837
189. Liu X, Guan Y, Wang L, Niu Y. MicroRNA-10b expression in node-negative breast cancer-correlation with metastasis and angiogenesis. Oncol Lett (2017) 14:5845–52. doi: 10.3892/ol.2017.6914
190. Lin J, Teo S, Lam DH, Jeyaseelan K, Wang S. MicroRNA-10b pleiotropically regulates invasion, angiogenicity and apoptosis of tumor cells resembling mesenchymal subtype of glioblastoma multiforme. Cell Death Dis (2012) 3:e398. doi: 10.1038/cddis.2012.134
191. Sun CY, She XM, Qin Y, Chu ZB, Chen L, Ai LS, et al. miR-15a and miR-16 affect the angiogenesis of multiple myeloma by targeting VEGF. Carcinogenesis (2013) 34:426–35. doi: 10.1093/carcin/bgs333
192. Ma H, Pan JS, Jin LX, Wu J, Ren YD, Chen P, et al. MicroRNA-17~92 inhibits colorectal cancer progression by targeting angiogenesis. Cancer Lett (2016) 376:293–302. doi: 10.1016/j.canlet.2016.04.011
193. Liu R, Zhang H, Wang X, Zhou L, Li H, Deng T, et al. The miR-24-Bim pathway promotes tumor growth and angiogenesis in pancreatic carcinoma. Oncotarget (2015) 6:43831–42. doi: 10.18632/oncotarget.6257
194. Dai D, Huang W, Lu Q, Chen H, Liu J, Hong B. miR24 regulates angiogenesis in gliomas. Mol Med Rep (2018) 18:358–68. doi: 10.3892/mmr.2018.8978
195. Tavazoie SF, Alarcon C, Oskarsson T, Padua D, Wang Q, Bos PD, et al. Endogenous human microRNAs that suppress breast cancer metastasis. Nature (2008) 451:147–52. doi: 10.1038/nature06487
196. Chen H, Li L, Wang S, Lei Y, Ge Q, Lv N, et al. Reduced miR-126 expression facilitates angiogenesis of gastric cancer through its regulation on VEGF-A. Oncotarget (2014) 5:11873–85. doi: 10.18632/oncotarget.2662
197. Huang TH, Chu TY. Repression of miR-126 and upregulation of adrenomedullin in the stromal endothelium by cancer-stromal cross talks confers angiogenesis of cervical cancer. Oncogene (2014) 33:3636–47. doi: 10.1038/onc.2013.335
198. Du C, Lv Z, Cao L, Ding C, Gyabaah OA, Xie H, et al. MiR-126-3p suppresses tumor metastasis and angiogenesis of hepatocellular carcinoma by targeting LRP6 and PIK3R2. J Transl Med (2014) 12:259. doi: 10.1186/s12967-014-0259-1
199. Gong C, Fang J, Li G, Liu HH, Liu ZS. Effects of microRNA-126 on cell proliferation, apoptosis and tumor angiogenesis via the down-regulating ERK signaling pathway by targeting EGFL7 in hepatocellular carcinoma. Oncotarget (2017) 8:52527–42. doi: 10.18632/oncotarget.17283
200. Sasahira T, Kurihara M, Bhawal UK, Ueda N, Shimomoto T, Yamamoto K, et al. Downregulation of miR-126 induces angiogenesis and lymphangiogenesis by activation of VEGF-A in oral cancer. Br J Cancer (2012) 107:700–6. doi: 10.1038/bjc.2012.330
201. Atanasov G, Dietel C, Feldbrugge L, Benzing C, Krenzien F, Brandl A, et al. Angiogenic miRNAs, the angiopoietin axis and related TIE2-expressing monocytes affect outcomes in cholangiocarcinoma. Oncotarget (2018) 9:29921–33. doi: 10.18632/oncotarget.25699
202. Santos PRB, Coutinho-Camillo CM, Soares FA, Freitas VS, Vilas-Boas DS, Xavier FCA, et al. MicroRNAs expression pattern related to mast cell activation and angiogenesis in paraffin-embedded salivary gland tumors. Pathol Res Pract (2017) 213:1470–6. doi: 10.1016/j.prp.2017.10.012
203. Luo J, Meng C, Tang Y, Zhang S, Wan M, Bi Y, et al. miR-132/212 cluster inhibits the growth of lung cancer xenografts in nude mice. Int J Clin Exp Med (2014) 7:4115–22.
