Analysis of the Prognostic Value and Gene Expression Mechanism of SHOX2 in Lung Adenocarcinoma
- 1Institute of Nephrology, Affiliated Hospital of Guangdong Medical University, Zhanjiang, China
- 2The Center of Pathological Diagnosis and Research, Affiliated Hospital of Guangdong Medical University, Zhanjiang, China
- 3Department of Respiration, The Second Affiliated Hospital of Guangdong Medical University, Zhanjiang, China
- 4Department of Pathology, Guangdong Medical University, Zhanjiang, China
Background: Detection of SHOX2 methylation has been used to assist in the early diagnosis of lung cancer in many hospitals as SHOX2 may be important in the tumorigenesis of lung cancer. However, there are few studies on the mRNA expression, methylation, and molecular mechanism of SHOX2 in lung cancer. We aimed to explore the role of SHOX2 in lung adenocarcinoma (LUAD).
Methods: First, we examined the differential expression of SHOX2 mRNA and methylation in cancerous and normal tissues using databases. Second, we analyzed the relationship between SHOX2 expression and common clinical parameters in LUAD patients. Third, we further explored the methylated level and its specific location of SHOX2 and the mainly factors of SHOX2 gene expression. Finally, we screened the correlatively expressed genes to analyze the pathways from the Gene Ontology and Kyoto Encyclopedia of Genes and Genomes using DAVID.
Results: We found that the mRNA expression of SHOX2 was higher in multiple cancers, including LUAD and lung squamous cell carcinoma (LUSC), than in normal tissues. Among LUAD patients, SHOX2 expression was higher in patients of middle–young age, with smoking history, in advanced stages, and with nodal distant metastasis. In addition, our results showed that patients with high expression of SHOX2 are prone to recurrence, poor differentiation, and poor prognosis. Thus, we identified that SHOX2 might be an oncogene for LUAD progression. The main factor influencing the high expression of SHOX2 mRNA may be DNA methylation, followed by copy number variation (CNV), but not by gene mutations in LUAD. Unexpectedly, we found that SHOX2 undergoes hypomethylation in the gene body instead of hypermethylation in the promoter. Additionally, SHOX2 has cross talk in the PI3K–Akt signaling pathway and ECM–receptor interaction.
Conclusion:SHOX2 is highly expressed in most cancers. SHOX2 gene expression might be mainly regulated by methylation of its gene body in LUAD, and its high expression or hypomethylation indicates poor differentiation and poor prognosis. SHOX2 could be involved in PI3K–Akt and other important cancer-related signaling pathways to promote tumorigenesis.
Introduction
Much progress has been made in the research field of lung cancer occurrence and progression. However, epidemiological investigations have shown that lung cancer has higher morbidity and mortality than other malignant cancers in China (Chen et al., 2016; Wu et al., 2019). The alarming statistical results of lung cancer remain mainly attributed to the late detection, late diagnosis, late treatment, and unsatisfactory therapeutic effect. In lung cancer, driver gene alterations are critical in the whole process of tumorigenesis, recurrence, and metastasis. More representative driver genes as biomarkers for accurate early diagnosis and prognostic evaluation and therapeutic targeted molecules could improve treatment efficiency and increase mortality. Therefore, the identification of these molecular biomarkers for lung cancer is of great clinical significance.
Short stature homeobox 2 (SHOX2) is highly orthologous to murine Shox2 and human SHOX (Blaschke et al., 1998; De Baere et al., 1998; Marchini et al., 2016; Hu et al., 2018). According to current studies on Shox2, SHOX, and SHOX2, SHOX2 is considered to play a critical role in idiopathic short stature and various types of cardiac arrhythmias (Mortensen et al., 2012; Marchini et al., 2016). However, in recent decades, many researchers have found that SHOX2 also plays an important role in multiple cancers, including lung cancer (Schneider et al., 2011; Hong et al., 2014; Sun et al., 2019). Interestingly, methylation detection of SHOX2 in sputum, blood, alveolar lavage fluid, and tissue of patients has been used to screen early lung cancer patients (Zhao et al., 2015; Weiss et al., 2017; Zhang et al., 2017; Shi et al., 2020). It is worth noting that there is little research on the potential prognostic influence and molecular mechanism of SHOX2 in lung cancer, especially in non-small-cell lung cancer (NSCLC) [mainly lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC)]. The clinical significance of SHOX2 research as an early diagnostic gene is beyond doubt.
In our research, data from The Cancer Genome Atlas (TCGA) were mined using several online analysis tools to evaluate the SHOX2 expression profile in NSCLC. The main factors of the SHOX2 gene expression mechanism, including mutation, copy number, and methylation, were then investigated. The relationship between SHOX2 expression and the clinical characteristics of LUAD was determined using publicly accessible databases. Finally, potential co-expressed genes and functional networks were analyzed to provide direction for further investigation into the mechanism of how SHOX2 works in lung cancer.
Methods
Oncomine Database Analysis
Oncomine is a large oncogene chip database that can be used to analyze and compare gene expression between tumors and normal tissues, as well as gene mutation profiles and their correlation with clinical characteristics (Rhodes et al., 2007). We used the Oncomine database to determine differences in the mRNA expression of the SHOX2 gene between tumors and normal tissues in various cancers. The results of this analysis are addressed with a p-value of 0.05, a fold change of 2, all gene rankings, and mRNA data type.
GEPIA
Gene expression profiling interactive analysis (GEPIA) is a web tool that provides key interactive analysis and customization capabilities, including tumor/normal differential expression profilometry, profile mapping, pathological staging, patient survival analysis, similar gene assay analysis, and dimensionality reduction analysis (Tang et al., 2017). We used GEPIA to address the differential expression of SHOX2 in all common cancers. We applied the following cut-off criteria: using the ANOVA method, |log2 FC| > 1, p-value < 0.01, and log2(TPM +1) for log-scale, matching TCGA and GTEx normal data, and adding all cancer tissue names.
GEO Microarray Analysis
The Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) is a database that stores chips, second-generation sequencing, and other high-throughput sequencing data worldwide (Edgar et al., 2002). An mRNA expression dataset (GSE33532) for early stage NSCLC was downloaded from the GEO database. GSE33532 stored mRNA expression information of 80 NSCLC tissue samples and 20 matched distant-normal samples from 20 patients. Microarray expression profiling was normalized by log2 transformation before analysis. These 20 patients were included in the validation dataset. The histological types of these NSCLC samples included adenocarcinoma, squamous cell carcinomas, and mixed carcinomas. Finally, we used GraphPad Prism 7 software to analyze and plot the collected data using unpaired t-test statistical methods. The p-value < 0.05 was considered statistically significant.
