- Department of Food Science, Institute for Postharvest and Food Sciences, Agriculture Research Organization Volcani Center, Rishon LeZion, Israel
Knowledge regarding bacterial dynamics during crop ensiling is important for understanding of the fermentation process and may facilitate the production of nutritious and stable silage. The objective of this study was to analyze the bacterial dynamics associated with whole crop wheat silage with and without inoculants. Whole crop wheat was ensiled in laboratory silos, with and without Lactobacillus inoculants (L. plantarum, L. buchneri), for 3 months. Untreated and L. plantarum-treated silages were sampled at several times during ensiling, while L. buchneri-treated silage was sampled only at 3 months. Bacterial composition was studied using next generation sequencing approach. Dominant bacteria, before ensiling, were Pantoea (34.7%), Weissella (28.4%) and Pseudomonas (10.4%), Exiguobacterium (7.8%), and Paenibacillus (3.4%). Exogenous inoculants significantly affected bacterial composition and dynamics during ensiling. At 3 months of ensiling, Lactobacillus dominated the silage bacterial population and reached an abundance of 59.5, 92.5, and 98.2% in untreated, L. plantarum- and L. buchneri-treated silages, respectively. The bacterial diversity of the mature silage was lower in both treated silages compared to untreated silage. Functional profiling of the bacterial communities associated with the wheat ensiling demonstrated that the abundant pathways of membrane transporters, carbohydrate and amino acids metabolisms followed different pattern of relative abundance in untreated and L. plantarum-treated silages. Only three pathways, namely base-excision repair, pyruvate metabolism and transcription machinery, were significantly different between untreated and L. buchneri-treated silages upon maturation. Lactic acid content was higher in L. plantarum-treated silage compared to untreated and L. buchneri-treated silage. Still, the pH of both treated silages was lower in the two Lactobacillus-treated silages compared to untreated silage. Aerobic stability test demonstrated that L. plantarum-, but not L. buchneri-supplement, facilitated silage deterioration. The lower aerobic stability of the L. plantarum-treated silage may be attributed to lower content of acetic acid and other volatile fatty acids which inhibit aerobic yeasts and molds. Indeed, high yeast count was recorded, following exposure to air, only in L. plantarum-treated silage, supporting this notion. Analysis of bacterial community of crop silage can be used for optimization of the ensiling process and the selection of appropriate inoculants for improving aerobic stability.
Introduction
Ensiling of forage crops is widely practiced worldwide in order to ensure continuous feed supply for ruminant livestock. The ensiling process comprises rapid achievement of anaerobic conditions and anaerobic fermentation of moist forage crops whereby lactic acid bacteria (LAB) convert water-soluble carbohydrates into organic acids, mainly lactic acid, resulting in forage acidification. Accumulation of LAB during the fermentation results in rapid pH decline that enables forage preservation for long time. Basically, ensiling fermentation process comprises four main stages: At initial stage, air is still present between the crop particles and the pH value is above 6.0. At this stage a variety of microorganisms are active. Secondly at the stage of fermentation, air is depleted through the activity of aerobic microorganisms and LAB become the dominant flora. At this stage the pH decreases to a value depending on forage composition and ensiling conditions. After successful fermentation, silage remains stable, as long as no air penetrates into it. At this stage sometimes, lactic acid is converted gradually to acetic acid by hetero-lactic bacteria, such as Lactobacillus buchneri (Weinberg and Chen, 2013). The last stage is unloading for feeding, when the silage is re-exposed to air and aerobic microorganisms, mainly yeasts and molds are activated (Weinberg and Muck, 1996). Anaerobic conditions and lactic acid formation, along with other volatile organic acids produced in the course of ensiling, restrict the growth of spoiling microbes e.g., yeasts, molds and other bacteria. LAB play a pivotal role in the ensiling process and many LAB containing additives are commercially available and used to enhance and improve the ensiling process, as well as, aerobic stability (Cai et al., 1998; Xie et al., 2012). LAB species commonly used to facilitate the ensiling fermentation belong to the genera Lactobacillus, Lactococcus, Enterococcus, and Pediococcus (Pahlow et al., 2003).
Ensiling has become a common global practice for feed preservation, especially in wet climates where the conservation of dried forage is problematic (Pahlow et al., 2003). The major forage crops worldwide, preserved in the form of silages, include whole-crop corn, alfalfa and various grasses (Wilkinson et al., 2003). Preservation of forages in the form of silages is a well-practiced in the temperate regions of Europe and North America, however, it is attaining a widespread popularity recently in tropical regions, too (Wilkins and Wilkinson, 2015). In Israel with semi-arid climate, wheat, which is grown on winter rains, is the major forage for ensiling. It is harvested in early spring at the milk stage of maturity when yields, ensiling characteristics and nutritional value are the optimum (Ashbell et al., 1997). It is recommended that the whole crop is chopped into 1–2 cm pieces and ensiled at 320–380 g/kg of dry matter (DM) (Weinberg et al., 2009). Ensiling of wheat is advantageous as wheat plant contains high dry matter and water soluble carbohydrate, and it can be grown under wider climatic and soil conditions (Ni et al., 2015). The wheat silages were reported to increase milk production of cows when used as an alternative of corn silages (Wang et al., 2013).
Ensiling is a natural preservation process, which involves complex microbial interactions. A significant effort is required for detailed characterization of the ecology of the microorganisms involved in the ensiling process and associated spoilage (McAllister et al., 2018). Most of the earlier studies related to microbial ecology associated with silages were performed using classical microbiological techniques. Later studies also used molecular techniques, e.g., Real-Time PCR, DGGE, 16S rRNA gene sequencing of isolates, and sequencing of 16S rRNA gene clone library to study the microbial ecology of silages (Stevenson et al., 2006; Parvin and Nishino, 2009, 2010; Pang et al., 2011; McGarvey et al., 2013). The application of next generation sequencing approach in the study of microbial ecology of silages is relatively new and enabled the identification of the microbial taxa associated with grasses (Eikmeyer et al., 2013; Li et al., 2015; Kasmaei et al., 2016; Zhao et al., 2017), oat (Romero et al., 2017), small grains forages (Duniere et al., 2017), soybean (Ni et al., 2017), alfalfa (Ogunade et al., 2018), corn (Kasmaei et al., 2016; Strauber et al., 2016; Zhao et al., 2016; Gharechahi et al., 2017; Keshri et al., 2018; Xu et al., 2018), and drumstick leaves (Wang et al., 2018). Compared to other silages, studies addressing the microbial populations associated with wheat silage are limited. Only few reports are available on the microbial community associated with wheat silages, which are based on cultured isolates and their molecular identification (Bal and Bal, 2012; Ni et al., 2015). In a more recent study, next generation sequencing approach has been applied for the analysis of the bacterial community of mature commercial wheat and corn silages in large bunker silos (Kraut-Cohen et al., 2016). Although, ensiling of wheat is attaining high popularity, in recent years, due to the high nutrient content of wheat and its wider availability (Ni et al., 2015), knowledge on the bacterial dynamics and the metabolic pathways associated with wheat ensiling process is still lacking. Furthermore, knowledge on the interplay between L. plantarum and L. buchneri, two common silage additives, and the bacterial population of wheat silage, is also missing.