204. Yang J, Gao T, Tang J, Cai H, Lin L, Fu S. Loss of microRNA-132 predicts poor prognosis in patients with primary osteosarcoma. Mol Cell Biochem (2013) 381:9–15. doi: 10.1007/s11010-013-1677-8
205. Zou C, Xu Q, Mao F, Li D, Bian C, Liu LZ, et al. MiR-145 inhibits tumor angiogenesis and growth by N-RAS and VEGF. Cell Cycle (2012) 11:2137–45. doi: 10.4161/cc.20598
206. Yang JY, Li Y, Wang Q, Zhou WJ, Yan YN, Wei WB. MicroRNA-145 suppresses uveal melanoma angiogenesis and growth by targeting neuroblastoma RAS viral oncogene homolog and vascular endothelial growth factor. Chin Med J (Engl) (2020). 113:1922–9. doi: 10.1097/CM9.0000000000000875
207. Wu SY, Rupaimoole R, Shen F, Pradeep S, Pecot CV, Ivan C, et al. A miR-192-EGR1-HOXB9 regulatory network controls the angiogenic switch in cancer. Nat Commun (2016) 7:11169. doi: 10.1038/ncomms11169
208. Wang Y, Zhang X, Zou C, Kung HF, Lin MC, Dress A, et al. miR-195 inhibits tumor growth and angiogenesis through modulating IRS1 in breast cancer. BioMed Pharmacother (2016) 80:95–101. doi: 10.1016/j.biopha.2016.03.007
209. Liu H, Chen Y, Li Y, Li C, Qin T, Bai M, et al. miR195 suppresses metastasis and angiogenesis of squamous cell lung cancer by inhibiting the expression of VEGF. Mol Med Rep (2019) 20:2625–32. doi: 10.3892/mmr.2019.10496
210. Cai C, He H, Duan X, Wu W, Mai Z, Zhang T, et al. miR-195 inhibits cell proliferation and angiogenesis in human prostate cancer by downregulating PRR11 expression. Oncol Rep (2018) 39:1658–70. doi: 10.3892/or.2018.6240
211. Forouzan Jahromi Z, Javeri A, Fakhr Taha M. Tumor suppressive effects of the pleiotropically acting miR-195 in colorectal cancer cells. EXCLI J (2019) 18:243–52. doi: 10.17179/excli2019-1166
212. Dai J, Wei R, Zhang P, Kong B. Overexpression of microRNA-195-5p reduces cisplatin resistance and angiogenesis in ovarian cancer by inhibiting the PSAT1-dependent GSK3beta/beta-catenin signaling pathway. J Transl Med (2019) 17:190. doi: 10.1186/s12967-019-1932-1
213. Fan J, Xu G, Chang Z, Zhu L, Yao J. miR-210 transferred by lung cancer cell-derived exosomes may act as proangiogenic factor in cancer-associated fibroblasts by modulating JAK2/STAT3 pathway. Clin Sci (Lond) (2020) 134:807–25. doi: 10.1042/CS20200039
214. Lin XJ, Fang JH, Yang XJ, Zhang C, Yuan Y, Zheng L, et al. Hepatocellular Carcinoma Cell-Secreted Exosomal MicroRNA-210 Promotes Angiogenesis In Vitro and In Vivo. Mol Ther Nucleic Acids (2018) 11:243–52. doi: 10.1016/j.omtn.2018.02.014
215. Wang Z, Liu Z, Liu B, Liu G, Wu S. Dissecting the roles of Ephrin-A3 in malignant peripheral nerve sheath tumor by TALENs. Oncol Rep (2015) 34:391–8. doi: 10.3892/or.2015.3966
216. Sobrinho Santos EM, Guimaraes TA, Santos HO, Cangussu LMB, de Jesus SF, Fraga CAC, et al. Leptin acts on neoplastic behavior and expression levels of genes related to hypoxia, angiogenesis, and invasiveness in oral squamous cell carcinoma. Tumour Biol (2017) 39:1010428317699130. doi: 10.1177/1010428317699130
