Standard operating procedures for biobank in oncology
- 1Biobank for Translational and Digital Medicine, IEO, European Institute of Oncology IRCCS, Milan, Italy
- 2Department of Oncology and Hemato-Oncology, University of Milan, Milan, Italy
- 3Division of Pathology, IEO, European Institute of Oncology IRCCS, Milan, Italy
- 4Technology Transfer Office, IEO, European Institute of Oncology IRCCS, Milan, Italy
- 5Patient Safety and Risk Management Service, IEO, European Institute of Oncology IRCCS, Milan, Italy
- 6Scientific Directorate, IEO, European Institute of Oncology IRCCS, Milan, Italy
Biobanks are biorepositories that collect, process, store, catalog, and distribute human biological samples, and record the associated data. The role and action field of these strategic infrastructures for implementing precision medicine in translational research is continuously evolving. To ensure the optimal quality at all stages of biobanking, specific protocols are required and should be elaborated according to updated guidelines, recommendations, laws, and rules. This article illustrates the standard operating procedures, including protocols, troubleshooting, and quality controls, of a fully certified biobank in a referral Cancer Center. This model involves all clinical departments and research groups to support the dual mission of academic cancer centers, i.e. to provide high-quality care and high-quality research. All biobanking activities based on the type of biological specimens are detailed and the most tricky methodological aspects are discussed, from patients’ informed consent to specimen management.
Introduction
Modern oncologic research requires that high-quality biological samples and the associated data are collected, tracked, processed, stored, cataloged, and distributed to research groups and collaborating partners (Kinkorová, 2015). This integrated biobanking approach has led to breakthroughs in both biomarker discovery and drug development (Pagni et al., 2019). Biobanks thus represent essential resources for basic, translational, and clinical research but they also act as key players linking academic research and the pharmaceutical biotechnology industry (Vaught et al., 2011; Hewitt and Watson, 2013; Jose et al., 2018; Coppola et al., 2019). Moreover, the ability to integrate not only clinical information but also biospecimens into big data repositories has intensified the centrality of biobanks (Margolis et al., 2014; Kinkorová, 2015; Drosou et al., 2017; Kinkorová and Topolčan, 2020). This is especially important as biorepositories have begun to incorporate patient information with comprehensive clinicopathologic, epidemiologic, and demographic data, together with multi-omics molecular information (Braun et al., 2014; Luo et al., 2014; Leff and Yang, 2015; Saifuddin et al., 2017; Bycroft et al., 2018; Hulsen et al., 2019; Bonnechère et al., 2021). The collection of this increasing amount of data requires strict quality controls and standard operating procedures (SOPs). The genomic and post-genomic field area has generated a high demand for high-quality biospecimen and data. Biorepositories in cancer research support scientists and clinicians to obtain disease-specific insights. For these reasons, biobanks should be established and updated following international guidelines, such as those from the International Agency for Research on Cancer (IARC), U.S. National Cancer Institute, United Kingdom. Confederation of Cancer Biobanks, and International Society for Biological and Environmental Repositories (ISBER), recommendations, laws, and rules (Vaught et al., 2009; Sanchini et al., 2016; Mendy et al., 2017). In this respect, networking is essential for sharing materials and data among institutions and research groups, particularly for the study of rare diseases (Montserrat and Taruscio, 2019).
Here, we present the organization of a fully certified (UNI EN ISO 9001:2015 - Certiquality) biobank in a referral Cancer Center, which is an integral part of the Italian node of the European Research Infrastructure on Biobanking (BBMRI-ERIC) (Salvaterra and Corfield, 2017). This facility works in compliance with the new standard ISO 20387:2018 “Biotechnology - Biobanking - General requirements for Biobanks”. All SOPs herein reported fulfilled the BBMRI-ERIC quality control and audits (https://www.bbmri-eric.eu/services/quality-management/). Protocols and best practices for the collection of surgical tissue samples, as well as biofluids (e.g., plasma, serum, blood, urine), cell cultures, and peripheral blood mononuclear cells (PBMC), are described in detail (Kanof et al., 2001; Elliott and Peakman, 2008; Guerin et al., 2010; Mallone et al., 2011; Fisher et al., 2018; Hojat et al., 2019; Rolfo et al., 2021). This model enables collaboration among research groups and industry, allowing patients to be an integral part of translational research.
Scientific/ethical approval and patients’ recruitment
All procedures involving biobanks must be approved by the local Scientific and Technical Committee, the Ethics Committee, and the directors of the involved clinical Units and surgical programs, according to the 1964 Declaration of Helsinki, the 2018 General Data Protection Regulation (GDPR), and subsequent amendments (Sanchini et al., 2016). In this prototype, the GDPR is represented by the Scientific Research Participation Agreement (SRPA), which is the standard informed consent that patients sign to donate biological samples, sensitive data, or genetic data at the European Institute of Oncology in Milan, Italy. The SRPA should be obtained from all patients for the storage, processing, and use of the data obtained for scientific purposes. Only the signed SRPA allows for biospecimen collection. Through this agreement, each patient can express his or her will and modalities of scientific research participation. Given the complexity of the concepts described in the SRPA, it is advisable to share educational material with the patients. For example, as a reminder of their first visit, patients can receive a text message whereby the SRPA information is provided. In addition, short engaging videos broadcasted in the waiting rooms can be employed to inform patients about the importance and implications of the SRPA. An example of a cartoon on biobank used at the European Institute of Oncology, Milan, Italy is freely available online (https://vimeo.com/679070846). During each phase of the hospitalization, SRPA can be administered to the patient by qualified professionals, including biobank staff, nurses, physicians, and biomedical personnel. If patients agree to participate in any study, they should receive all the specific study information approved by the Ethics Committee. Informed consent in the form of SRPA is obtained from all patients for their material to be stored in the biobank and used for further studies. Hence, SOPs, guidelines, and recommendations do not permit the collection and storage of biospecimens in the absence of patients’ consent. Therefore, SRPA should be continuously updated to inform patients properly and comprehensively. To solve any patient’s withdrawal from the previous SRPA, a patient sample take-out methodology should be implemented (Schmanski et al., 2021). To obtain the collected samples, researchers, or external collaborators (for-profit or non-profit) should apply to the Biobank Scientific-Technical Committee and/or the Institutional Ethics Committee. A specific form has to be filled by the applicant for evaluation and linked to an approved evaluation of the project.