SurvivalMeth Analysis
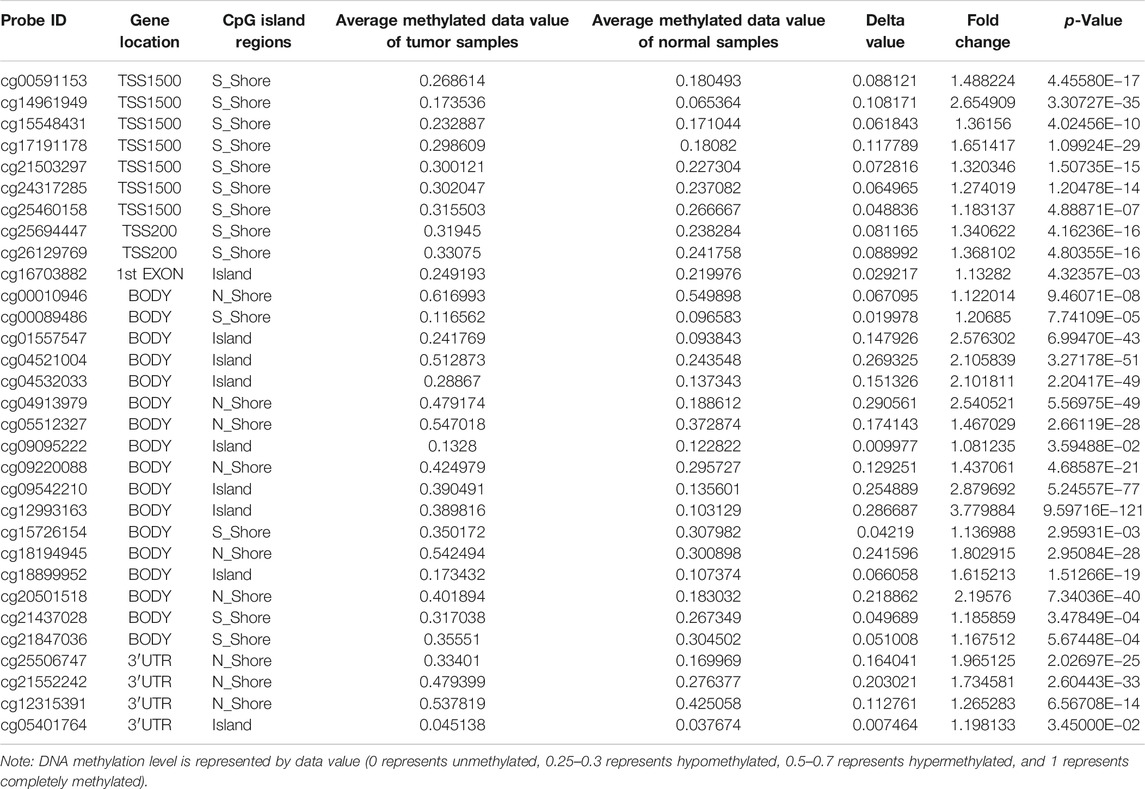
SurvivalMeth (http://bio-bigdata.hrbmu.edu.cn/survivalmeth/) is a web server that is freely available to address the prognostic information of cancer-associated methylation, based on TCGA, CCLE, and GEO (Zhang et al., 2020). We used SurvivalMeth to investigate the effect of SHOX2 DNA methylation–related functional elements on protein expression and LUAD prognosis. We applied the following cut-off criteria: the chosen disease of lung adenocarcinoma, the chosen experimental platform of 450 K (Illumine Infinium HumanMethylation450 BeadChip), the chosen gene symbol of SHOX2, transcript-related elements including “1st exon, 3′UTR, 5′UTR, gene body, TSS1500, and TSS200,” CpG island–related elements including “Island, N_Shelf, N_Shore, S_Shelf, and S_Shore,” unrestricted repeat element–related element and CTCF-binding region, statistical methods using t-test, a threshold value of 0.05, and median group strategy.
UCSC Xena Analysis
The University of California Santa Cruz Cancer Genomics Browser (UCSC Xena) (http://xena.ucsc.edu/) is an online database of genomic, transcriptomic, and clinical and phenotypic data (Kent et al., 2002; Zweig et al., 2008; Navarro Gonzalez et al., 2021). To investigate the dominant factors influencing SHOX2 expression in LUAD, UCSC Xena was used to analyze the relationship between the methylation level, copy number, mutation, Kaplan–Meier survival analysis, and mRNA expression of SHOX2 in TCGA LUAD samples. We set the following conditions: sample types including solid tissue normal and primary tumor, gene expression using RNAseq-IlluminaHiSeq, DNA methylation using Methylation 450K, copy number using (gene-level)-gistic2, and somatic mutation using (SNP and INDEL)-MC3 public version. Additionally, we used this tool to analyze SHOX2 expression patterns in different pathological types.
UALCAN Analysis
UALCAN (http://ualcan.path.uab.edu) is an online database that provides free visual figures of gene expression, survival analysis, correlation analysis, and gene DNA promoter region methylation data, grouped by clinicopathological features between normal and tumor tissues, based on TCGA data (Chandrashekar et al., 2017). To explore the relationship between SHOX2 and the clinicopathological features of NSCLC patients, the expression and methylation levels of SHOX2 in NSCLC and adjacent normal tissues were identified using UALCAN.
MethSurv Analysis
MethSurv (https://biit.cs.ut.ee/methsurv/) used methylation group data from the “Cancer Genome Map” to perform survival analysis on the methylation patterns of CpG to achieve a preliminary assessment of tumor biomarkers based on methylation (Modhukur et al., 2018). We used MethSurv to further explore the relationship between SHOX2 methylation and several clinical characterizations in a heat map.
cBioPortal Database Analysis
The cBioPortal database (v3.5.4) (http://www.cbioportal.org) was used to perform the cancer genomic analysis (Gao et al., 2013; Jing et al., 2019). We chose two datasets, “Lung Adenocarcinoma (TCGA, Firehose Legacy)” and “Lung Adenocarcinoma (TCGA, Nature 2014),” to analyze the co-expressed SHOX2 genes. We then set the following parameters: select genomic profiles [mutations, putative copy number alterations from GISTIC, and mRNA expression z-scores relative to diploid samples (RNA Seq V2 RSEM)], select patient/case set (all samples), and enter genes (user-defined list, SHOX2). Subsequently, we entered the co-expression interface and acquired the genes correlated with SHOX2 in satisfactory samples from these datasets. These data were downloaded, and the next operating steps were followed.
Using the Venn Diagram Tool
We selected the first 300 SHOX2 correlated genes from the previous steps with a higher Spearman correlation coefficient and p < 0.05. According to the applied cBioPortal database, one gene list was named “TCGA, Firehose Legacy” and the other gene list was named “TCGA, Nature 2014.” We input the two lists into the upload lists. Then, we acquired the Venn diagram (http://bioinformatics.psb.ugent.be/webtools/Venn/) of SHOX2 co-expressed related genes.
Functional and Pathway Enrichment Analysis
The Database for Annotation, Visualization and Integrated Discovery (DAVID) (version 6.8; https://david.ncifcrf.gov/) is a biological information database that integrates biological data and analysis tools to provide systematic and comprehensive annotated biological function information for large-scale gene or protein lists to help users extract biological information from them (Huang da et al., 2009). We used DAVID to perform Gene Ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis of the selected common SHOX2 co-expressed related genes screened by the two datasets. GO enrichment analysis consisted of cellular component (CC), biological process (BP), and molecular function (MF) analyses. Following these steps, we uploaded these genes, selected the identifier “OFFICIAL_GENE_SYMBOL,” selected “Homo spaiens,” selected “Gene List,” submitted the list, and finally obtained functional annotation charts. We then acquired the GO functional enrichment and KEGG pathway enrichment results. Calculated via Fisher’s exact test, a p-value < 0.05 and a count >2 were considered statistically significant.
Results
SHOX2 mRNA Expression Profile in Multiple Cancers
To determine the expression level of SHOX2 in various types of tumors, we examined the differential expression of SHOX2 between tumor tissue and paired normal tissue using Oncomine and GEPIA online analysis tools. The Oncomine database has a total of 320 unique analyses for SHOX2. There are 52 significant unique analyses among them that showed SHOX2 upexpression, while 15 analyses showed SHOX2 downexpression (Figure 1A). SHOX2 expression was higher in most cancers, including sarcoma, brain and CNS cancer, head and neck squamous carcinoma (HNSC), and lung cancer. Meanwhile, it was downregulated in breast cancer, leukemia, and many other cancers (Figure 1A). For further verification, we used the GEPIA web tool to compare the differential expression of SHOX2 between cancerous and normal tissues. As shown in Figure 1B, higher SHOX2 expression was observed in glioblastoma (GBM), HNSC, and LUSC. In addition, lower SHOX2 expression was observed in breast invasive carcinoma (BRCA), acute myeloid leukemia (LAML), and testicular germ cell tumors (TGCTs). In summary, the results from the two databases were consistent, supporting the evidence that SHOX2 should be an oncogene.