Materials and Methods
Ensiling Experiment
Whole-plant wheat harvested at the soft dough maturity stage was chopped to 2 cm pieces and ensiled in 1.5 l anaerobic jars (Weck, Wehr, Germany). Silage treatments included natural microbiome without additives (Untreated) and addition of L. plantarum MTD1 (Ecosyl, Stokesley, United Kingdom) (Lp-Treated). The inoculant was applied at a final concentration of ca. 106 CFU/g crop by suspending an adequate amount of the bacteria (based on its concentration in the inoculant product) in 25 ml distilled water and spraying the suspension on 10 kg chopped wheat spread on 2 × 1 m plastic sheeting, followed by thorough mixing. Untreated freshly chopped wheat crops were sampled in triplicates, before ensiling (time 0). Triplicate jars of the untreated and Lp-treated silages were sampled, at 6 h after ensiling, and on days 1, 2, 7, 15, 30, and 90. A similar triplicates of ensiling jars was inoculated with L. buchneri (Ecosyl, Stokesley, United Kingdom) (Lb-treated), at a final concentration of ca. 106 CFU/g. Lb-treated- jars were sampled only after 90 days of ensiling in order to compare the effects of L. plantarum and L. buchneri inoculants on the quality and bacterial community of mature silage. Following sampling on day 90, the silages were subjected to an aerobic stability test for five additional days, as described before (Weinberg et al., 2009). Visual appraisal, production of CO2, change in pH and numbers of yeasts and molds served as spoilage indicators.
Analyses of Silage Characteristics
Dry matter (DM) was determined in triplicate by oven drying for 48 h at 60°C. A mixture 40 g of silages in 360 ml of distilled water was stomached for 3 min (Bagmixer, Interscience, France), the filtered aqueous extract was used for estimation of lactic acid content, ethanol and acetic acid. Lactic acid content was determined by the spectrophotometric method of Barker and Summerson (1941). Ethanol and acetic acid were determined with a gas chromatograph equipped with a semi capillary FFAP (nitroterephthalic acid-modified polyethylene glycol) column (Hewlett Packard, Waldborn, Germany), over a temperature range of 40–230°C. Microbiological evaluation included enumeration of LAB on Rogosa and MRS agar plates (Oxoid, Basingstoke, United Kingdom), yeasts and molds were enumerated on malt extract agar plates (Difco, Detroit, MI, United States) acidified to pH 4.0 with lactic acid. The plates were incubated at 30°C for 3 days.
Bacterial DNA Isolation
Untreated and inoculant-treated samples, collected at the different time points, were stored at −20°C before further analysis. Isolation of bacterial DNA and purification were performed as described previously (Keshri et al., 2018). Silage samples (10 g) were mixed with 40 ml each of sterilized double-distilled water (SDDW) and vortexed for 2 min. The mixture was filtered using cotton mesh to remove larger plant material and the liquor was centrifuged at 12,800 × g for 30 min at 4°C. The supernatant was discarded and pellet was used for microbial DNA isolation using PowerSoil DNA Isolation Kit (Mo Bio Laboratories, Inc., United States), following manufacturer’s protocol. The quality and quantity of the DNA were assessed at A260/280 nm by NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, United States). The purified DNA was stored at −20°C until used.
Library Preparation and Sequencing
Bacterial identification was performed using amplicon sequencing of 16S rRNA gene. PCR amplification was carried out to amplify the V4–V5 conserved regions of bacterial 16S rRNA gene sequences using primers 515F and 926R (Caporaso et al., 2011). Library preparation, pooling and sequencing were performed at the DNA Services facility, Research Resources Center, University of Illinois at Chicago. Briefly, genomic DNA was PCR amplified using a two-stage targeted amplicon sequencing protocol (Green et al., 2015). DNA library preparation and sequencing were performed as previously described (Keshri et al., 2018). Sequence data were received as fastq files and submitted to the National Center for Biotechnology Sequence Read Archive under Bio Project accession number PRJNA497711.
Sequence Data Analysis
Sequence processing was performed using Mothur software, version 1.39.1 (Schloss et al., 2009). Sequence analysis of each combined single fastq file was processed using the Mothur MiSeq SOP accessed on the 3/1/18 (Kozich et al., 2013). The paired-end MiSeq Illumina reads targeting bacterial community from the 57 wheat silage samples were aligned and converted to contigs yielding 3,360,079 reads. Sequences were quality checked and sequences having ambiguous characters, homopolymers longer than 8 bp were removed. Unique sequences and their frequency in each sample were identified and then a pre-clustering algorithm was used to further denoise sequences within each sample. Singletons were removed from the dataset and chimera removal was performed using the UCHIME algorithm (Edgar et al., 2011). Finally, classification of the high quality filtered bacterial sequences (1,869,232) was done by the Mothur version of Bayesian classifier (Wang et al., 2007) with the full length sequences and taxonomy references from NR SILVA database of release 123 (Pruesse et al., 2007). The operational taxonomic unit (OTU) was assigned at 97% identity of the sequences. Rarefaction was performed on each sample to assess sampling adequacy, using a 100 sequence increment. After normalizing the number of sequences in each sample (based on rarefied or subsampled data i.e., the minimum number of remaining sequences in any of the samples; 2200 sequences); alpha diversity was assessed by calculating the richness estimator Chao1 and the Shannon diversity index. Library coverage and species evenness were also calculated. The taxonomic classification and diversity indices of samples from respective replicates were averaged. Beta diversity (variation in community structure between samples) was determined using the same subsampling approach, an index that accounts for proportional abundances of both shared and non-shared OTUs.