217. Giannakakis A, Sandaltzopoulos R, Greshock J, Liang S, Huang J, Hasegawa K, et al. miR-210 links hypoxia with cell cycle regulation and is deleted in human epithelial ovarian cancer. Cancer Biol Ther (2008) 7:255–64. doi: 10.4161/cbt.7.2.5297
218. Wu XG, Zhou CF, Zhang YM, Yan RM, Wei WF, Chen XJ, et al. Cancer-derived exosomal miR-221-3p promotes angiogenesis by targeting THBS2 in cervical squamous cell carcinoma. Angiogenesis (2019) 22:397–410. doi: 10.1007/s10456-019-09665-1
219. Santhekadur PK, Das SK, Gredler R, Chen D, Srivastava J, Robertson C, et al. Multifunction protein staphylococcal nuclease domain containing 1 (SND1) promotes tumor angiogenesis in human hepatocellular carcinoma through novel pathway that involves nuclear factor kappaB and miR-221. J Biol Chem (2012) 287:13952–8. doi: 10.1074/jbc.M111.321646
220. Wu YY, Chen YL, Jao YC, Hsieh IS, Chang KC, Hong TM. miR-320 regulates tumor angiogenesis driven by vascular endothelial cells in oral cancer by silencing neuropilin 1. Angiogenesis (2014) 17:247–60. doi: 10.1007/s10456-013-9394-1
221. Zhu H, Jiang X, Zhou X, Dong X, Xie K, Yang C, et al. Neuropilin-1 regulated by miR-320 contributes to the growth and metastasis of cholangiocarcinoma cells. Liver Int (2018) 38:125–35. doi: 10.1111/liv.13495
222. Fish JE, Santoro MM, Morton SU, Yu S, Yeh RF, Wythe JD, et al. miR-126 regulates angiogenic signaling and vascular integrity. Dev Cell (2008) 15:272–84. doi: 10.1016/j.devcel.2008.07.008
223. Chen LT, Xu SD, Xu H, Zhang JF, Ning JF, Wang SF. MicroRNA-378 is associated with non-small cell lung cancer brain metastasis by promoting cell migration, invasion and tumor angiogenesis. Med Oncol (2012) 29:1673–80. doi: 10.1007/s12032-011-0083-x
224. Chan JK, Kiet TK, Blansit K, Ramasubbaiah R, Hilton JF, Kapp DS, et al. MiR-378 as a biomarker for response to anti-angiogenic treatment in ovarian cancer. Gynecol Oncol (2014) 133:568–74. doi: 10.1016/j.ygyno.2014.03.564
225. Lee DY, Deng Z, Wang CH, Yang BB. MicroRNA-378 promotes cell survival, tumor growth, and angiogenesis by targeting SuFu and Fus-1 expression. Proc Natl Acad Sci USA (2007) 104:20350–5. doi: 10.1073/pnas.0706901104
226. Gerhardt H, Golding M, Fruttiger M, Ruhrberg C, Lundkvist A, Abramsson A, et al. VEGF guides angiogenic sprouting utilizing endothelial tip cell filopodia. J Cell Biol (2003) 161:1163–77. doi: 10.1083/jcb.200302047
227. Rosano S, Cora D, Parab S, Zaffuto S, Isella C, Porporato R, et al. A regulatory microRNA network controls endothelial cell phenotypic switch during sprouting angiogenesis. Elife (2020) 9:e48095. doi: 10.7554/eLife.48095
228. Tsang JS, Ebert MS, van Oudenaarden A. Genome-wide dissection of microRNA functions and cotargeting networks using gene set signatures. Mol Cell (2010) 38:140–53. doi: 10.1016/j.molcel.2010.03.007
229. Chamorro-Jorganes A, Araldi E, Penalva LO, Sandhu D, Fernandez-Hernando C, Suarez Y. MicroRNA-16 and microRNA-424 regulate cell-autonomous angiogenic functions in endothelial cells via targeting vascular endothelial growth factor receptor-2 and fibroblast growth factor receptor-1. Arterioscler Thromb Vasc Biol (2011) 31:2595–606. doi: 10.1161/ATVBAHA.111.236521