Management of biological samples
After previous verification of the patients’ SRPA, each biological sample can be collected and treated. Samples may include: fresh and frozen tissue samples related to the patients who underwent surgery; fresh and frozen tissue biopsies; cytological samples (e.g., needle aspirations, excreted, ascites, pleural fluids) from needle aspiration or brushing/scraping, and from affixing to surgically removed tissues for small lesions; biofluids (blood, serum, plasma, PBMC, oral swab, urine, feces, ascites) of patients in pre-hospitalization, patients enrolled in clinical trials, and any other subjects involved in screening projects. Each phase of samples collection should be compliant with the latest ISO standards (e.g. ISO 20387:2018).
Check-in, aliquoting, and distribution
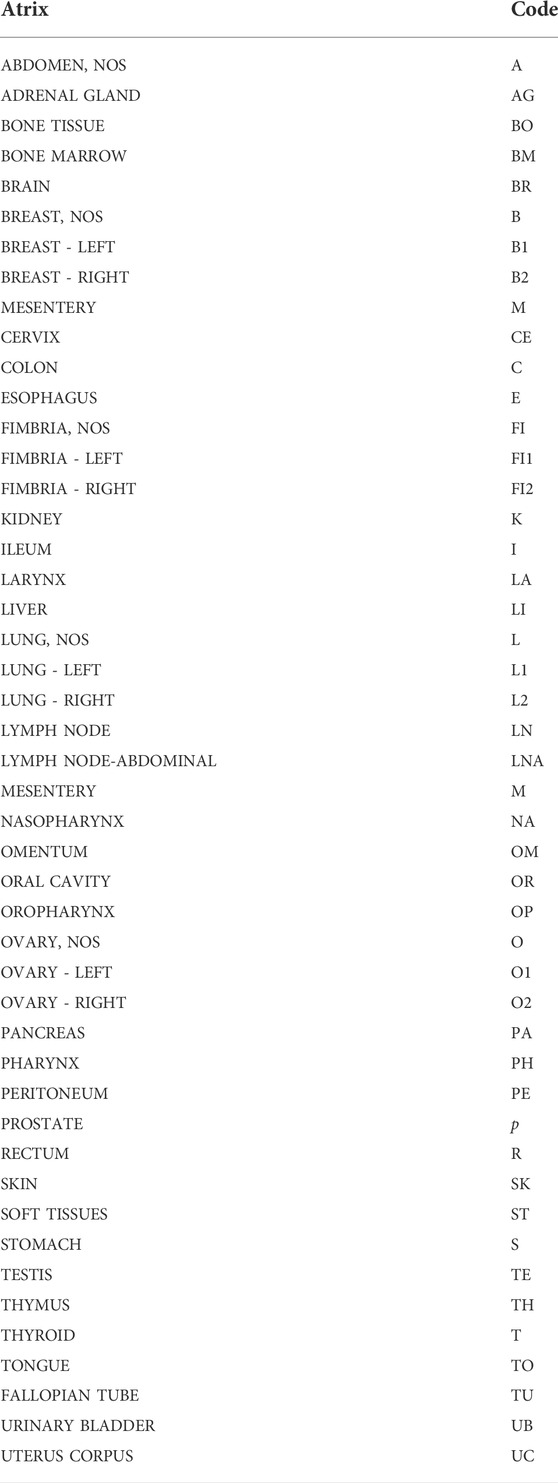
When surgical intervention is scheduled, the Biobank data manager should check in the Surgery Plan if the patient is eligible for samples collection by the signed SRPA presence. If eligible, a surgery plan for biobank must be prepared to check the correspondence of the patient’s inclusion/exclusion criteria, and the patient’s consent for clinical trial or research project. To store and track the significant amount of data generated from the processing and analysis of patients’ biological samples, the use a Laboratory Information Management System (LIMS) software named is highly recommended. This software is considered the biobank neural network because it should ideally interact bidirectionally with all the other softwares and applications used in the Institution, as portrayed in Figure 1. All processing and analysis are tracked by the LIMS, allowing the biobank operators to minimize possible errors, as each type of aliquot is electronically recorded using barcodes (Figure 2). Further, all samples should be divided into subcategories according to the processing, such as the fresh sample, and the frozen sample. The aliquots’ ID is a sequential series of numbers and letters generated by LIMS, according to SOPs, as exemplified for biofluids in Table 1 and tissue samples in Table 2. An example of an ID code is: “12-B1-00100-01” where “12” indicates the year (2012), “B1” indicates the anatomical origin (left breast), and “00100-01” identifies the sample. Once an aliquot is requested, a database query allows the retrieval of the requested samples. This process needs a tracked form with information, a short description of the approved research project, and information on the principal investigator (PI). Consequently, the LIMS generates the requested ID for each aliquot sorted into a picklist. Later, the biobank technicians can check the ID list to retrieve the requested aliquots. Last, barcoded tubes (e.g. Nunc™ Coded Cryobank Vial Systems, Thermo Scientific, Waltham, Massachusetts, US) and relative data can be delivered.