Statistical Analysis
Statistical analysis of silage parameters, relative abundances and diversity indices was performed by Analysis of Variance (ANOVA) and pairs Tukey HSD test using JMP Pro 13.0.0 software (SAS, Cary, NC, United States). Two dimensional non-metric multidimensional scaling (NMDS) plots were generated based on the weighted unifrac distance matrix calculated between different samples using the OTU table with OTU clustering at the species level (Lozupone et al., 2006). Differences in bacterial community composition among the samples were assessed using Unifrac weighted distance metric.
Functional Profiling of the Bacterial Community Associated With Wheat Silage
Phylogenetic investigation of communities by reconstruction of unobserved states (PICRUSt, release 1.0.0) was used to predict the functional profiling based on the 16S rRNA gene sequences of the bacterial communities associated with the wheat ensiling process (Langille et al., 2013). For this analysis, OTUs were closed-reference picked against the Greengenes database by Mothur (v.1.39.1). The functional taxonomies were normalized, predicted, and categorized according to online protocols of PICRUSt1 to generate the predicted KEGGs (Kyoto encyclopedia of genes and genomes) pathways Orthology (KO) classification (Kanehisa et al., 2008). The final predicted metagenome (a biom table), is analyzed with a software package, Statistical Analysis of Taxonomic and Functional Profiles (STAMP) v.2.0.9 (Parks et al., 2014) to test and visualize significant predicted functional differences in bacterial communities between the different ensiling times and type of treatments.
Results
Silage Characteristics
The DM content of fresh wheat was 456 ± 6 g/kg and decreased after 90 days of ensiling to 443 ± 4 g/kg. The DM content of Lp- and Lb-treated silages at 90 days, were 455 ± 5 g/kg and 420 ± 9 g/kg, respectively. The initial pH of the freshly chopped wheat samples was 6.5, and it decreased to 4.7 in untreated samples after 90 days of ensiling. However, the pH values of the Lp-treated silages decreased faster and reached a value of 4.2 as early as 7 days and further decreased to pH 4.0 following 15 days of ensiling (Figure 1). The pH of Lb-treated silages was 4.3 after 90 days of ensiling. Microbial, chemical and visual parameters of the silages after 90 d of ensiling as well as after air exposure are summarized in Table 1. Lactic acid content were significantly different among the three silages and was highest in the Lp-treated silages. Acetic acid contents were significantly higher in Lb-treated silages as compared with both untreated and Lp-treated silages. Ethanol concentration was similar in untreated and Lb-treated silages, but was significantly lower in Lp-treated silages. Lactobacillus counts were significantly higher in Lb-treated silages and molds were not detected in any of the treatments. Yeast counts were significantly higher in untreated than the Lp-treated silages, and were below the detection level in Lb-treated silages (Table 1). The aerobic stability test indicated that untreated- and Lb-treated silages were more stable than Lp-treated silage, as evident by the higher counts of yeast and molds, as well as CO2 produced in the latter case.
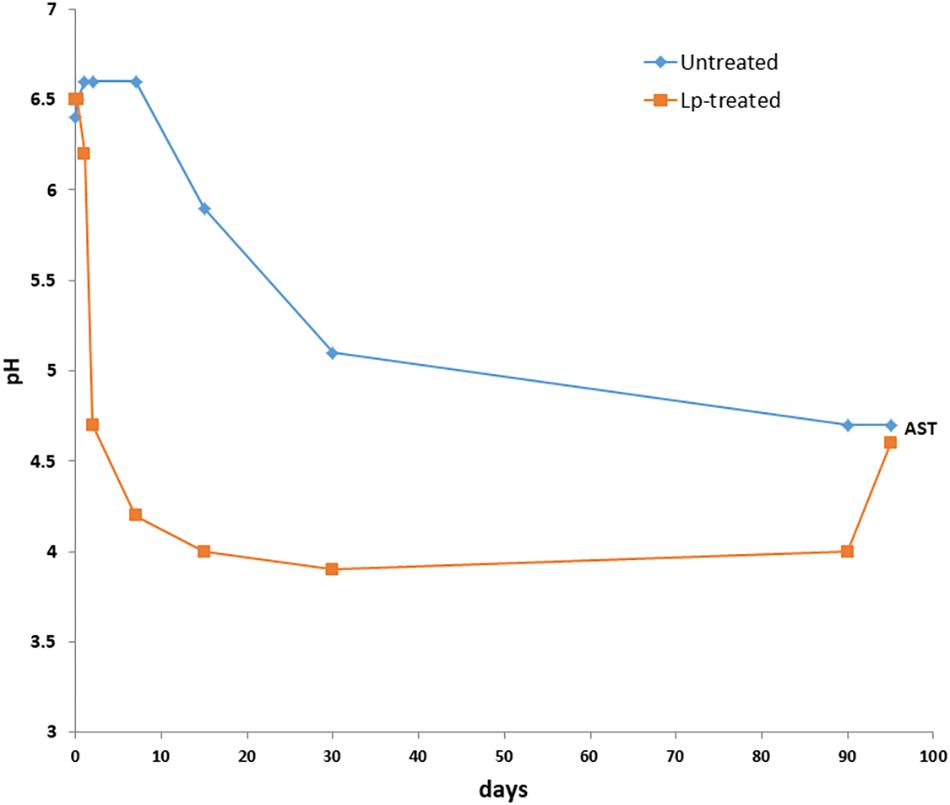
Figure 1. Changes in pH of wheat silage during the course of ensiling with and without L. plantarum-supplement. AST denotes 90 days mature silages followed by 5 days aerobic stability test.