230. Yang Z, Wu L, Zhu X, Xu J, Jin R, Li G, et al. MiR-29a modulates the angiogenic properties of human endothelial cells. Biochem Biophys Res Commun (2013) 434:143–9. doi: 10.1016/j.bbrc.2013.03.054
231. Voutouri C, Kirkpatrick ND, Chung E, Mpekris F, Baish JW, Munn LL, et al. Experimental and computational analyses reveal dynamics of tumor vessel cooption and optimal treatment strategies. Proc Natl Acad Sci USA (2019) 116:2662–71. doi: 10.1073/pnas.1818322116
232. Frentzas S, Simoneau E, Bridgeman VL, Vermeulen PB, Foo S, Kostaras E, et al. Vessel co-option mediates resistance to anti-angiogenic therapy in liver metastases. Nat Med (2016) 22:1294–302. doi: 10.1038/nm.4197
233. Kuczynski EA, Yin M, Bar-Zion A, Lee CR, Butz H, Man S, et al. Co-option of Liver Vessels and Not Sprouting Angiogenesis Drives Acquired Sorafenib Resistance in Hepatocellular Carcinoma. J Natl Cancer Inst (2016) 108:djw030. doi: 10.1093/jnci/djw030
234. Liu W, Lv C, Zhang B, Zhou Q, Cao Z. MicroRNA-27b functions as a new inhibitor of ovarian cancer-mediated vasculogenic mimicry through suppression of VE-cadherin expression. RNA (2017) 23:1019–27. doi: 10.1261/rna.059592.116
235. Mao XG, Xue XY, Wang L, Zhang X, Yan M, Tu YY, et al. CDH5 is specifically activated in glioblastoma stemlike cells and contributes to vasculogenic mimicry induced by hypoxia. Neuro Oncol (2013) 15:865–79. doi: 10.1093/neuonc/not029
236. Seftor RE, Hess AR, Seftor EA, Kirschmann DA, Hardy KM, Margaryan NV, et al. Tumor cell vasculogenic mimicry: from controversy to therapeutic promise. Am J Pathol (2012) 181:1115–25. doi: 10.1016/j.ajpath.2012.07.013
237. Zhao N, Sun H, Sun B, Zhu D, Zhao X, Wang Y, et al. miR-27a-3p suppresses tumor metastasis and VM by down-regulating VE-cadherin expression and inhibiting EMT: an essential role for Twist-1 in HCC. Sci Rep (2016) 6:23091. doi: 10.1038/srep23091
238. Xu S, Zhang J, Xue H, Guo X, Han X, Li T, et al. MicroRNA-584-3p reduces the vasculogenic mimicry of human glioma cells by regulating hypoxia-induced ROCK1 dependent stress fiber formation. Neoplasma (2017) 64:13–21. doi: 10.4149/neo_2017_102
239. Dong B, Mu L, Qin X, Qiao W, Liu X, Yang L, et al. Stathmin expression in glioma-derived microvascular endothelial cells: a novel therapeutic target. Oncol Rep (2012) 27:714–8. doi: 10.3892/or.2011.1525
240. Song Y, Mu L, Han X, Li Q, Dong B, Li H, et al. MicroRNA-9 inhibits vasculogenic mimicry of glioma cell lines by suppressing Stathmin expression. J Neurooncol (2013) 115:381–90. doi: 10.1007/s11060-013-1245-9
241. Xue H, Gao X, Xu S, Zhang J, Guo X, Yan S, et al. MicroRNA-Let-7f reduces the vasculogenic mimicry of human glioma cells by regulating periostin-dependent migration. Oncol Rep (2016) 35:1771–7. doi: 10.3892/or.2016.4548