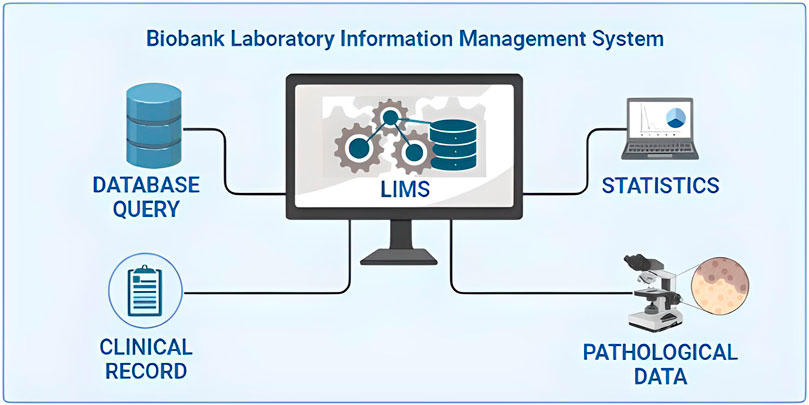
FIGURE 1. Integration of the laboratory information management system (LIMS) of biobanks in the critical junction of data from different sources.
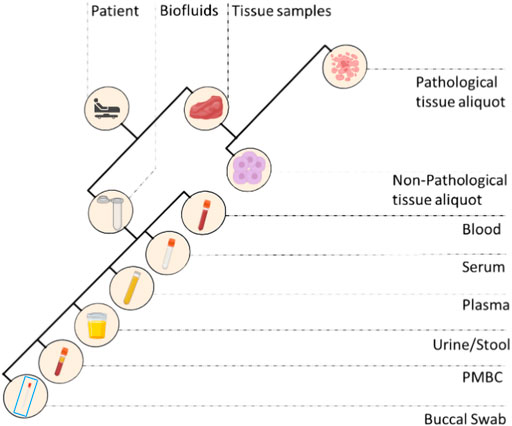
FIGURE 2. Different types of biospecimens collected in a standard biobank. PMBC, peripheral blood mononuclear cells.
Sample types
Tissue
During the gross examination of the surgical sample, the pathologist determines whether there is sufficient material (i.e. exceeding diagnosis) for research purposes. When available/possible, the non-pathological counterpart is also collected. Normal and tumor samples are labelled and placed in sterile Petri dishes on ice and divided into fresh and/or optimal cutting temperature (OCT) compound aliquots. Samples are then frozen for 3 min at −120°C in isopentane before transfer to cryopreservation rooms and stored at −80°C. It is important to perform a quality check for each frozen aliquot before distribution and use it to obtain a histological assessment of the cellularity on hematoxylin and eosin (H&E) cryosections (Table 3).
Whole blood
For whole blood samples collection, 6 ml labeled vacutainer tubes, containing anti-coagulant Na2 ethylenediaminetetraacetic acid (EDTA), are used by the nurses at the assessment centers. Following the collection, the biobank technicians process the sample using its medical record number and the episode code and register each aliquot in the biobank software. For clinical studies, or project-specific requirements, the blood is prepared for shipment, otherwise it is firstly collected using a blood amount of 900 μL in 1 ml barcoded cryotubes for one or more aliquots, and stored at −80°C.
Blood serum
For blood serum samples collection, 6 ml labeled vacutainer tubes containing a thixotropic barrier gel at the bottom of the tube, are initially used. Tubes are left for clotting for 3 h at room temperature (RT), and then centrifugated on a refrigerated centrifuge at 828 x g for 10 min. Depending on the initial amount of whole blood taken, the biobank technicians’ rate 450 μL of serum in the 0.5 ml barcoded cryotubes for the maximum amount of aliquots, and stored at −80°C. When the last aliquot serum volume is < 450 μL, the sample is registered as “leftover”.
Blood plasma and cf-DNA/RNA
To separate the plasma from the whole blood, blood-filled vacutainers need to be centrifuged at 2,000 x g for 10 min at RT. After centrifugation, the upper plasma layer should be removed and transferred to a sterile 15 ml conical tube for a second centrifugation at 16,000 x g for 10 min at RT to remove contaminating blood cells. Then, the obtained plasma can be transferred to a barcoded cryotube for the maximum amount of aliquots, depending on the volume. The remaining blood is collected for subsequent cf-DNA and/or cf-RNA purification. Aliquots should be stored at −80°C.
Peripheral blood mononuclear cells
For peripheral blood mononuclear cells (PBMC) isolation, the blood is drawn in 7.5 ml labeled vacutainer tubes, containing Na2 EDTA as an anticoagulant reagent. The blood is subsequently transferred into an empty 50 ml conical tube and diluted in a 1:1 ratio using phosphate buffered saline (PBS) 1X (e.g., 15 ml of blood +15 ml of PBS 1X). Again, the diluted blood is layered on the top of a clean 50 ml conical tube containing 15 ml of Ficoll, without mixing the two solutions. After centrifugation at 400 x g for 30 min at RT, the white layer containing PBMC is collected and placed in a new sterile 50 ml conical tube. PBMCs are washed by adding 45 ml of PBS, mixed, and centrifuged at 400 x g for 10 min at 4°C. After discarding the supernatant, the pellet containing PBMCs is resuspended in PBS and counted using Tuerks solution and single-use slide for counting cells (e.g. Biosigma S.P.A. Cat. no. 347143/001). Cells are washed once more with PBS and resuspended at 2-3x10ˆ6 cells/mL of FSB +10% dimethyl sulfoxide (DMSO) to be frozen. Finally, 1 ml of resuspended cells are transferred in cryotubes for storage at −80°C.