FIGURE 1. SHOX2 mRNA expression in multiple cancers. (A) The number of datasets with SHOX2 mRNA differential expression between tumor tissues and normal tissues. The left column (red) represents overexpression, and the right column (blue) represents downexpression. This visualized graphic is available from Oncomine, and the threshold parameters were set as follows: p-value = 0.05, fold change = 2, all gene rankings, and mRNA data type. (B)SHOX2 gene expression profile in various tumor samples and normal samples by GEPIA. SHOX2: short stature homeobox 2; GEPIA: gene expression profiling interactive analysis.
Analysis of SHOX2 Expression in LUAD/LUSC and Normal Tissues
In order to explore SHOX2 mRNA expression and its relationship with clinical features, we addressed the GSE33532 data using GraphPad Prism 7 and used UALCAN to analyze the relative clinical data of LUAD/LUSC patients from TCGA. The SHOX2 mRNA expression profiling dataset of NSCLC showed that SHOX2 is highly expressed in both LUAD and LUSC tissues compared to paired normal tissues (Figure 2A). The difference in SHOX2 expression was statistically significant. This result was verified by an expanded sample size analysis using the online analysis database UALCAN. As shown in Figure 2B, we obtained consistent results.
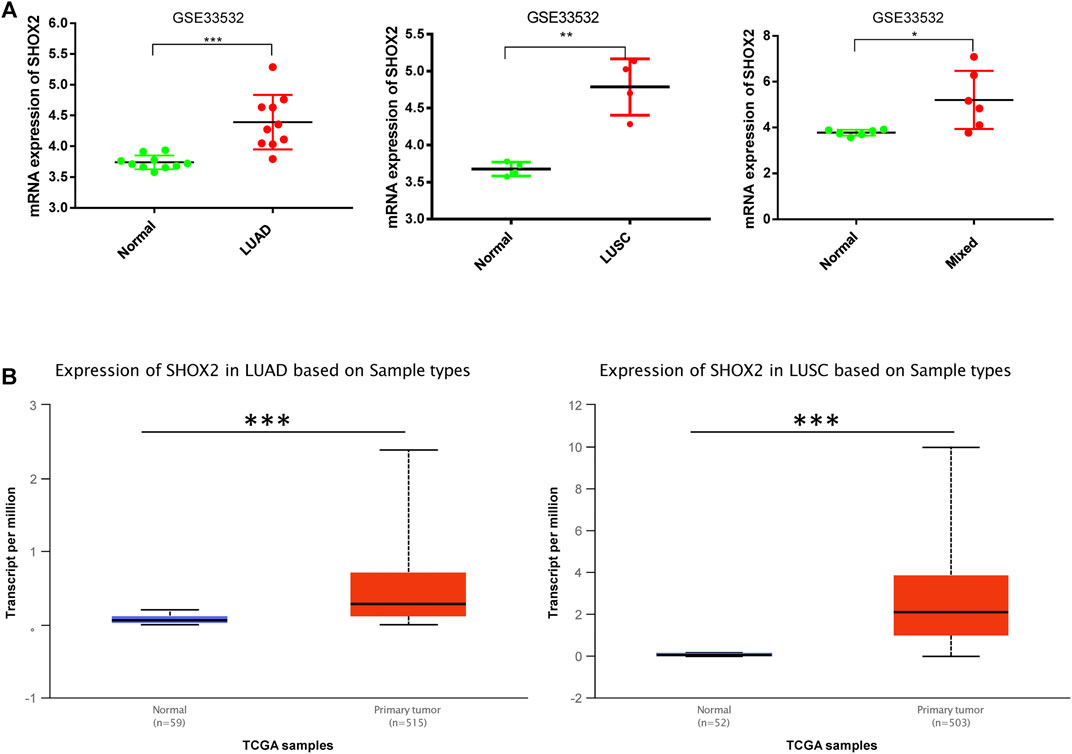
FIGURE 2. Box plot showing the relative expression of SHOX2 in normal and NSCLC samples. (A)SHOX2 expression is shown by microarray analysis of 20 NSCLC patients in GSE33532 including 10 LUAD patients, 4 LUSC patients, and 6 mixed patients. These data are compared with those of paired normal tissues. The data are addressed by GraphPad Prism 7.0 and applied paired t-test. (B) The expression levels of SHOX2 between normal and LUAD or LUSC tissues are shown in the box plot by UALCAN online analysis tool application. *** represents p < 0.001 (highly significant); ** represents p < 0.01 (very significant); * represents p < 0.05 (significant); LUAD: lung adenocarcinoma; LUSC: lung squamous carcinoma; NSCLC: non-small-cell lung cancer.
The Relationship of SHOX2 Expression With the Clinical Characteristics of LUAD Patients
In recent decades, the incidence of LUAD has been higher than that of LUSC. Moreover, there are few reports on the relationship between SHOX2 expression and the clinicopathological features of LUAD. Therefore, we used the UALCAN online analysis tool to address SHOX2 mRNA expression in different subgroups having multiple clinicopathological features from LUAD samples from TCGA. The SHOX2 mRNA expression level was significantly higher in LUAD patients than in healthy people in subgroup analyses based on sex, age, smoking habits, stage, nodal metastasis, and TP-53 mutation status (Figures 3A–F). In addition, subgroup analyses showed that SHOX2 expression was higher in patients aged 20–40 years, with a history of smoking, in stage 2, and with nodal distant metastasis, compared to other clinical features (Figure 3). The pathology of disease can help clinicians to clearly diagnose the disease and take reasonable and effective treatment measures. UCSC Xena was used to analyze the changes of SHOX2 expression in different histological subtypes of invasive LUAD from a cohort of TCGA LUAD (n = 706) (Figure 3G). The results showed that SHOX2 expression varied significantly among different histological subtypes of LUAD (one-way ANOVA, p = 0.0001271, f = 4.098). SHOX2 expression was higher in subtypes prone to lymph node metastasis, prone to recurrence, with poor differentiation, and with poor prognosis.