242. Pan Z, Tian Y, Niu G, Cao C. Role of microRNAs in remodeling the tumor microenvironment (Review). Int J Oncol (2020) 56:407–16. doi: 10.3892/ijo.2019.4952
243. Du YE, Tu G, Yang G, Li G, Yang D, Lang L, et al. MiR-205/YAP1 in Activated Fibroblasts of Breast Tumor Promotes VEGF-independent Angiogenesis through STAT3 Signaling. Theranostics (2017) 7:3972–88. doi: 10.7150/thno.18990
244. Ribatti D, Annese T, Ruggieri S, Tamma R, Crivellato E. Limitations of Anti-Angiogenic Treatment of Tumors. Transl Oncol (2019) 12:981–6. doi: 10.1016/j.tranon.2019.04.022
245. Matejuk A, Collet G, Nadim M, Grillon C, Kieda C. MicroRNAs and tumor vasculature normalization: impact on anti-tumor immune response. Arch Immunol Ther Exp (Warsz) (2013) 61:285–99. doi: 10.1007/s00005-013-0231-4
246. Freedman AS JC, Mauch P, Aster JC. Non-Hodgkin’s lymphoma. In: DeVita VT, Lawrence TS, Rosenberg SA, Wilkins LW, editors. Principles and Practice of Oncology, 10th ed. Lippincott Williams & Wilkins (2015).
247. Shanbhag S, Ambinder RF. Hodgkin lymphoma: A review and update on recent progress. CA Cancer J Clin (2018) 68:116–32. doi: 10.3322/caac.21438
248. Swerdlow SH, Campo E, Pileri SA, Harris NL, Stein H, Siebert R, et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood (2016) 127:2375–90. doi: 10.1182/blood-2016-01-643569
249. Georgantas RW,3, Hildreth R, Morisot S, Alder J, CG L, Heimfeld S, et al. CD34+ hematopoietic stem-progenitor cell microRNA expression and function: a circuit diagram of differentiation control. Proc Natl Acad Sci USA (2007) 104:2750–5. doi: 10.1073/pnas.0610983104
250. Liu CG, Calin GA, Meloon B, Gamliel N, Sevignani C, Ferracin M, et al. An oligonucleotide microchip for genome-wide microRNA profiling in human and mouse tissues. Proc Natl Acad Sci USA (2004) 101:9740–4. doi: 10.1073/pnas.0403293101
251. Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature (2005) 435:834–8. doi: 10.1038/nature03702
252. Nelson PT, Baldwin DA, Scearce LM, Oberholtzer JC, Tobias JW, Mourelatos Z. Microarray-based, high-throughput gene expression profiling of microRNAs. Nat Methods (2004) 1:155–61. doi: 10.1038/nmeth717
253. Schmittgen T, Jiang J, Liu Q, Yang L. A High-Throughput Method to Monitor the Expression of microRNA Precursors. Nucleic Acids Res (2004) 32:e43–e. doi: 10.1093/nar/gnh040
254. Metzler M, Wilda M, Busch K, Viehmann S, Borkhardt A. High expression of precursor microRNA-155/BIC RNA in children with Burkitt lymphoma. Genes Chromosomes Cancer (2004) 39:167–9. doi: 10.1002/gcc.10316
255. Guo S, Lu J, Schlanger R, Zhang H, Wang JY, Fox MC, et al. MicroRNA miR-125a controls hematopoietic stem cell number. Proc Natl Acad Sci USA (2010) 107:14229–34. doi: 10.1073/pnas.0913574107
256. Koralov SB, Muljo SA, Galler GR, Krek A, Chakraborty T, Kanellopoulou C, et al. Dicer ablation affects antibody diversity and cell survival in the B lymphocyte lineage. Cell (2008) 132:860–74. doi: 10.1016/j.cell.2008.02.020
257. Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science (2004) 303:83–6. doi: 10.1126/science.1091903