Stool and buccal swab
Feces and buccal swabs are collected in 15 ml tubes (e.g. Stool Sample Collection and Stabilization Kit Canvax Cat. no 0013), containing 50 mM Tris-HCl, 10 mM NaCl, and 10 mM EDTA pH 7.5 and stored at −80°C.
Disaster recovery plan
Biobanks are dedicated to managing valuable and possibly irreplaceable biological specimens. Therefore, biomaterials and associated data must be managed and protected carefully as their loss can destroy years of research efforts and costs, and potentially result in damage to the Institution (Eng and Tan, 2019). For this reason, risk management and practical crisis management plans must be established for any biobank (Parry-Jones et al., 2017). It is essential to define a data protection program that must satisfy various needs that may range from remote data only (backup) to disaster recovery (as a set of technological measures and organizational processes aimed at restoring systems, data, and infrastructures necessary to provide biobank services during emergencies) and to ensure continuity of service and recovery of materials and data during emergencies (Cicek and Olson, 2020).
Staff training programs
The quality and quantity of samples and data stored in a biobank directly depend on the biobank personnel, including data managers and technicians (Hartung et al., 2021). Modern biobanking must rely on high-level training programs for biobank employees not only to allow harmonization of correct sample handling but also to ensure safety and quality (Kinkorová and Topolčan, 2020). Not surprisingly, training certificates of biobank employees are needed for the accreditation process (Williams et al., 2019). Types of training programs include master’s programs, certificate courses, and workshops. Due to the paucity of available formal training programs, biobanks often train most of their new staff on site (Castellanos-Uribe et al., 2020). Learning about teamwork, personnel safety, patient privacy, biospecimen quality, and SOPs is crucial not only for efficiency and productivity but also for the personnel’s career success. A well-designed training program should include helpful tips, tricks, and troubleshooting. International collaboration and exchange programs might facilitate the process of creating next-generation biobanking staff.
Representative results
A total of 38,446 annotated biofluids and a total of 10,205 tissue samples were collected by the Biobank for Translational and Digital Medicine Unit at the IEO, European Institute of Oncology, Milan, Italy from April 2012 to December 2021 (Figure 3A). The highest number of samples were related to breast cancer, urological malignancies, tumors of the female genital tract, head and neck carcinomas, and lung cancer (Figures 3B,C). The cumulative analysis of plasma, buccal swab, urine, and stool samples revealed a significant increase in the number of collected samples, particularly for the urine in patients with urological malignancies, reaching a total number of 726 urine samples (Figure 3D). These samples have been divided into multiple aliquots related to specific projects or clinical trials and to the institutional universal collection of samples. Taken together, 28 different projects were responsible for the vast majority of aliquot distribution, for a total number of 28,852 aliquots, as detailed in Table 4. The total number of tissue samples whose aliquots were employed for research purposes were 8,383/10,205 (82%). These data confirm the fundamental role of certified biobanks not only for samples collection but also for samples distribution and use by research groups.

FIGURE 3. Types and number of samples collected by the biobank of the European Institute of Oncology by year. (A) Cumulative collection of tissue and blood/serum samples; (B) Cumulative collection of breast tissue samples; (C) Cumulative collection of tumor samples from the ovary, prostate, lung, and colon; (D) Cumulative collection of non-tissue samples, i.e. feces, saliva/swab, plasma, and urine.
Discussion
The transversal role of biobanks in scientific research, particularly in oncologic pathology, and basic and clinical sciences, has put these important infrastructures on the front line of personalized medicine evolution (Kinkorová, 2015; Coppola et al., 2019). Indeed, cancer is still a leading cause of morbidity and mortality worldwide (Sung et al., 2021). For these reasons, the understanding of cancer pathogenesis, mechanisms of disease, and biomarkers discovery at the multi-omics level is becoming an urgent clinical need, akin the support in drug discovery (Kinkorová, 2015; Yan et al., 2018; Szustakowski et al., 2021). When a biobank is established several challenges, from methodological to operative and ethical issues need to be assessed. The first important point is to manage the existing institution data. This can be done by using a laboratory information management system software able to receive and integrate different types of information, from clinical to pathological and digital data. In literature, several valuable softwares have been used to implement biobank databases (Tukacs et al., 2012; Paul et al., 2017; Fthenou et al., 2019; Im et al., 2019), and some freeware can be obtained for biobank management (Voegele et al., 2013; Willers et al., 2021). A biobank consent form is another critical step in data acquisition and management (Beskow et al., 2017; Kinkorová et al., 2019; Kasperbauer et al., 2021; Schmanski et al., 2021). The application of an adequate SRPA is a necessary agreement on legal and ethical aspects of the patient’s data storage and usage (D'Abramo et al., 2015; Sanchini et al., 2016). All SOPs described in this work are continuously under evaluation and improvement and they are currently compliant with the ISO 20387:2018 standards. It should be noted, however, that standardization and improvement of pre-analytical procedures for in-vitro diagnostics is a continuous process. The most updated high-priority pre-analytical CEN and ISO standard documents as well as corresponding External Quality Assessment (EQA) schemes and implementation tools are detailed in Table 5. Not only following adequate SOPs is essential to secure research achievements, but also have qualified personnel, aware of the biobank’s role and potential (Caixeiro et al., 2016; Kintossou et al., 2020). Another important aspect related to the multidisciplinary collaboration in biobanking is represented by the pathologist (Angerilli et al., 2021). Pathologists are the only professionals able to ensure both the tissue sampling for diagnosis and the biobank. Finally, it should be mentioned that the efforts and resources invested to set up and sustain a biobank are significant and such work should be traced and, most importantly, recognized in scientific publications (Howard et al., 2018). In this respect, the Bioresource Research Impact Factor/Framework (BRIF) initiative was proposed for transparency and to promote the responsible and effective use of biomaterials (Cambon-Thomsen, 2003). Another point that is worth mentioning is related to the integration of artificial intelligence (AI) and machine learning into modern biobanks (Kinkorová and Topolčan, 2020; Eccher et al., 2021; Narita et al., 2021; Rizzo et al., 2022). This would allow for the integration of a digitalized database with digital pathology and high throughput molecular data, potentially representing a quantum leap in biobanking.