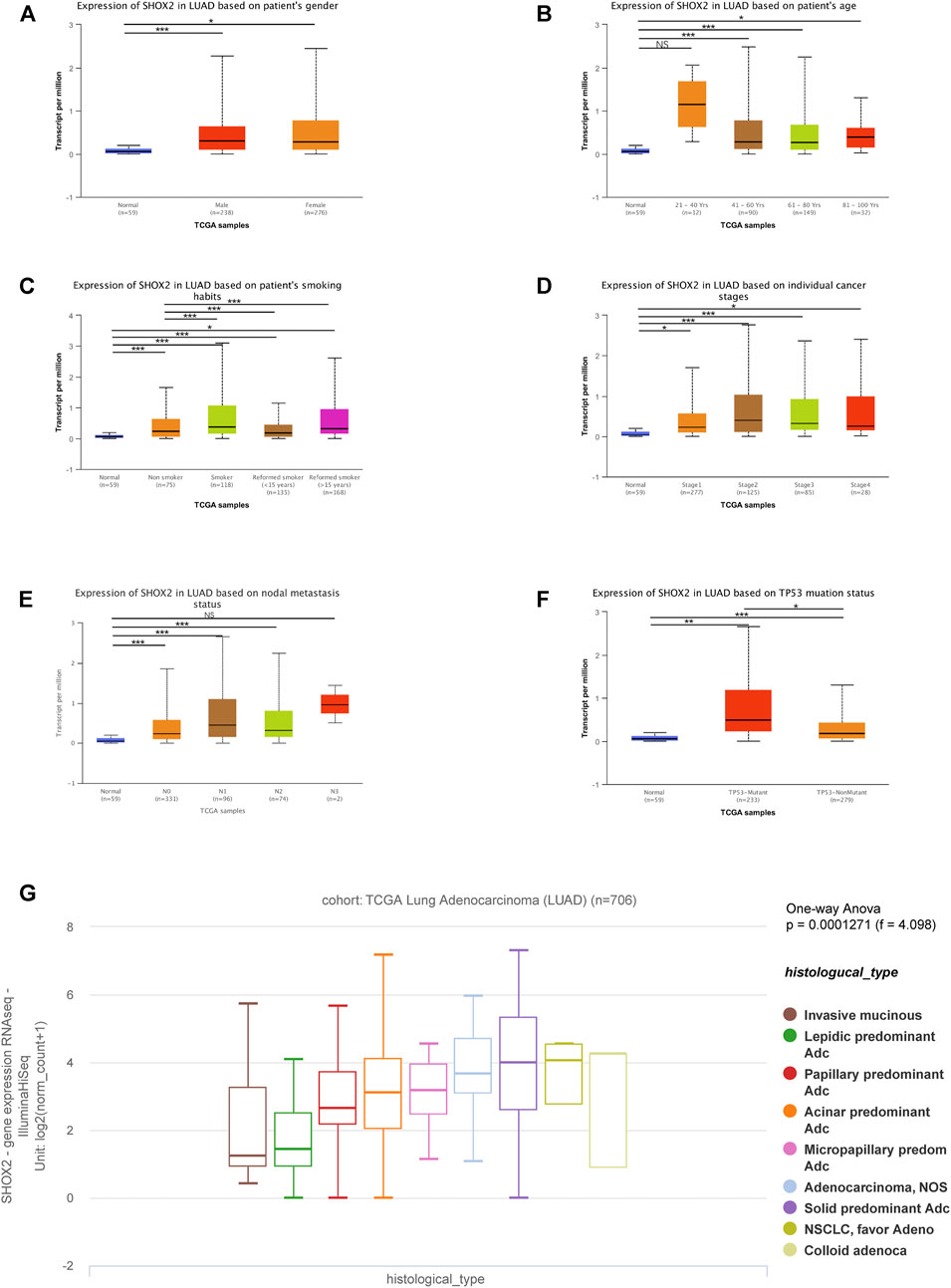
FIGURE 3. Box plot showing the characteristic relationships of (A) gender, (B) age, (C) smoking habits, (D) stage, (E) nodal metastasis, (F) TP-53 mutation status, and (G) histological subtypes with SHOX2 mRNA expression in LUAD analyzed using the online tools UALCAN and UCSC Xena. Data are represented as mean ± SE. *** represents p < 0.001 (highly significant); ** represents p < 0.01 (very significant); * represents p < 0.05 (significant); LUAD: lung adenocarcinoma.
Furthermore, to confirm the prognostic value of SHOX2 in LUAD, UCSC Xena was searched to investigate the effects of SHOX2 expression, methylation, and CNV on overall survival (OS) and disease-specific survival (DSS). It was confirmed that high SHOX2 mRNA expression was significantly associated with decreased OS and DSS time in LUAD (p < 0.05); DNA methylation and copy number variation had no significant effect on OS (p > 0.05), and a higher copy number indicated a shorter DSS (p < 0.05). The SHOX2 expression level and DNA methylation level had no significant correlation with DSS (p > 0.05) (Figure 4).
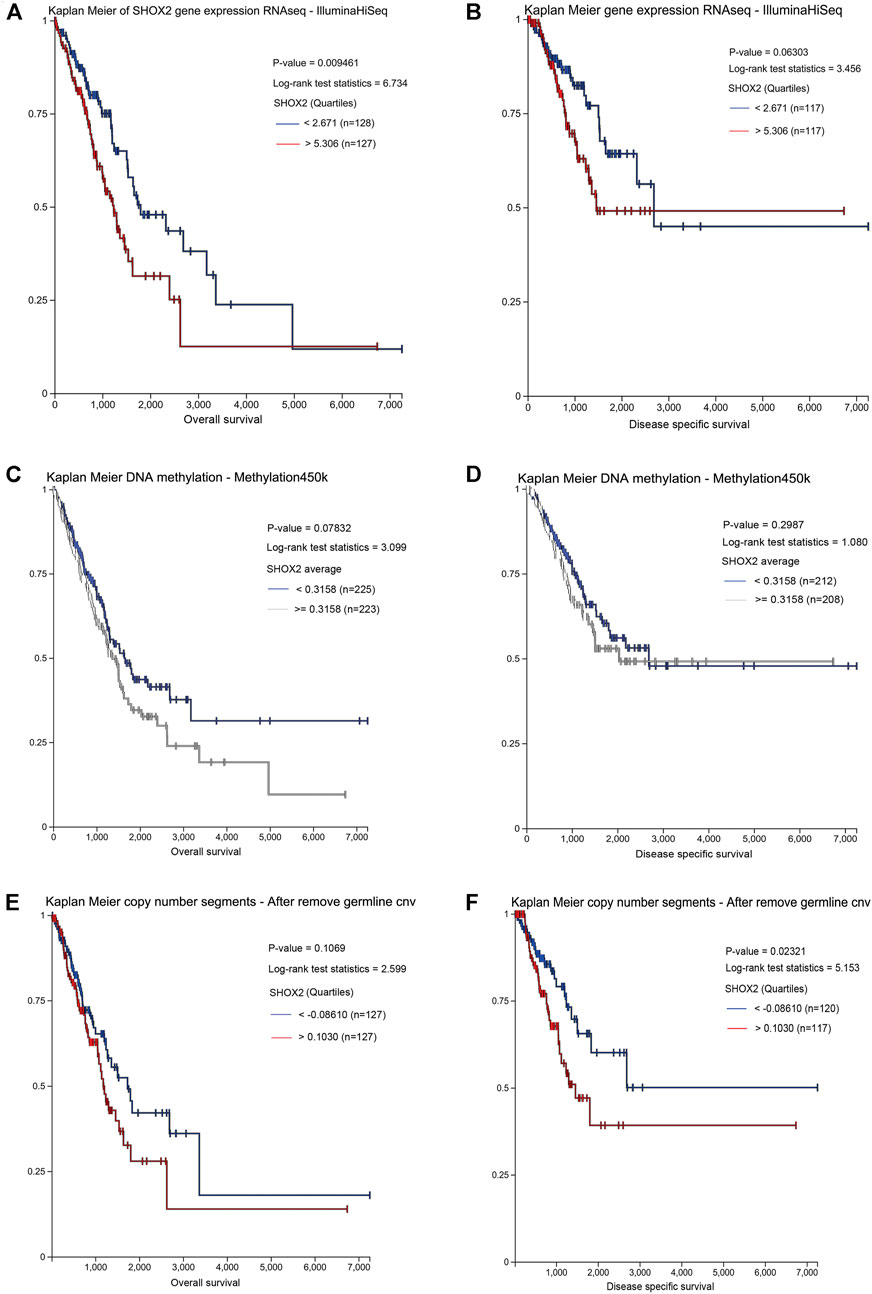
FIGURE 4. Prognostic value of the SHOX2 mRNA expression level in LUAD patients by UCSC Xena (Kaplan–Meier plotter). (A) Relationship between SHOX2 expression and OS in LUAD. (B) Relationship between SHOX2 expression and DSS in LUAD. (C) Relationship between SHOX2 DNA methylation and OS in LUAD. (D) Relationship between SHOX2 DNA methylation and DSS in LUAD. (E) Relationship between SHOX2 copy number segments and OS in LUAD. (F) Relationship between SHOX2 copy number segments and DSS in LUAD. p < 0.05 indicates statistical significance. LUAD: lung adenocarcinoma; OS: overall survival; DSS: disease-specific survival.