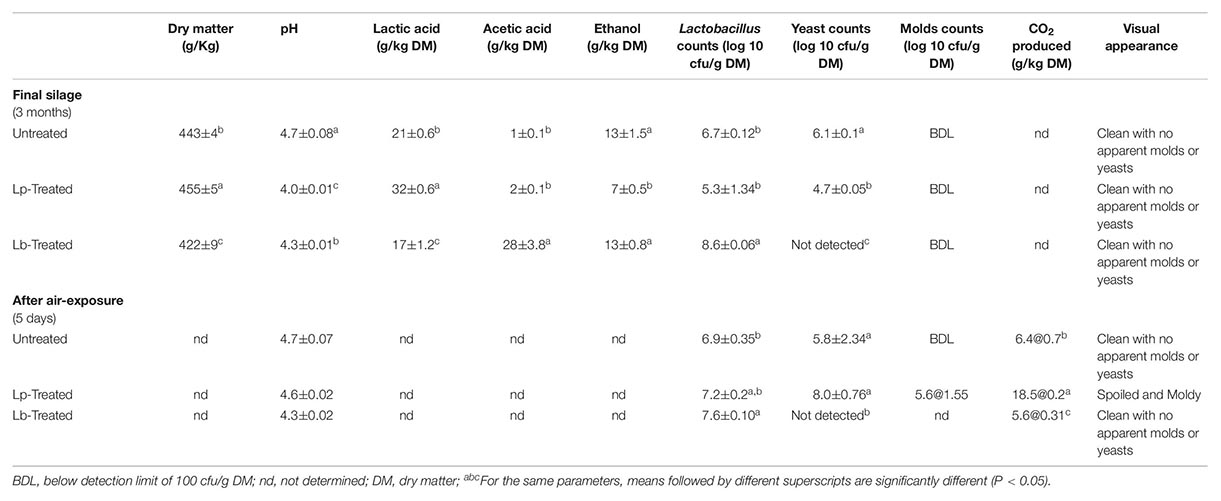
Table 1. Characteristics of the final wheat silages (90 days) and after aerobic exposure for 5 days.
Bacterial Richness and Diversity During Wheat Ensiling
Overall, 1,869,232 quality filtered 16S rRNA sequences were clustered in to 645 OTUs. The library coverage was more than 99% in all samples. In untreated silage, the richness initially increased and reached its maximum value (91.7 ± 2.1) at 6 h of ensiling (Table 2). The richness then declined and reached its lowest value (33.2 ± 4.07) at day 7, followed by gradual increment up to day 30 (57.2 ± 1.34). A similar initial trend is observed in Lp-treated silage; the relatively high initial richness (70.4 ± 4.64 at 6 h) is declined until day 7 (36.5 ± 1.4). However, the richness then sharply increased to a peak (75.9 ± 1.35) at day 15 and then sharply decline up to day 30 (44 ± 1.17), and stays almost constant until maturation at day 90 (Table 2). In both untreated and Lp-treated silages, richness were almost same at the terminal stage, but the diversity was significantly higher (P < 0.05) in case of untreated silages (Table 2). In Lb-treated silages, the richness and diversity at the terminal stage were significantly lower (P < 0.05) than both untreated and Lp-treated silages (Table 2).
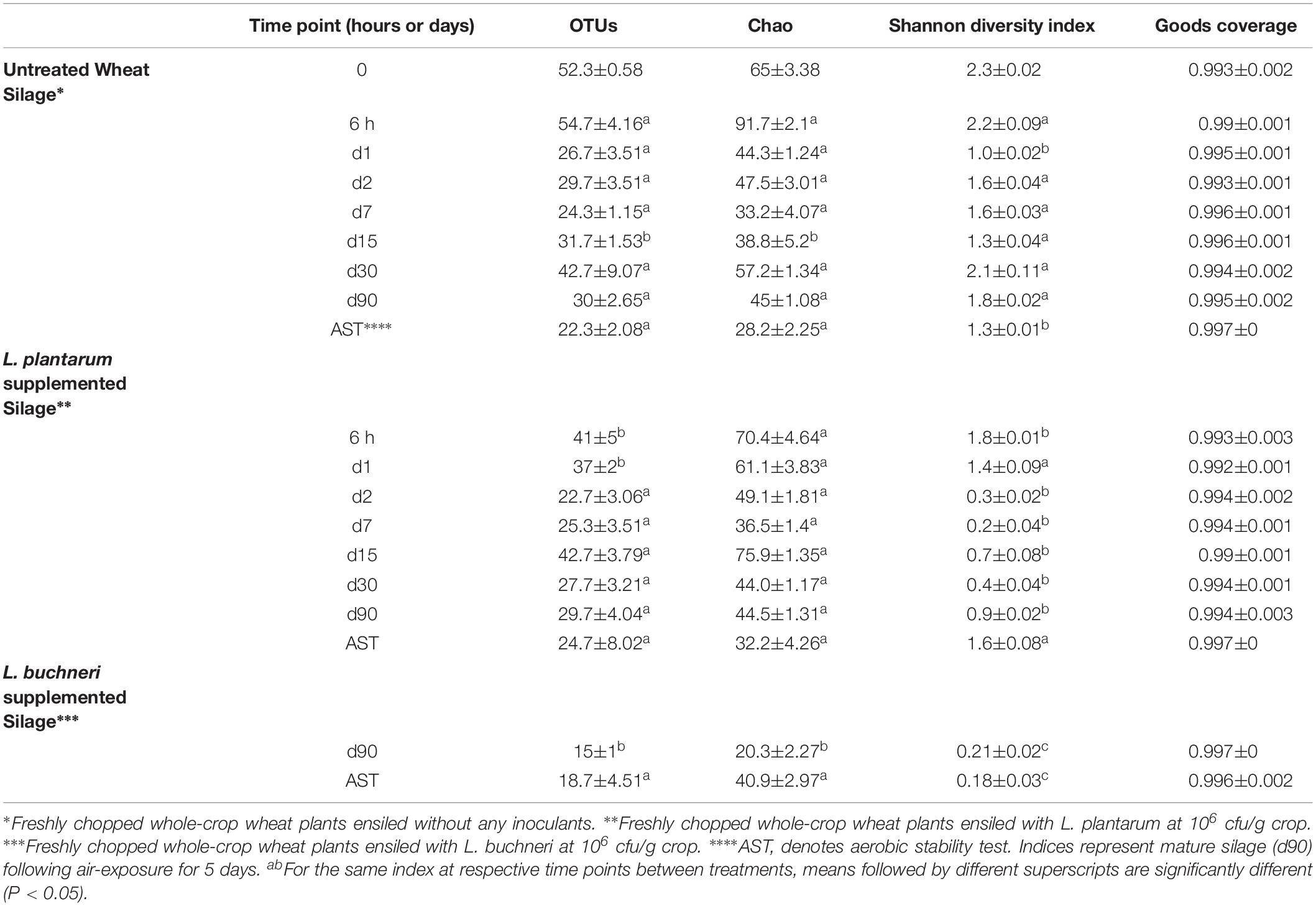
Table 2. Diversity indices for bacterial communities associated with Wheat silage samples at different time points.