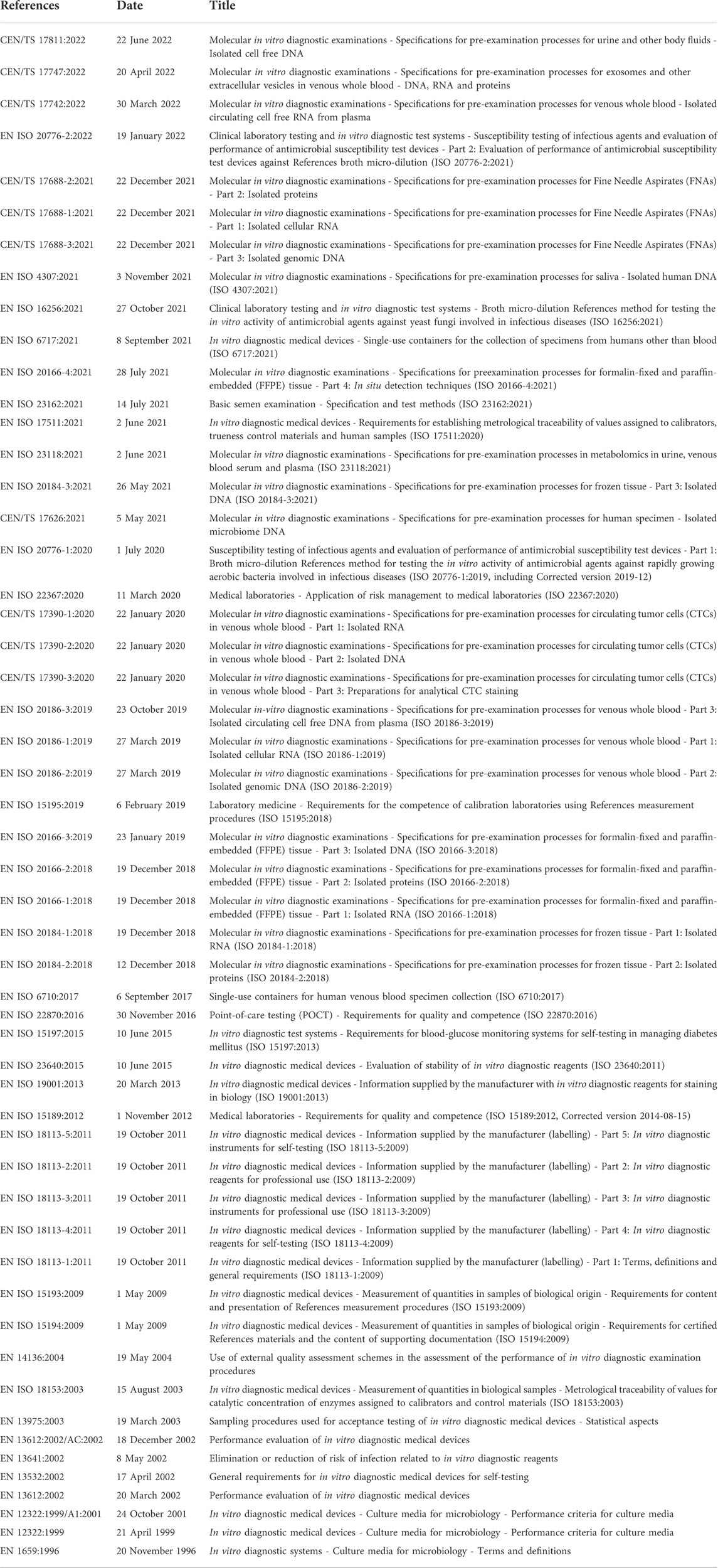
TABLE 5. European Committee for Standardization Technical Committee (CEN/TC) 140 in vitro diagnostic medical devices published standards. All projects are sorted by date and available at https://www.spidia.eu/projects/standard-documents (Accessed 28 July 2022).
Data availability statement
Requests to access the datasets should be directed to B4M= ED@ieo.it.
Ethics statement
The studies involving human participants were reviewed and approved by IEO IRB. The patients/participants provided their written informed consent to participate in this study.
Author contributions
Methodology, GB, MC, GT, CC, EG-R, MM, NF, Investigation, NF, GB, MC Writing—original draft, GB, LZ, Writing—Review and Editing, GB, MI, MF, AA, RO, GV, and NF.