258. Li QJ, Chau J, Ebert PJ, Sylvester G, Min H, Liu G, et al. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell (2007) 129:147–61. doi: 10.1016/j.cell.2007.03.008
259. Neilson JR, Zheng GX, Burge CB, Sharp PA. Dynamic regulation of miRNA expression in ordered stages of cellular development. Genes Dev (2007) 21:578–89. doi: 10.1101/gad.1522907
260. Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, et al. Requirement of bic/microRNA-155 for normal immune function. Science (2007) 316:608–11. doi: 10.1126/science.1139253
261. Thai TH, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y, et al. Regulation of the germinal center response by microRNA-155. Science (2007) 316:604–8. doi: 10.1126/science.1141229
262. Seto AG, Beatty X, Lynch JM, Hermreck M, Tetzlaff M, Duvic M, et al. Cobomarsen, an oligonucleotide inhibitor of miR-155, co-ordinately regulates multiple survival pathways to reduce cellular proliferation and survival in cutaneous T-cell lymphoma. Br J Haematol (2018) 183:428–44. doi: 10.1111/bjh.15547
263. Moehlera T.M. HAD, Goldschmidta H, Barlogie B. Angiogenesis in hematologic malignancies. Crit Rev Oncology/Hematology (2003) 45:227–44. doi: 10.1016/S1040-8428(02)00135-X
264. Getaneh Z, Asrie F, Melku M. MicroRNA profiles in B-cell non-Hodgkin lymphoma. EJIFCC (2019) 30:195–214.
265. Paydas S, Acikalin A, Ergin M, Celik H, Yavuz B, Tanriverdi K. Micro-RNA (miRNA) profile in Hodgkin lymphoma: association between clinical and pathological variables. Med Oncol (2016) 33:34. doi: 10.1007/s12032-016-0749-5
266. Sandhu SK, Croce CM, Garzon R. Micro-RNA Expression and Function in Lymphomas. Adv Hematol (2011) 2011:347137. doi: 10.1155/2011/347137
267. Matsuyama H, Suzuki HI, Nishimori H, Noguchi M, Yao T, Komatsu N, et al. miR-135b mediates NPM-ALK-driven oncogenicity and renders IL-17-producing immunophenotype to anaplastic large cell lymphoma. Blood (2011) 118:6881–92. doi: 10.1182/blood-2011-05-354654
268. Ji Y, Zhang W. Th17 cells: positive or negative role in tumor? Cancer Immunol Immunother (2010) 59:979–87. doi: 10.1007/s00262-010-0849-6
269. Dejean E, Renalier MH, Foisseau M, Agirre X, Joseph N, de Paiva GR, et al. Hypoxia-microRNA-16 downregulation induces VEGF expression in anaplastic lymphoma kinase (ALK)-positive anaplastic large-cell lymphomas. Leukemia (2011) 25:1882–90. doi: 10.1038/leu.2011.168
270. He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, et al. A microRNA polycistron as a potential human oncogene. Nature (2005) 435:828–33. doi: 10.1038/nature03552
271. Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell (2008) 132:875–86. doi: 10.1016/j.cell.2008.02.019
272. Dews M, Homayouni A, Yu D, Murphy D, Sevignani C, Wentzel E, et al. Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nat Genet (2006) 38:1060–5. doi: 10.1038/ng1855
273. Doebele C, Bonauer A, Fischer A, Scholz A, Reiss Y, Urbich C, et al. Members of the microRNA-17-92 cluster exhibit a cell-intrinsic antiangiogenic function in endothelial cells. Blood (2010) 115:4944–50. doi: 10.1182/blood-2010-01-264812