The diversity of untreated silage was maximal (2.3 ± 0.02) at time 0 and lowest at day 1 (1 ± 0.02). The diversity then increased to (1.6 ± 0.04) at day 2, decreased up to day 15 and sharply increased up to day 30 of ensiling. The diversity of Lp-treated silage was lower than the untreated silage at 6 h, and continued to decrease up to day 7 (0.2 ± 0.04). The diversity then increased up to day 15 (0.7 ± 0.08) when it decreased until day 30 (0.4 ± 0.04). A gradual increase in the diversity (from 0.4 ± 0.04 to 0.9 ± 0.02) was observed until day 90. The diversity of untreated silage is significantly higher (P < 0.05) than that of Lp-treated silage during the entire ensiling from day 2 to day 90 (Table 2).
The two-dimensional NMDS plot depicts different clusters of similar bacterial community composition (BCC) during various ensiling times (Figure 2). A remarkable spatial variation exists in untreated silage between pre-ensiled (time 0) and mature silage (day 90). There is also a significant variation in BCC of mature silage in untreated, Lp- and Lb-treated silage. Exposure of the mature silage to air, for 5 days, resulted in considerable changes in the BCC of untreated and Lp-treated samples, but not in the BCC of Lb-treated silage. Weighted Unifrac metric analysis (Supplementary Table S1), based on OTU abundance, shows no significant changes in BCC of untreated silage from time 0 (fresh chopped wheat) to day 1. Significant changes in BCC was observed during ensiling of both untreated and Lp-treated silages (day 2, 7, 15, 30, 90, and 6 h, day 1, 7, 30, 90, respectively). The BCC of mature silages at 90 day, differed significantly in untreated, Lp- and Lb-treated silages.
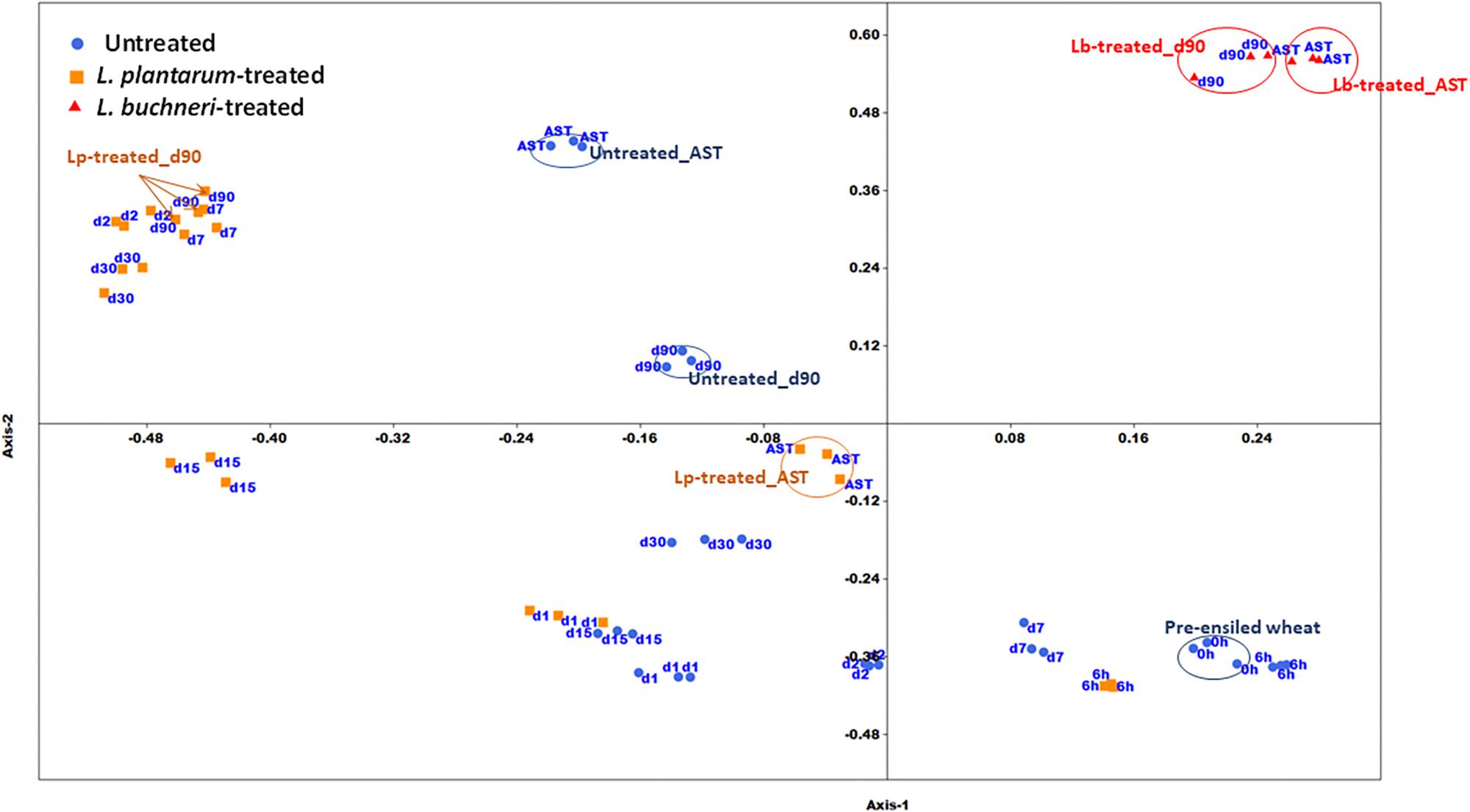
Figure 2. Non-metric Multidimensional Scaling plot of bacterial community composition in untreated and inoculants’ treated silages. AST denotes mature silage samples after aerobic exposure test for 5 days; h and d represents hour and days of ensiling, respectively. Arrows and circles indicate similar treatments/time points.