Acknowledgments
The authors would like to thank all the patients that during the last decade actively participated in our research programs through the donation of their biospecimens. Without them, this research would not be possible. We are also grateful to all the personnel working at IEO, nurses, technicians, biologists, doctors, and the Directors of all the clinical and research units. The authors are grateful to Pier Paolo Di Fiore and Giancarlo Pruneri for their guidance since the very beginning of our Biobank Unit. Finally, we dedicate this work to Umberto Veronesi, the founder of IEO, and his pioneering approach to integrating cancer research and patient care.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Angerilli, V., Galuppini, F., Pagni, F., Fusco, N., Malapelle, U., and Fassan, M. (2021). The role of the pathologist in the next-generation era of tumor molecular characterization. Diagnostics 11 (2), 339. doi:10.3390/diagnostics11020339
Beskow, L. M., Lin, L., Dombeck, C. B., Gao, E., and Weinfurt, K. P. (2017). Improving biobank consent comprehension: A national randomized survey to assess the effect of a simplified form and review/retest intervention. Genet. Med. 19 (5), 505–512. doi:10.1038/gim.2016.157
Bonnechère, B., Liu, J., Thomson, A., Amin, N., and Van Duijn, C. M. (2021). Ethnicity influences risk of dementia in the UK Biobank. Alzheimer's. Dementia 17 (10), e056077. doi:10.1002/alz.056077
Braun, K. L., Tsark, J. U., Powers, A., Croom, K., Kim, R., Gachupin, F. C., et al. (2014). Cancer patient perceptions about biobanking and preferred timing of consent. Biopreserv. Biobank. 12 (2), 106–112. doi:10.1089/bio.2013.0083
Bycroft, C., Freeman, C., Petkova, D., Band, G., Elliott, L. T., Sharp, K., et al. (2018). The UK Biobank resource with deep phenotyping and genomic data. Nature 562 (7726), 203–209. doi:10.1038/s41586-018-0579-z
Caixeiro, N. J., Byun, H. L., Descallar, J., Levesque, J. V., de Souza, P., and Soon Lee, C. (2016). Health professionals' opinions on supporting a cancer biobank: Identification of barriers to combat biobanking pitfalls. Eur. J. Hum. Genet. 24 (5), 626–632. doi:10.1038/ejhg.2015.191
Cambon-Thomsen, A. (2003). Assessing the impact of biobanks. Nat. Genet. 34 (1), 25–26. doi:10.1038/ng0503-25b
Castellanos-Uribe, M., Gormally, E., Zhou, H., Matzke, E., and Watson, P. H. (2020). Biobanking education. Biopreserv. Biobank. 18 (1), 1–3. doi:10.1089/bio.2019.29062.mjc
Cicek, M. S., and Olson, J. E. (2020). Mini-Review of laboratory operations in biobanking: Building biobanking resources for translational research. Front. Public Health 8, 362. doi:10.3389/fpubh.2020.00362
Coppola, L., Cianflone, A., Grimaldi, A. M., Incoronato, M., Bevilacqua, P., Messina, F., et al. (2019). Biobanking in health care: Evolution and future directions. J. Transl. Med. 17 (1), 172. doi:10.1186/s12967-019-1922-3
D'Abramo, F., Schildmann, J., and Vollmann, J. (2015). Research participants' perceptions and views on consent for biobank research: A review of empirical data and ethical analysis. BMC Med. Ethics 16, 60. doi:10.1186/s12910-015-0053-5
Drosou, M., Jagadish, H. V., Pitoura, E., and Stoyanovich, J. (2017). Diversity in big data: A review. Big Data 5 (2), 73–84. doi:10.1089/big.2016.0054
Eccher, A., Fontanini, G., Fusco, N., Girolami, I., Graziano, P., Rocco, E. G., et al. (2021). Digital slides as an effective tool for programmed death ligand 1 combined positive score assessment and training: Lessons learned from the "programmed death ligand 1 key learning program in head-and-neck squamous cell carcinoma. J. Pathol. Inf. 12, 1. doi:10.4103/jpi.jpi_63_20
Elliott, P., and Peakman, T. C. (2008). The UK Biobank sample handling and storage protocol for the collection, processing and archiving of human blood and urine. Int. J. Epidemiol. 37 (2), 234–244. doi:10.1093/ije/dym276
Eng, C. B., and Tan, W. L. (2019). Disaster prevention and recovery. Methods Mol. Biol. 1897, 31–41. doi:10.1007/978-1-4939-8935-5_4
Fisher, W. E., Cruz-Monserrate, Z., McElhany, A. L., Lesinski, G. B., Hart, P. A., Ghosh, R., et al. (2018). Standard operating procedures for biospecimen collection, processing, and storage: From the consortium for the study of chronic pancreatitis, diabetes, and pancreatic cancer. Pancreas 47 (10), 1213–1221. doi:10.1097/mpa.0000000000001171
Fthenou, E., Al Emadi, A., Mahal, F. F., Chettupuzhakaran, L. T., Al Thani, A., and Afifi, N. (2019). Conception, implementation, and integration of heterogenous information technology infrastructures in the Qatar biobank. Biopreserv. Biobank. 17 (6), 494–505. doi:10.1089/bio.2019.0067