Therefore, these results suggest that SHOX2 might be a driving factor in the development of LUAD and may act as a potential diagnostic and prognostic indicator in LUAD.
Influencing-Factor Analysis of SHOX2 Gene Expression in LUAD
To determine the main influencing factors of SHOX2 gene expression, we used several online tools performing mutation, methylation, and CNV alteration analysis. We used three methylation analysis tools (UCSC Xena, SurvivalMeth, and MethSurv) to probe the SHOX2 methylation level in LUAD patients, from different perspectives. The methylation level of SHOX2 was examined in TCGA LUAD patients, based on age, race, ethnicity, alive or dead events, SHOX2 mRNA expression, CpG island–related elements, and transcript-related elements. From Figure 5 and Table 1, we can conclude that DNA methylation of SHOX2 was elevated in LUAD tissues compared with that in normal lung tissues. As shown in Figure 5, SHOX2 DNA was only locally methylated. To explore the DNA-methylated specific site of SHOX2 and verify the acquired results, we conducted an in-depth study using TCGA LUAD patient data in SurvivalMeth and MethSurv. In LUAD tissues, SHOX2 DNA methylation mainly occurred in the gene body, at the hypomethylation level (p < 0.05) (Figures 5 and 6, Tables 1 and 2). According to previous research results, hypomethylation of the gene body leads to the high expression of oncogenes, which is opposite to the hypermethylation of the gene promoter for tumor suppressive genes (Yang et al., 2014). Therefore, hypomethylation of the SHOX2 gene body might be one of the main factors causing mRNA expression. In conclusion, in LUAD tissue, the mRNA expression of SHOX2 might be correlated with the DNA methylation level, followed by CNV, but not with gene mutation (Figure 5).
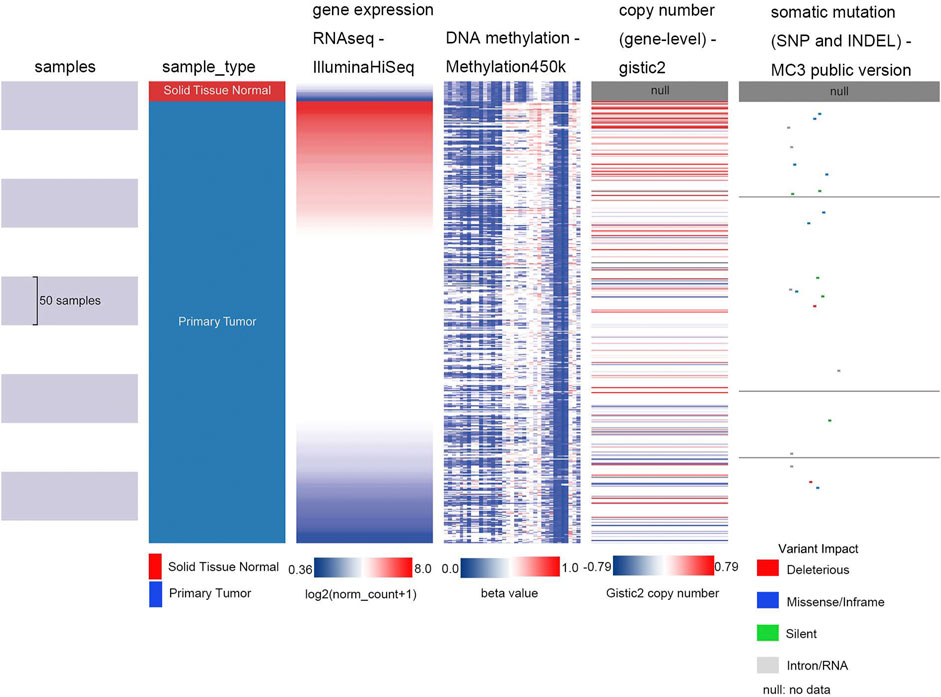
FIGURE 5. Heat map of SHOX2 mRNA expression, methylation, copy number, and somatic mutation in patients with primary LUAD and normal tissue. The data are obtained from TCGA-LUAD with a total of 706 samples. LUAD: lung adenocarcinoma.
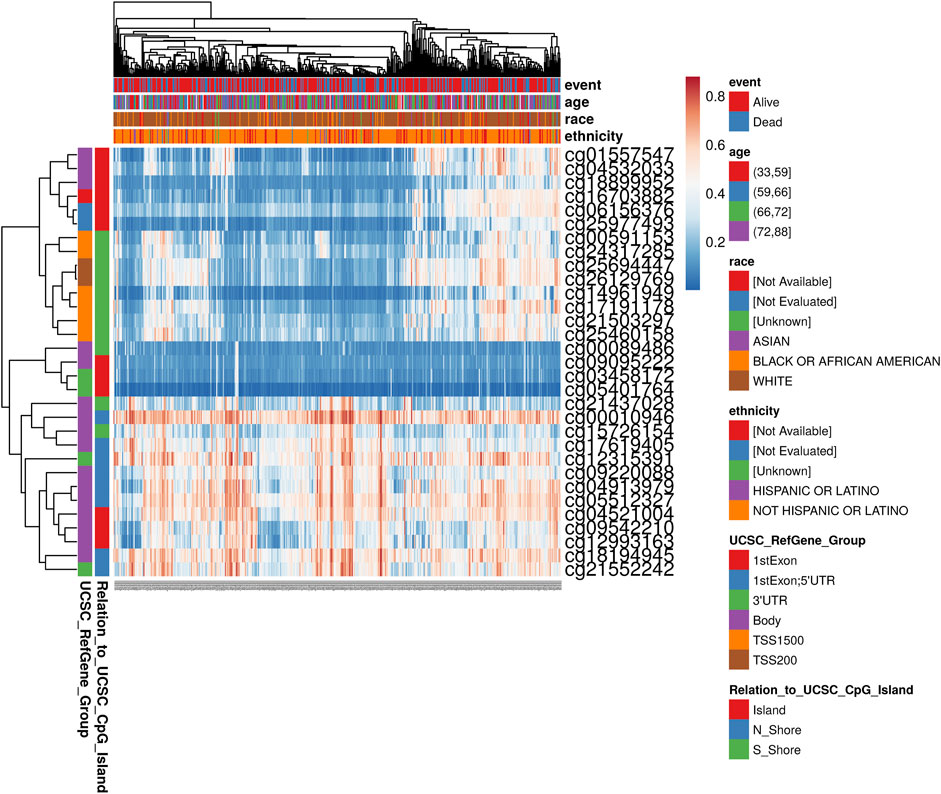
FIGURE 6. Heat map of methylation levels of the SHOX2 methylated probe. Red to blue: high expression to low expression. Various colorful side-boxes are used to characterize the event, age, race, ethnicity, UCSC_refGene_Group, and relation to UCSC_CpG_island.