274. Rahman MM, Brane AC, Tollefsbol TO. MicroRNAs and Epigenetics Strategies to Reverse Breast Cancer. Cells (2019) 8:1214. doi: 10.3390/cells8101214
275. Tomasetti M, Gaetani S, Monaco F, Neuzil J, Santarelli L. Epigenetic Regulation of miRNA Expression in Malignant Mesothelioma: miRNAs as Biomarkers of Early Diagnosis and Therapy. Front Oncol (2019) 9:1293. doi: 10.3389/fonc.2019.01293
276. Li MH, Fu SB, Xiao HS. Genome-wide analysis of microRNA and mRNA expression signatures in cancer. Acta Pharmacol Sin (2015) 36:1200–11. doi: 10.1038/aps.2015.67
277. Wang T, Wang G, Zhang X, Wu D, Yang L, Wang G, et al. The expression of miRNAs is associated with tumour genome instability and predicts the outcome of ovarian cancer patients treated with platinum agents. Sci Rep (2017) 7:14736. doi: 10.1038/s41598-017-12259-w
278. van Beijnum JR, Nowak-Sliwinska P, van Berkel M, Wong TJ, Griffioen AW. A genomic screen for angiosuppressor genes in the tumor endothelium identifies a multifaceted angiostatic role for bromodomain containing 7 (BRD7). Angiogenesis (2017) 20:641–54. doi: 10.1007/s10456-017-9576-3
279. Plotnikova O, Baranova A, Skoblov M. Comprehensive Analysis of Human microRNA-mRNA Interactome. Front Genet (2019) 10:933. doi: 10.3389/fgene.2019.00933
280. van Beijnum JR, Griffioen AW. In silico analysis of angiogenesis associated gene expression identifies angiogenic stage related profiles. Biochim Biophys Acta (2005) 1755:121–34. doi: 10.1016/j.bbcan.2005.06.001
281. Poliseno L, Tuccoli A, Mariani L, Evangelista M, Citti L, Woods K, et al. MicroRNAs modulate the angiogenic properties of HUVECs. Blood (2006) 108:3068–71. doi: 10.1182/blood-2006-01-012369
282. Babae N, Bourajjaj M, Liu Y, Van Beijnum JR, Cerisoli F, Scaria PV, et al. Systemic miRNA-7 delivery inhibits tumor angiogenesis and growth in murine xenograft glioblastoma. Oncotarget (2014) 5:6687–700. doi: 10.18632/oncotarget.2235
283. Ramjiawan RR, Griffioen AW, Duda DG. Anti-angiogenesis for cancer revisited: Is there a role for combinations with immunotherapy? Angiogenesis (2017) 20:185–204. doi: 10.1007/s10456-017-9552-y
284. Wang WX, Wilfred BR, Baldwin DA, Isett RB, Ren N, Stromberg A, et al. Focus on RNA isolation: obtaining RNA for microRNA (miRNA) expression profiling analyses of neural tissue. Biochim Biophys Acta (2008) 1779:749–57. doi: 10.1016/j.bbagrm.2008.01.005
285. Mase M, Grasso M, Avogaro L, D’Amato E, Tessarolo F, Graffigna A, et al. Selection of reference genes is critical for miRNA expression analysis in human cardiac tissue. A focus on atrial fibrillation. Sci Rep (2017) 7:41127. doi: 10.1038/srep41127
286. Witwer KW. Circulating microRNA biomarker studies: pitfalls and potential solutions. Clin Chem (2015) 61:56–63. doi: 10.1373/clinchem.2014.221341
287. Pogribny IP. MicroRNAs as biomarkers for clinical studies. Exp Biol Med (Maywood) (2018) 243:283–90. doi: 10.1177/1535370217731291