Guerin, J. S., Murray, D. W., McGrath, M. M., Yuille, M. A., McPartlin, J. M., and Doran, P. P. (2010). Molecular medicine Ireland guidelines for standardized biobanking. Biopreserv. Biobank. 8 (1), 3–63. doi:10.1089/bio.2010.8101
Hartung, M. L., Baber, R., Herpel, E., Specht, C., Brucker, D. P., Schoneberg, A., et al. (2021). Harmonization of biobank education for biobank technicians: Identification of learning objectives. BioTech. 10 (2), 7. doi:10.3390/biotech10020007
Hewitt, R., and Watson, P. (2013). Defining biobank. Biopreserv. Biobank. 11 (5), 309–315. doi:10.1089/bio.2013.0042
Hojat, A., Wei, B., Olson, M. G., Mao, Q., and Yong, W. H. (2019). Procurement and storage of surgical biospecimens. Methods Mol. Biol. 1897, 65–76. doi:10.1007/978-1-4939-8935-5_7
Howard, H. C., Mascalzoni, D., Mabile, L., Houeland, G., Rial-Sebbag, E., and Cambon-Thomsen, A. (2018). How to responsibly acknowledge research work in the era of big data and biobanks: Ethical aspects of the Bioresource research impact factor (BRIF). J. Community Genet. 9 (2), 169–176. doi:10.1007/s12687-017-0332-6
Hulsen, T., Jamuar, S. S., Moody, A. R., Karnes, J. H., Varga, O., Hedensted, S., et al. (2019). From big data to precision medicine. Front. Med. 6, 34. doi:10.3389/fmed.2019.00034
Im, K., Gui, D., and Yong, W. H. (2019). An introduction to hardware, software, and other information technology needs of biomedical biobanks. Methods Mol. Biol. 1897, 17–29. doi:10.1007/978-1-4939-8935-5_3
Jose, R., Rooney, R., Nagisetty, N., Davis, R., and Hains, D. (2018). Biorepository and integrative genomics initiative: Designing and implementing a preliminary platform for predictive, preventive and personalized medicine at a pediatric hospital in a historically disadvantaged community in the USA. Epma J. 9 (3), 225–234. doi:10.1007/s13167-018-0141-y
Kanof, M. E., Smith, P. D., and Zola, H. (2001). Isolation of whole mononuclear cells from peripheral blood and cord blood. Curr. Protoc. Immunol. 7, 1. doi:10.1002/0471142735.im0701s19
Kasperbauer, T. J., Schmidt, K. K., Thomas, A., Perkins, S. M., and Schwartz, P. H. (2021). Incorporating biobank consent into a healthcare setting: Challenges for patient understanding. AJOB Empir. Bioeth. 12 (2), 113–122. doi:10.1080/23294515.2020.1851313
Kinkorová, J., and Topolčan, O. (2020). Biobanks in the era of big data: Objectives, challenges, perspectives, and innovations for predictive, preventive, and personalised medicine. EPMA J. 11 (3), 333–341. doi:10.1007/s13167-020-00213-2
Kinkorová, J., Topolčan, O., and Kučera, R. (2019). Informed consent in the newly established biobank. Int. J. Environ. Res. Public Health 16 (20), 3943. doi:10.3390/ijerph16203943
Kinkorová, J. (2015). Biobanks in the era of personalized medicine: Objectives, challenges, and innovation: Overview. Epma J. 7 (1), 4. doi:10.1186/s13167-016-0053-7
Kintossou, A. K., N’dri, M. K., Money, M., Cissé, S., Doumbia, S., Soumahoro, M.-K., et al. (2020). Study of laboratory staff’ knowledge of biobanking in Côte d’Ivoire. BMC Med. Ethics 21 (1), 88. doi:10.1186/s12910-020-00533-y
Leff, D. R., and Yang, G.-Z. (2015). Big data for precision medicine. Engineering 1 (3), 277–279. doi:10.15302/J-ENG-2015075
Luo, J., Guo, X. R., Tang, X. J., Sun, X. Y., Yang, Z. S., Zhang, Y., et al. (2014). Intravital biobank and personalized cancer therapy: The correlation with omics. Int. J. Cancer 135 (7), 1511–1516. doi:10.1002/ijc.28632
Mallone, R., Mannering, S. I., Brooks-Worrell, B. M., Durinovic-Belló, I., Cilio, C. M., Wong, F. S., et al. (2011). Isolation and preservation of peripheral blood mononuclear cells for analysis of islet antigen-reactive T cell responses: Position statement of the T-cell workshop committee of the immunology of diabetes society. Clin. Exp. Immunol. 163 (1), 33–49. doi:10.1111/j.1365-2249.2010.04272.x
Margolis, R., Derr, L., Dunn, M., Huerta, M., Larkin, J., Sheehan, J., et al. (2014). The national institutes of health's big data to knowledge (BD2K) initiative: Capitalizing on biomedical big data. J. Am. Med. Inf. Assoc. 21 (6), 957–958. doi:10.1136/amiajnl-2014-002974
Mendy, M., Caboux, E., Lawlor, R. T., Wright, J., and Wild, C. P. (2017). Common minimum technical standards and protocols for biobanks dedicated to cancer research. Lyon FR: IARC Technical Publications International Agency for Research on Cancer.