Enrichment Analysis of Correlated Genes With SHOX2 in LUAD
Finally, to explore the possible signaling pathways from the genes correlated with SHOX2, we analyzed mRNA sequencing data from 816 LUAD patients in two TCGA research cohorts: Firehose Legacy (586 samples) and Nature 2014 (230 samples). As shown in the Venn diagram (Figure 7A), 139 significantly correlated genes with SHOX2 appeared in both cohorts (p < 0.05). This suggests that SHOX2 has a widespread impact on the transcriptome. We then performed functional enrichment analysis of SHOX2 and these 139 common significantly correlated genes using the DAVID database. The top 10 GO terms of these genes, according to the gene counts, are shown in Figure 7B, including biological processes (BPs), cellular components (CCs), and molecular functions (MFs). The BP of SHOX2 was mainly associated with positive regulation of cell migration, cell division, skeletal system development, and mitotic nuclear division. The CC of SHOX2 was mainly located in the extracellular space. The MF of SHOX2 mainly focuses on the structural constituents of the extracellular matrix and serine-type endopeptidase activity. Surprisingly, the significant KEGG pathways included the PI3K–Akt signaling pathway, ECM–receptor interaction, protein digestion and absorption, and amebiasis (p < 0.05).
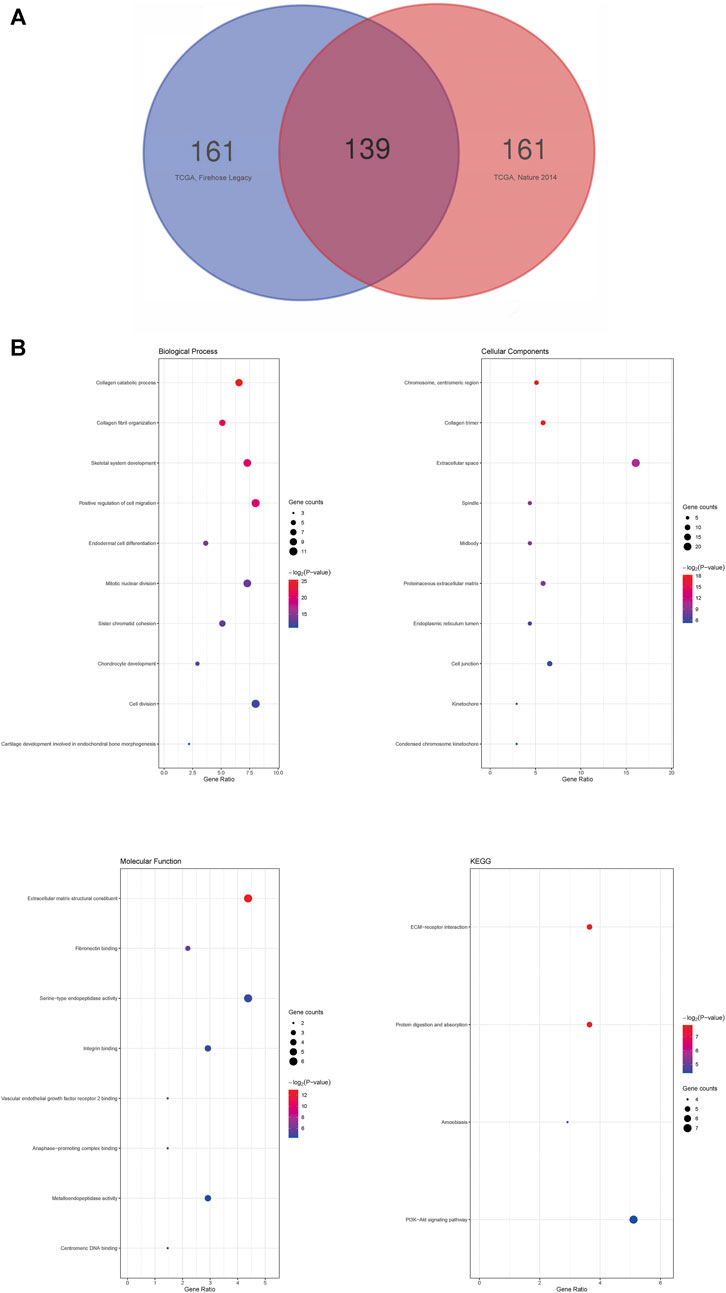
FIGURE 7. Enrichment analysis of correlated genes with SHOX2 in LUAD. (A) Venn diagram of SHOX2 correlated genes in LUAD. The 139 significantly correlated genes with SHOX2 appeared in two TCGA research cohorts, Firehose Legacy (586 samples) and Nature 2014 (230 samples). (B) GO and KEGG analyses of SHOX2 and the significantly correlated genes by DAVID database. The node size represents the gene ratio; the node color represents the p-value. LUAD: lung adenocarcinoma; GO: Gene Ontology; KEGG: Kyoto Encyclopedia of Genes and Genomes.
Discussion
Early detection, early diagnosis, and early treatment are of key importance in preventing disease development. At present, conventional diagnostic methods for lung cancer include computed tomography imaging and cellular/histopathological examination. Emerging molecular diagnostic methods are being increasingly applied in clinical practice because they are more sensitive and objective. DNA methylation alteration is considered an early predictor of cancer and can be detected during the early stages of tumorigenesis (Zhang Y. et al., 2016; Dirks et al., 2016; Wang et al., 2018; Srisuttee et al., 2020). Because of the hypermethylation of SHOX2 in lung cancer, methylation detection has been applied to assist in the early diagnosis of unknown pulmonary nodules (Kneip et al., 2011; Konecny et al., 2016; Weiss et al., 2017; Shi et al., 2020). Many studies have reported that SHOX2 is highly expressed in many cancers. The specific role of SHOX2 in lung cancer patients and the molecular mechanism of its action are unknown.
As reported in detail in our previous review (Li et al., 2020), we synthesized the results of existing studies on SHOX2 and its related homonymous genes in mice, lung cancer and other tumors and concluded that SHOX2 might play an important role in tumorigenesis, metastasis, and recurrence in lung cancer. Here, we used bioinformatics analysis to verify the above conclusions.
In our study, we found that the mRNA expression of SHOX2 was higher in multiple cancers than in paired normal tissues, including LUAD and LUSC tissues. Among the LUAD patients, SHOX2 expression was higher in patients of middle–young age, with smoking history, in advanced stages, and with nodal distant metastasis. In addition, our results showed that patients with high expression of SHOX2 are prone to recurrence, poor differentiation, and poor prognosis. Here, we identified that SHOX2 plays a negative role in LUAD progression and could act as an oncogene. Next, we performed SHOX2 methylation analysis. Finally, we found that SHOX2 undergoes hypomethylation in the gene body. The main factors for SHOX2 mRNA expression are the DNA methylation and CNV, but not gene mutations in LUAD. Additionally, SHOX2 has cross talk in the PI3K−Akt signaling pathway and in ECM–receptor interactions.
The potential role of SHOX2 in patients with lung cancer may have been explained by many previous studies. We identified that the mRNA expression of SHOX2 could assist in indicating subsets of LUAD with worse prognosis and survival. Zhang and Zhou also reported that SHOX2 was an indicator for identifying subgroups with worse prognosis in lower-grade gliomas, which is consistent with our result (Zhang Y.A. et al., 2016). Additionally, there have been other similar reports of SHOX2 methylation as a predictor of malignancy (Leiro et al., 2019; Xu et al., 2020).
Tumor cell genomes showed global hypomethylation and local hypermethylation. Local hypermethylation may occur on the promoter CpG island of a tumor suppressor gene to inhibit the expression of this gene, or on the gene body CpG island of an oncogene to induce high expression of this gene (Yang et al., 2014; Gagliardi et al., 2018). Downregulated TSGs or upregulated oncogenes are critical in tumorigenesis. According to recent research, SHOX2 is considered an oncogene. Unexpectedly, the present majority of related reports declare that SHOX2 methylation occurs on the promoter CpG island, which can seem incomprehensible. To explain the discrepancy of high methylation of SHOX2 in cancer tissue and to identify whether methylation occurs at the gene promoter, we conducted further research to try to explain this phenomenon. We used UCSC XENA, SurvivalMeth, and MethSurv to obtain methylation data for all methylation sites of SHOX2 and then analyzed them statistically using Excel. Interestingly, we observed that the SHOX2 methylation is in a hypomethylated state, and methylation mainly occurs at the gene body instead of at the gene promoter. In summary, SHOX2 is highly expressed, and its genomes are hypomethylated in LUAD, which might be the main mechanism of gene expression. Therefore, the current description of SHOX2 promoter hypermethylation may not be rigid.