Bacterial Population Dynamics During the Course of Ensiling
Proteobacteria were the dominant population in pre-ensiled wheat (time 0), with an abundance of 53.6%, and their abundance decreased during ensiling down to 0.12, 2, and 0.7% in untreated, Lp- and Lb-treated silages, respectively. The Firmicutes dominated the mature silage population with an abundance of 99.8, 97.9, and 99.2% in untreated and Lp- and Lb-treated silages, respectively (Figure 3). Overall, the Firmicutes and Proteobacteria were the most abundant phyla, at any time point during ensiling, and constituted together more than 98% of relative abundances.
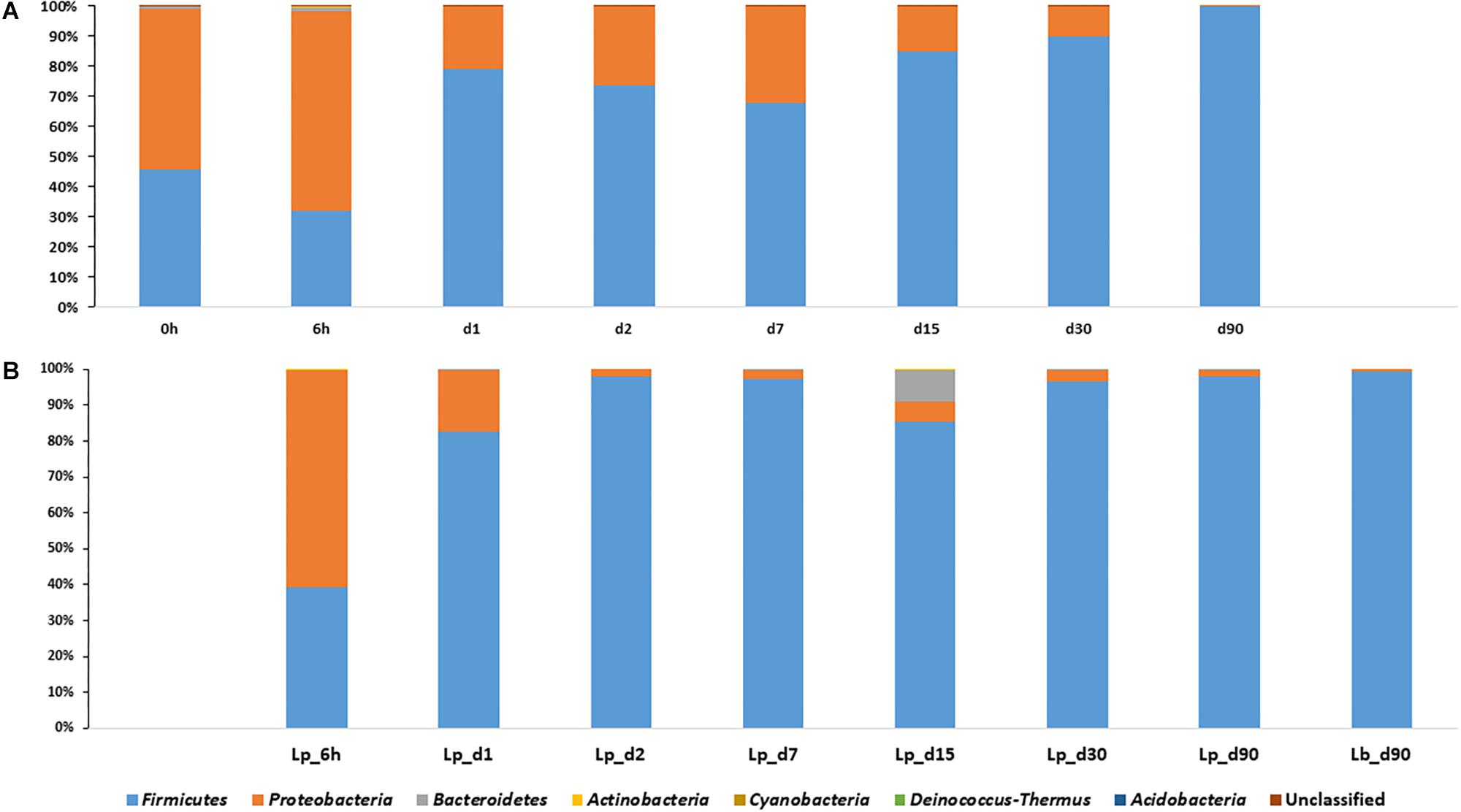
Figure 3. Dynamics of bacterial phyla in course of wheat ensiling without inoculant (A), and with L. plantarum- and L. buchneri-supplement (B).
The dominant genera in freshly chopped, pre-ensiled wheat plants, were Pantoea (34.7%), Weissella (28.4%), Pseudomonas (10.4%), Exiguobacterium (7.4%), and Paenibacillus (3.4%). After 6 h of ensiling, the abundance of Pantoea increased to 46.3%, but then decreased continuously during silage maturation and at the terminal stage of 90 days they represented only 0.02% of the overall population. The relative abundance of Weissella kept on fluctuating during the course of ensiling. Pseudomonas decreased continuously during ensiling and was not found in mature silage. Enterococcus increased significantly at day 7 where it represented 33.9% of relative abundance and then decreased to reach 1.8% in mature silage. Lactobacillus was initially presented at very low abundance (0.53%), which decreased after 6 h of ensiling to 0.09%, but then increased continuously up to 59.5% relative abundance in mature silage (Figure 4A).