Montserrat, A., and Taruscio, D. (2019). Policies and actions to tackle rare diseases at European level. Ann. Ist. Super. Sanita 55 (3), 296–304. doi:10.4415/ann_19_03_17
Narita, A., Ueki, M., and Tamiya, G. (2021). Artificial intelligence powered statistical genetics in biobanks. J. Hum. Genet. 66 (1), 61–65. doi:10.1038/s10038-020-0822-y
Pagni, F., Guerini-Rocco, E., Schultheis, A. M., Grazia, G., Rijavec, E., Ghidini, M., et al. (2019). Targeting immune-related biological processes in solid tumors: We do need biomarkers. Int. J. Mol. Sci. 20 (21), E5452. doi:10.3390/ijms20215452
Parry-Jones, A., Hansen, J., Simeon-Dubach, D., and Bjugn, R. (2017). Crisis management for biobanks. Biopreserv. Biobank. 15 (3), 253–263. doi:10.1089/bio.2016.0048
Paul, S., Gade, A., and Mallipeddi, S. (2017). The state of cloud-based biospecimen and biobank data management tools. Biopreserv. Biobank. 15 (2), 169–172. doi:10.1089/bio.2017.0019
Rizzo, P. C., Girolami, I., Marletta, S., Pantanowitz, L., Antonini, P., Brunelli, M., et al. (2022). Technical and diagnostic issues in whole slide imaging published validation studies. Front. Oncol. 12, 918580. doi:10.3389/fonc.2022.918580
Rolfo, C., Mack, P., Scagliotti, G. V., Aggarwal, C., Arcila, M. E., Barlesi, F., et al. (2021). Liquid biopsy for advanced nsclc: A consensus statement from the international association for the study of lung cancer. J. Thorac. Oncol. 16 (10), 1647–1662. doi:10.1016/j.jtho.2021.06.017
Saifuddin, S. R., Devlies, W., Santaolalla, A., Cahill, F., George, G., Enting, D., et al. (2017). King's health partners' prostate cancer biobank (KHP PCaBB). BMC Cancer 17 (1), 784. doi:10.1186/s12885-017-3773-8
Salvaterra, E., and Corfield, J. (2017). Advances in biobanking practice through public and private collaborations. Sharjah United Arab Emirates: Bentham Science Publishers.
Sanchini, V., Bonizzi, G., Disalvatore, D., Monturano, M., Pece, S., Viale, G., et al. (2016). A trust-based pact in research biobanks. From theory to practice. Bioethics 30 (4), 260–271. doi:10.1111/bioe.12184
Schmanski, A., Roberts, E., Coors, M., Wicks, S. J., Arbet, J., Weber, R., et al. (2021). Research participant understanding and engagement in an institutional, self-consent biobank model. J. Genet. Couns. 30 (1), 257–267. doi:10.1002/jgc4.1316
Sung, H., Ferlay, J., Siegel, R. L., Laversanne, M., Soerjomataram, I., Jemal, A., et al. (2021). Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Ca. Cancer J. Clin. 71 (3), 209–249. doi:10.3322/caac.21660
Szustakowski, J. D., Balasubramanian, S., Kvikstad, E., Khalid, S., Bronson, P. G., Sasson, A., et al. (2021). Advancing human genetics research and drug discovery through exome sequencing of the UK Biobank. Nat. Genet. 53 (7), 942–948. doi:10.1038/s41588-021-00885-0
Tukacs, E., Korotij, A., Maros-Szabo, Z., Molnar, A. M., Hajdu, A., and Torok, Z. (2012). Model requirements for biobank software systems. Bioinformation 8 (6), 290–292. doi:10.6026/97320630008290
Vaught, J., Kelly, A., and Hewitt, R. (2009). A review of international biobanks and networks: Success factors and key benchmarks. Biopreserv. Biobank. 7 (3), 143–150. doi:10.1089/bio.2010.0003
Vaught, J., Rogers, J., Myers, K., Lim, M. D., Lockhart, N., Moore, H., et al. (2011). An NCI perspective on creating sustainable biospecimen resources. J. Natl. Cancer Inst. Monogr. 2011 (42), 1–7. doi:10.1093/jncimonographs/lgr006
Voegele, C., Bouchereau, B., Robinot, N., McKay, J., Damiecki, P., and Alteyrac, L. (2013). A universal open-source electronic laboratory notebook. Bioinformatics 29 (13), 1710–1712. doi:10.1093/bioinformatics/btt253
Willers, C., Lynch, T., Chand, V., Islam, M., Lassere, M., and March, L. (2021). A versatile, secure, and sustainable all-in-one biobank-registry data solution: The A3BC REDCap model. Biopreserv. Biobank. 20 (3), 244–259. doi:10.1089/bio.2021.0098
Williams, R. R., Gupta, D., and Yong, W. H. (2019). Orientation and training of new biobank personnel. Methods Mol. Biol. 1897, 51–63. doi:10.1007/978-1-4939-8935-5_6
Keywords: biobank, translational research, biomarkers, cancer research, tissue samples, liquid biopsy, standard operating procedures, quality control
Citation: Bonizzi G, Zattoni L, Capra M, Cassi C, Taliento G, Ivanova M, Guerini-Rocco E, Fumagalli M, Monturano M, Albini A, Viale G, Orecchia R and Fusco N (2022) Standard operating procedures for biobank in oncology. Front. Mol. Biosci. 9:967310. doi: 10.3389/fmolb.2022.967310
Received: 12 June 2022; Accepted: 01 August 2022;
Published: 26 August 2022.
Edited by:
Fernando Schmitt, University of Porto, PortugalReviewed by:
Zisis Kozlakidis, International Agency For Research On Cancer (IARC), FranceSerena Bonin, University of Trieste, Italy
Copyright © 2022 Bonizzi, Zattoni, Capra, Cassi, Taliento, Ivanova, Guerini-Rocco, Fumagalli, Monturano, Albini, Viale, Orecchia and Fusco. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nicola Fusco, nicola.fusco@ieo.it
 Giuseppina Bonizzi1
Giuseppina Bonizzi1  Mariia Ivanova
Mariia Ivanova Elena Guerini-Rocco
Elena Guerini-Rocco Roberto Orecchia
Roberto Orecchia Nicola Fusco
Nicola Fusco