288. Chen Y, Gelfond JA, McManus LM, Shireman PK. Reproducibility of quantitative RT-PCR array in miRNA expression profiling and comparison with microarray analysis. BMC Genomics (2009) 10:407. doi: 10.1186/1471-2164-10-407
289. Liu CG, Calin GA, Volinia S, Croce CM. MicroRNA expression profiling using microarrays. Nat Protoc (2008) 3:563–78. doi: 10.1038/nprot.2008.14
290. Moldovan L, Batte KE, Trgovcich J, Wisler J, Marsh CB, Piper M. Methodological challenges in utilizing miRNAs as circulating biomarkers. J Cell Mol Med (2014) 18:371–90. doi: 10.1111/jcmm.12236
291. Kuo TY, Hsi E, Yang IP, Tsai PC, Wang JY, Juo SH. Computational analysis of mRNA expression profiles identifies microRNA-29a/c as predictor of colorectal cancer early recurrence. PLoS One (2012) 7:e31587. doi: 10.1371/journal.pone.0031587
292. Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature (2008) 455:58–63. doi: 10.1038/nature07228
293. Uhlmann S, Mannsperger H, Zhang JD, Horvat EA, Schmidt C, Kublbeck M, et al. Global microRNA level regulation of EGFR-driven cell-cycle protein network in breast cancer. Mol Syst Biol (2012) 8:570. doi: 10.1038/msb.2011.100
294. Garzon R, Marcucci G, Croce CM. Targeting microRNAs in cancer: rationale, strategies and challenges. Nat Rev Drug Discovery (2010) 9:775–89. doi: 10.1038/nrd3179
295. Ribatti D, Vacca A, Dammacco F, English D. Angiogenesis and anti-angiogenesis in hematological malignancies. J Hematother Stem Cell Res (2003) 12:11–22. doi: 10.1089/152581603321210091
296. Annese T, Tamma R, De Giorgis M, Ruggieri S, Maiorano E, Specchia G, et al. RNAscope dual ISH-IHC technology to study angiogenesis in diffuse large B-cell lymphomas. Histochem Cell Biol (2020) 153:185–92. doi: 10.1007/s00418-019-01834-z
297. Parilla M, Kadri S, Patil SA, Fitzpatrick C, Ritterhouse L, Segal J, et al. Integrating a Large Next-Generation Sequencing Panel into the Clinical Diagnosis of Gliomas Provides a Comprehensive Platform for Classification from FFPE Tissue or Smear Preparations. J Neuropathol Exp Neurol (2019) 78:257–67. doi: 10.1093/jnen/nly130
Keywords: angiogenesis, lymphoma, microenvironment, microRNA, tumor progression
Citation: Annese T, Tamma R, De Giorgis M and Ribatti D (2020) microRNAs Biogenesis, Functions and Role in Tumor Angiogenesis. Front. Oncol. 10:581007. doi: 10.3389/fonc.2020.581007
Received: 07 July 2020; Accepted: 27 October 2020;
Published: 27 November 2020.
Edited by:
Zohreh Amoozgar, Massachusetts General Hospital and Harvard Medical School, United StatesReviewed by:
Andrea Caporali, University of Edinburgh, United KingdomIgor Lucas Gomes dos Santos, Massachusetts General Hospital and Harvard Medical School, United States
Copyright © 2020 Annese, Tamma, De Giorgis and Ribatti. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tiziana Annese, tiziana.annese@uniba.it; Domenico Ribatti, domenico.ribatti@uniba.it
 Tiziana Annese
Tiziana Annese Roberto Tamma
Roberto Tamma Domenico Ribatti
Domenico Ribatti