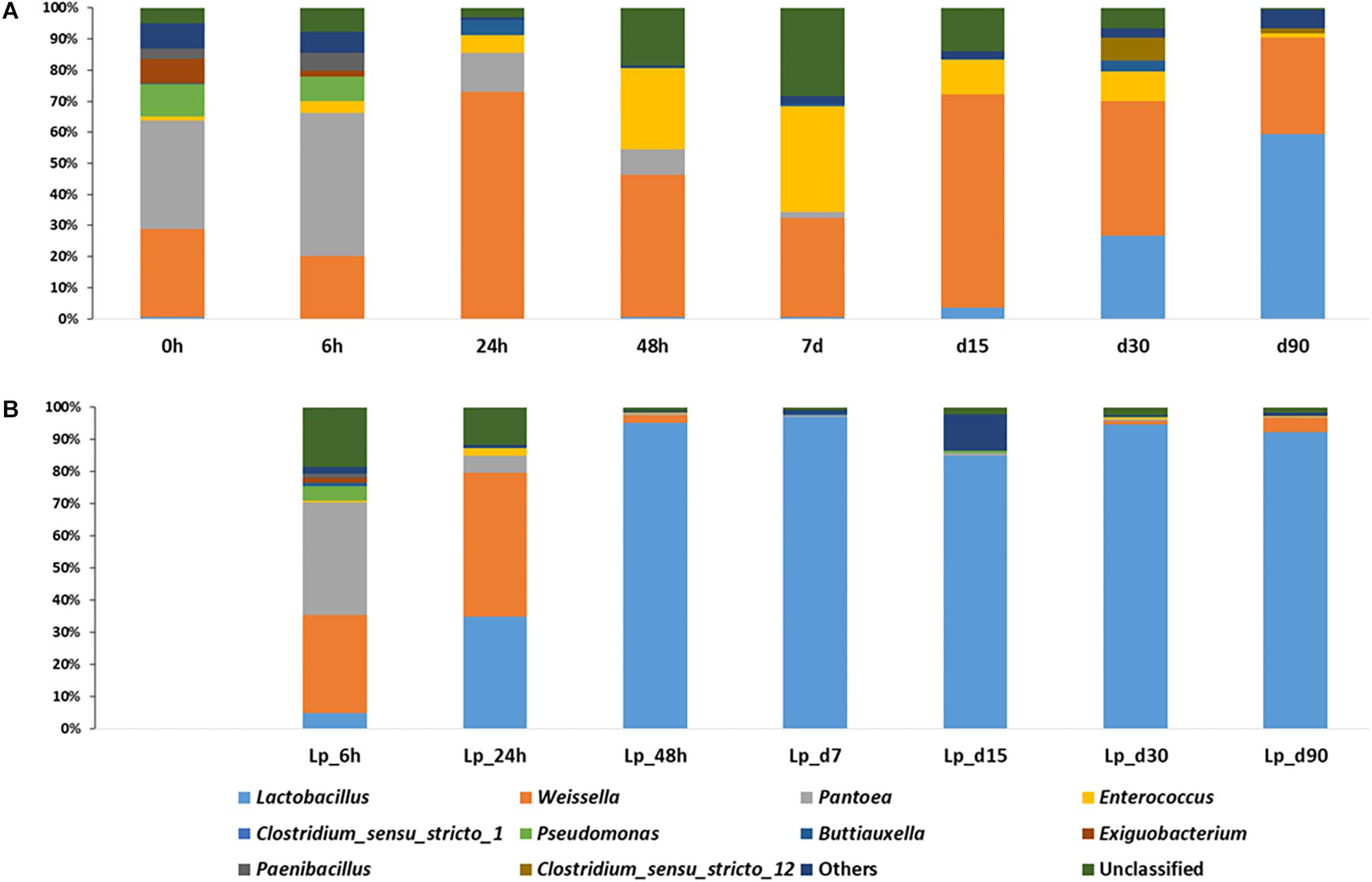
Figure 4. Dynamics of abundant bacterial genera during the course of ensiling in (A) untreated silage and (B) L. plantarum-treated silage. Ten most abundant genera are presented, whereas all other identified genera were presented as “Others.”
In Lp-treated silage, Pantoea remained the most abundant genus (34.9%) after 6 h of ensiling, then it decreased during ensiling up to 0.16% at the terminal stage. Weissella abundance was increased at 6 h (30.5%) and on day 1 (44.7%), then declined and in the mature silage it was represented by 4.4% of the bacterial populations. Similar to untreated samples, Pseudomonas decreased during ensiling and was not found at 90 days of ensiling. The relative abundance of Enterococcus was maximum at day 1 (2.3%), and then decreased to reach 0.28% at 90 days. Lactobacillus species were in low abundance (4.8%), at 6 h of incubation, and rapidly increased to 34.8% at day 1, and at day 2, they already encompassed more than 95% of the population. Their relative abundance was maximum at day 7 (96.8%), and dropped to 92.4% at 90 days (Figure 4B).
Effect of Inoculants on Bacterial Composition in Mature Silage
There was a significant difference in the relative abundance of Lactobacillus sp. between Lp- and Lb-treated and untreated silages at the terminal stage of ensiling (P < 0.05) (Figure 5A). Although Lactobacillus was the most abundant genus in all treatments, it encompassed only 59.5% of the population, in untreated silage, followed by Weissella (31%), Pediococcus (5.8%), Clostridium_sensu_stricto_12 (1.6%), Enterococcus (1.1%). In contrast, the abundance of Lactobacillus in treated silages was much higher. In Lp-treated silage, Lactobacillus species represented 92.4% of the entire bacterial population followed by Weissella (4.4%) and Enterococcus (0.28%). In Lb-treated silages, Lactobacillus were the most dominant genus covering 98.2% of the population followed by Clostridium_sensu_stricto_12 (0.7%) and Enterococcus (0.1%) (Figure 5A). Only 21 genera were detected in untreated mature silage, while 31 and 38 genera were identified in Lp- and Lb-treated silages, respectively (Supplementary Table S2).