The regulation of gene expression is the molecular basis of cell differentiation, morphogenesis, and ontogenesis in vivo. The influencing factors of gene expression include one or two of the following alterations in most cases: local mutation, CNV, DNA methylation, and the expression level of master transcription factors on another chromosome. We found that hypomethylation of the SHOX2 gene body may be one of the main mechanisms driving the upregulation of SHOX2, and CNV is also one of them. Schneider et al. (2011) and Zhang Y.-A. et al. (2016) may agree with our findings.
According to our enrichment analysis of SHOX2, the BP of SHOX2 was mainly associated with positive regulation of cell migration, cell division, skeletal system development, and mitotic nuclear division, and the significant KEGG pathways included the PI3K−Akt signaling pathway, ECM−receptor interaction, protein digestion and absorption, and amebiasis. These findings are consistent with the fact that SHOX2 is considered an oncogene and a diagnostic and predicted biomarker. This is critical for understanding how SHOX2 expression alter and how SHOX2 lead to cancers such as LUAD.
In this study, we combined multiple online bioinformatics data analysis platforms and tools to provide a statistical analysis of the correlation between SHOX2 expression and clinical characteristic parameters, clinical prognosis, DNA methylation, CNV, mutations, co-expressed genes, and related signaling pathways in LUAD. SHOX2 is highly expressed in most cancers. SHOX2 gene expression may be mainly regulated by methylation of the gene body in LUAD, and its high expression or hypomethylation indicates poor differentiation and poor prognosis. SHOX2 is involved in PI3K−Akt and other important cancer-related signaling pathways to promote tumorigenesis.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
NL, YZ, MT, and JH conceptualized and designed the idea and analyzed and interpreted the data. JH provided administrative support. NL, YZ, MT, BL, DZ, YL, XR, XZ, LL, and HW contributed to the provision of study materials or patients and collected and assembled the data. NL and YZ wrote the manuscript. All authors approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (81572610) and the “Yangfan Plan” for Outstanding Scholars in Guangdong Province (4YF16002G) which all are from JH.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Thanks are due to Bo Zhao (Official Wechat Account: SCIPhD) of ShengXinZhuShou for suggestions on the manuscript.
References
Blaschke, R. J., Monaghan, A. P., Schiller, S., Schechinger, B., Rao, E., Padilla-Nash, H., et al. (1998). SHOT, a SHOX-Related Homeobox Gene, Is Implicated in Craniofacial, Brain, Heart, and Limb Development. Proc. Natl. Acad. Sci. 95, 2406–2411. doi:10.1073/pnas.95.5.2406
Chandrashekar, D. S., Bashel, B., Balasubramanya, S. A. H., Creighton, C. J., Ponce-Rodriguez, I., Chakravarthi, B. V. S. K., et al. (2017). UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 19, 649–658. doi:10.1016/j.neo.2017.05.002
Chen, W., Zheng, R., Baade, P. D., Zhang, S., Zeng, H., Bray, F., et al. (2016). Cancer Statistics in China, 2015. CA: A Cancer J. Clin. 66, 115–132. doi:10.3322/caac.21338
De Baere, E., Speleman, F., Van Roy, N., De Paepe, A., and Messiaen, L. (1998). Assignment1 of SHOX2 (Alias OG12X and SHOT) to Human Chromosome Bands 3q25→q26.1 by In Situ Hybridization. Cytogenet. Genome Res. 82, 228–229. doi:10.1159/000015108
Dirks, R. A., Stunnenberg, H. G., and Marks, H. (2016). Genome-wide Epigenomic Profiling for Biomarker Discovery, Clin. Epigenetics 8, 122. doi:10.1186/s13148-016-0284-4
Edgar, R., Domrachev, M., and Lash, A. E. (2002). Gene Expression Omnibus: NCBI Gene Expression and Hybridization Array Data Repository. Nucl. Acids Res. 30, 207–210. doi:10.1093/nar/30.1.207
Gagliardi, M., Strazzullo, M., and Matarazzo, M. R. (2018). DNMT3B Functions: Novel Insights from Human Disease, Front Cell Dev. Biol. 6, 140. doi:10.3389/fcell.2018.00140
Gao, J., Aksoy, B. A., Dogrusoz, U., Dresdner, G., Gross, B., Sumer, S. O., et al. (2013). Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the cBioPortal. Sci. Signal. 6, pl1. doi:10.1126/scisignal.2004088
Hong, S., Noh, H., Teng, Y., Shao, J., Rehmani, H., Ding, H.-F., et al. (2014). SHOX2 Is a Direct miR-375 Target and a Novel Epithelial-To-Mesenchymal Transition Inducer in Breast Cancer Cells. Neoplasia 16, 279–290. e1-5. doi:10.1016/j.neo.2014.03.010
Hu, W., Xin, Y., Zhao, Y., and Hu, J. (2018). Shox2: The Role in Differentiation and Development of Cardiac Conduction System. Tohoku J. Exp. Med. 244, 177–186. doi:10.1620/tjem.244.177
Huang, D. W., Sherman, B. T., and Lempicki, R. A. (2009). Systematic and Integrative Analysis of Large Gene Lists Using DAVID Bioinformatics Resources. Nat. Protoc. 4, 44–57. doi:10.1038/nprot.2008.211
Jing, X., Liang, H., Hao, C., Yang, X., and Cui, X. (2019). Overexpression of MUC1 Predicts Poor Prognosis in Patients with Breast Cancer, Oncol. Rep. 41, 801–810. doi:10.3892/or.2018.6887
Kent, W. J., Sugnet, C. W., Furey, T. S., Roskin, K. M., Pringle, T. H., Zahler, A. M., et al. (2002). The Human Genome Browser at UCSC. Genome Res. 12, 996–1006. doi:10.1101/gr.229102
Kneip, C., Schmidt, B., Seegebarth, A., Weickmann, S., Fleischhacker, M., Liebenberg, V., et al. (2011). SHOX2 DNA Methylation Is a Biomarker for the Diagnosis of Lung Cancer in Plasma. J. Thorac. Oncol. 6, 1632–1638. doi:10.1097/jto.0b013e318220ef9a
Konecny, M., Markus, J., Waczulikova, I., Dolesova, L., Kozlova, R., Repiska, V., et al. (2016). The Value of SHOX2 Methylation Test in Peripheral Blood Samples Used for the Differential Diagnosis of Lung Cancer and Other Lung Disorders. Neoplasma 63, 246–253. doi:10.4149/210_150419n208
Leiro, V., De Chiara, L., Rodríguez-Girondo, M., Botana-Rial, M., Valverde, D., Núñez-Delgado, M., et al. (2019) Methylation Assessment for the Prediction of Malignancy in Mediastinal Adenopathies Obtained by Endobronchial Ultrasound-Guided Transbronchial Needle Aspiration in Patients with Lung Cancer, Cancers (Basel) 11:1408. doi:10.3390/cancers11101408
Li, N., Zeng, Y., and Huang, J. (2020). Signaling Pathways and Clinical Application of RASSF1A and SHOX2 in Lung Cancer. J. Cancer Res. Clin. Oncol. 146, 1379–1393. doi:10.1007/s00432-020-03188-9
Marchini, A., Ogata, T., and Rappold, G. A. (2016). A Track Record on SHOX: From Basic Research to Complex Models and Therapy. From Basic Res. Complex Models Ther. Endocr Rev 37, 417–448. doi:10.1210/er.2016-1036
Modhukur, V., Iljasenko, T., Metsalu, T., Lokk, K., Laisk-Podar, T., and Vilo, J. (2018). MethSurv: a Web Tool to Perform Multivariable Survival Analysis Using DNA Methylation Data. Epigenomics 10, 277–288. doi:10.2217/epi-2017-0118
Mortensen, K. H., Andersen, N. H., and Gravholt, C. H. (2012). Cardiovascular Phenotype in Turner Syndrome-Integrating Cardiology, Genetics, and Endocrinology. Endocr. Rev. 33, 677–714. doi:10.1210/er.2011-1059
Navarro Gonzalez, J., Zweig, A. S., Speir, M. L., Schmelter, D., Rosenbloom, K. R., Raney, B. J., et al. (2021). The UCSC Genome Browser Database: 2021 Update. Nucl. Acids Res. 49, D1046–d1057. doi:10.1093/nar/gkaa1070
Rhodes, D. R., Kalyana-Sundaram, S., Mahavisno, V., Varambally, R., Yu, J., Briggs, B. B., et al. (2007). Oncomine 3.0: Genes, Pathways, and Networks in a Collection of 18,000 Cancer Gene Expression Profiles. Neoplasia 9, 166–180. doi:10.1593/neo.07112
Schneider, K. U., Dietrich, D., Fleischhacker, M., Leschber, G., Merk, J., Schäper, F., et al. (2011). Correlation of SHOX2 Gene Amplification and DNA Methylation in Lung Cancer Tumors. BMC Cancer. 11, 102. doi:10.1186/1471-2407-11-102
Shi, J., Chen, X., Zhang, L., Fang, X., Liu, Y., Zhu, X., et al. (2020). Performance Evaluation of SHOX2 and RASSF1A Methylation for the Aid in Diagnosis of Lung Cancer Based on the Analysis of FFPE Specimen. Front. Oncol. 10, 565780. doi:10.3389/fonc.2020.565780
Srisuttee, R., Arayataweegool, A., Mahattanasakul, P., Tangjaturonrasme, N., Kerekhanjanarong, V., Keelawat, S., et al. (2020). Evaluation of NID2 Promoter Methylation for Screening of Oral Squamous Cell Carcinoma. BMC Cancer 20, 218. doi:10.1186/s12885-020-6692-z
Sun, C., Liu, X. H., and Sun, Y. R. (2019). MiR-223-3p Inhibits Proliferation and Metastasis of Oral Squamous Cell Carcinoma by Targeting SHOX2. Eur. Rev. Med. Pharmacol. Sci. 23, 6927–6934. doi:10.26355/eurrev_201908_18732
Tang, Z., Li, C., Kang, B., Gao, G., Li, C., and Zhang, Z. (2017). GEPIA: a Web Server for Cancer and normal Gene Expression Profiling and Interactive Analyses. Nucl. Acids Res. 45, W98–w102. doi:10.1093/nar/gkx247
Wang, Y., Zhang, L., Yang, J., Li, B., and Wang, J. (2018). CDH13 Promoter Methylation Regulates Cisplatin Resistance of Non-small Cell Lung Cancer Cells. Oncol. Lett. 16, 5715–5722. doi:10.3892/ol.2018.9325
Weiss, G., Schlegel, A., Kottwitz, D., König, T., and Tetzner, R. (2017). Validation of the SHOX2/PTGER4 DNA Methylation Marker Panel for Plasma-Based Discrimination between Patients with Malignant and Nonmalignant Lung Disease. J. Thorac. Oncol. 12, 77–84. doi:10.1016/j.jtho.2016.08.123
Wu, C., Li, M., Meng, H., Liu, Y., Niu, W., Zhou, Y., et al. (2019). Analysis of Status and Countermeasures of Cancer Incidence and Mortality in China. Sci. China Life Sci. 62, 640–647. doi:10.1007/s11427-018-9461-5
Xu, Z., Wang, Y., Wang, L., Xiong, J., Wang, H., Cui, F., et al. (2020). The Performance of the SHOX2/PTGER4 Methylation Assay Is Influenced by Cancer Stage, Age, Type and Differentiation. Biomarkers Med. 14, 341–351. doi:10.2217/bmm-2019-0325
Yang, X., Han, H., De Carvalho, D. D., Lay, F. D., Jones, P. A., and Liang, G. (2014). Gene Body Methylation Can Alter Gene Expression and Is a Therapeutic Target in Cancer. Cancer Cell 26, 577–590. doi:10.1016/j.ccr.2014.07.028
Zhang, C., Yu, W., Wang, L., Zhao, M., Guo, Q., Lv, S., et al. (2017). DNA Methylation Analysis of the SHOX2 and RASSF1A Panel in Bronchoalveolar Lavage Fluid for Lung Cancer Diagnosis. J. Cancer 8, 3585–3591. doi:10.7150/jca.21368
Zhang, C., Zhao, N., Zhang, X., Xiao, J., Li, J., Lv, D., et al. (2020) SurvivalMeth: a Web Server to Investigate the Effect of DNA Methylation-Related Functional Elements on Prognosis, Brief Bioinform. 22(3):bbaa162. doi:10.5194/essd-2020-182-rc1
Zhang, Y.-A., Zhou, Y., Luo, X., Song, K., Ma, X., Sathe, A., et al. (2016). SHOX2 Is a Potent Independent Biomarker to Predict Survival of WHO Grade II-III Diffuse Gliomas. EBioMedicine 13, 80–89. doi:10.1016/j.ebiom.2016.10.040
Zhang, Y., Breitling, L. P., Balavarca, Y., Holleczek, B., Schöttker, B., and Brenner, H. (2016). Comparison and Combination of Blood DNA Methylation at Smoking-Associated Genes and at Lung Cancer-Related Genes in Prediction of Lung Cancer Mortality. Int. J. Cancer 139, 2482–2492. doi:10.1002/ijc.30374
Zhao, Q. T., Guo, T., Wang, H-E., Zhang, X-P., Zhang, H., Wang, Z-K., et al. (2015). Diagnostic Value of SHOX2 DNA Methylation in Lung Cancer: a Meta-Analysis, Onco Targets Ther. 8, 3433–3439. doi:10.2147/ott.S94300
Keywords: SHOX2, methylation, expression, clinical parameter, LUAD, bioinformatic platform analysis
Citation: Li N, Zeng Y, Tai M, Lin B, Zhu D, Luo Y, Ren X, Zhu X, Li L, Wu H and Huang J (2021) Analysis of the Prognostic Value and Gene Expression Mechanism of SHOX2 in Lung Adenocarcinoma. Front. Mol. Biosci. 8:688274. doi: 10.3389/fmolb.2021.688274
Received: 30 March 2021; Accepted: 26 May 2021;
Published: 28 June 2021.
Edited by:
Hossain Shekhar, University of Dhaka, BangladeshReviewed by:
Magdalena Julita Orzechowska, Medical University of Lodz, PolandShilpita Karmakar, Independent Researcher, Rocky Hill, CT, United States
Copyright © 2021 Li, Zeng, Tai, Lin, Zhu, Luo, Ren, Zhu, Li, Wu and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jian Huang, 18665763598@163.com
†These authors have contributed equally to this work
 Nanhong Li
Nanhong Li Yu Zeng
Yu Zeng Min Tai4
Min Tai4  Biyun Lin
Biyun Lin Hongrong Wu
Hongrong Wu