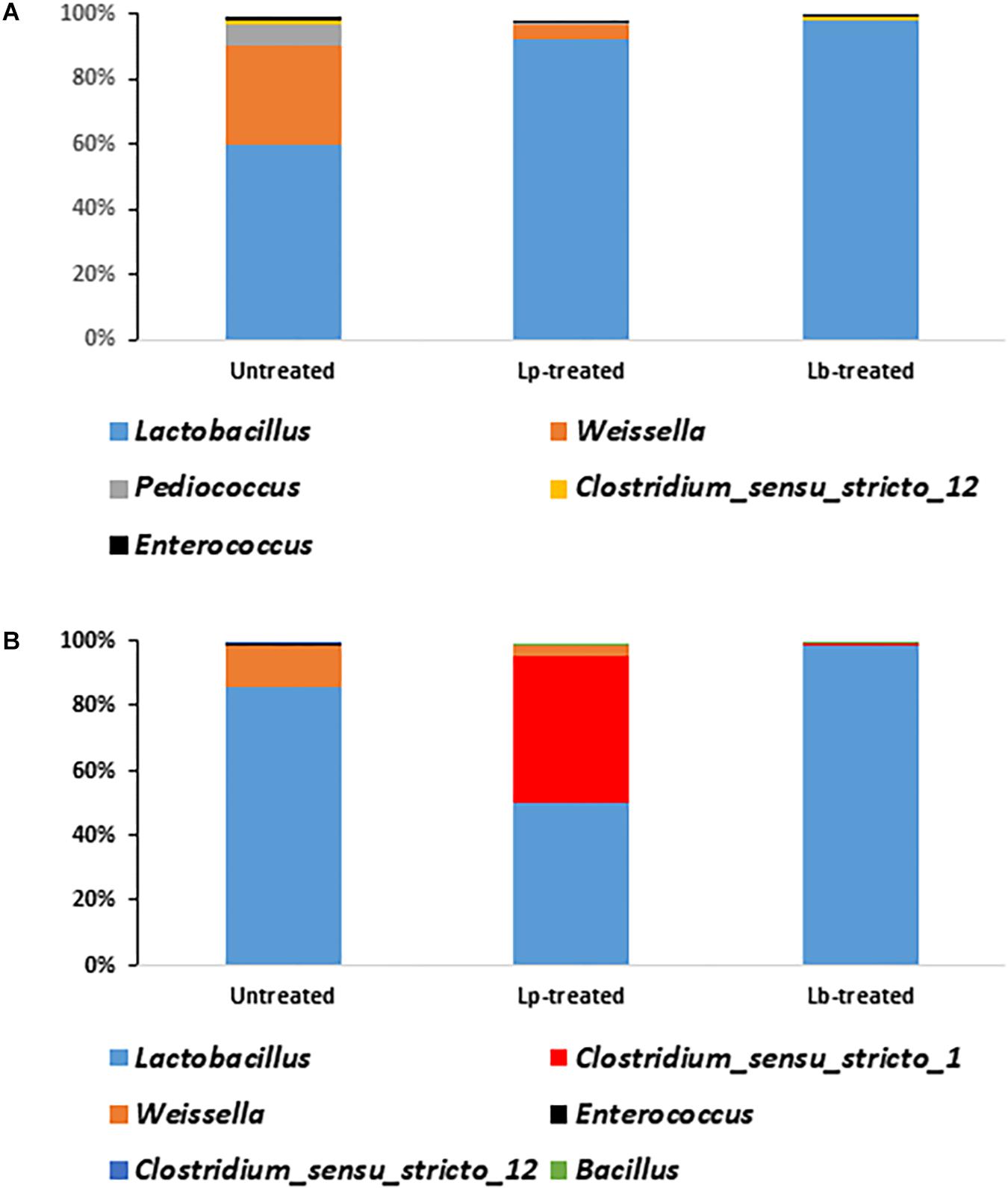
Figure 5. Dominant bacterial genera in wheat silages (A) after 3 months of ensiling and (B) after aerobic stability test. Untreated denotes wheat silage ensiled with no supplement, Lp-treated and Lb-treated denotes wheat silage ensiled in the presence of L. plantarum, and L. buchneri, respectively. Only genera with relative abundance > 0.1% are presented.
Bacterial Dynamics During Aerobic Stability Test
To test silage stability, Lp-, Lb-treated and untreated mature silages, were air-exposed for 5 days and their quality parameters were analyzed. The bacterial richness of the untreated and Lp-treated silages slightly decreased during exposure to air, whereas it increased in Lb-treated silage (Table 2). The diversity decreased in the untreated and Lb-treated silages, while it increased in Lp-treated silage (Table 2). After exposure to air, Lactobacillus remained the most dominant population in all silages (Figure 5B). In untreated silage, Lactobacillus significantly increased from 59.5 to 85% (P < 0.05), while in Lb-treated silage, its abundance remained constant (98%) after air exposure. A significant change was observed in Lp-treated silages, where Lactobacillus dropped from 92.4% in mature silage to 49.8% after air-exposure, whereas the genus Clostridium_sensu_stricto_1 increased from 0.03 to 45.3%. Other prevalent genera that were detected, in air-exposed untreated silage were Weissella (12.7%), Enterococcus (0.5%) and 12 other genera with relative abundance below 0.05%. In total, 16 and 17 genera were identified in untreated and Lp-treated silages, respectively, while 30 genera were detected in Lb-treated silages (Supplementary Table S3).
Predicted Function of Bacterial Community Associated With Ensiling
Understanding the metabolic pathways during crop’s ensiling is important for the optimization of the process. In order to gain knowledge regarding potential metabolic pathways that are involved in ensiling, the bacterial community data was analyzed by PICRUSt. Overall a total of 268 metabolic pathways were predicted in all silages using the Kyoto Encyclopedia of Genes and Genomes (KEGG). The most abundant predicted pathways, in pre-ensiled wheat, were (general) transporters (7.7%), ABC transporters (4.3%), DNA repair and recombination proteins (2.5%), two-component systems (2.4%), purine metabolism (2.1%), secretion systems (2.1%), transcription factors (2%), other ion-coupled transporters (1.7%), bacterial motility proteins (1.6%) and ribosome (1.6%). Variations in the abundance of the pathways were observed during ensiling and after aerobic exposure in both untreated and treated silages (Supplementary Table S4), and were visualized using heat map plot (Supplementary Figure S1).
The abundant pathways of membrane transporters (general and ABC transporters) and metabolisms (carbohydrate and amino acids) followed different pattern of relative abundance in untreated and Lp-treated silages (Figures 6–8). Comparison of mature silages (d90), showed 32 significantly different pathways among untreated, and Lp-treated silages, which have more than 0.001% of difference in their relative abundance at d90. These pathways belonged to the functions of membrane transporters, cell motility, carbohydrate-, amino acids-, energy-, nucleotide-, terpenoids and polyketides- metabolisms, glycan biosynthesis and metabolisms, signal transduction, folding, sorting and degradation, replication and repair, and translation (Supplementary Figure S2). Comparison of Lb-treated to non-treated control after 90 days showed only 3 pathways that were significantly different (>0.001%) between the two treatments (Supplementary Figure S3). The pathways belonged to base excision repair, pyruvate metabolism and transcription machinery.
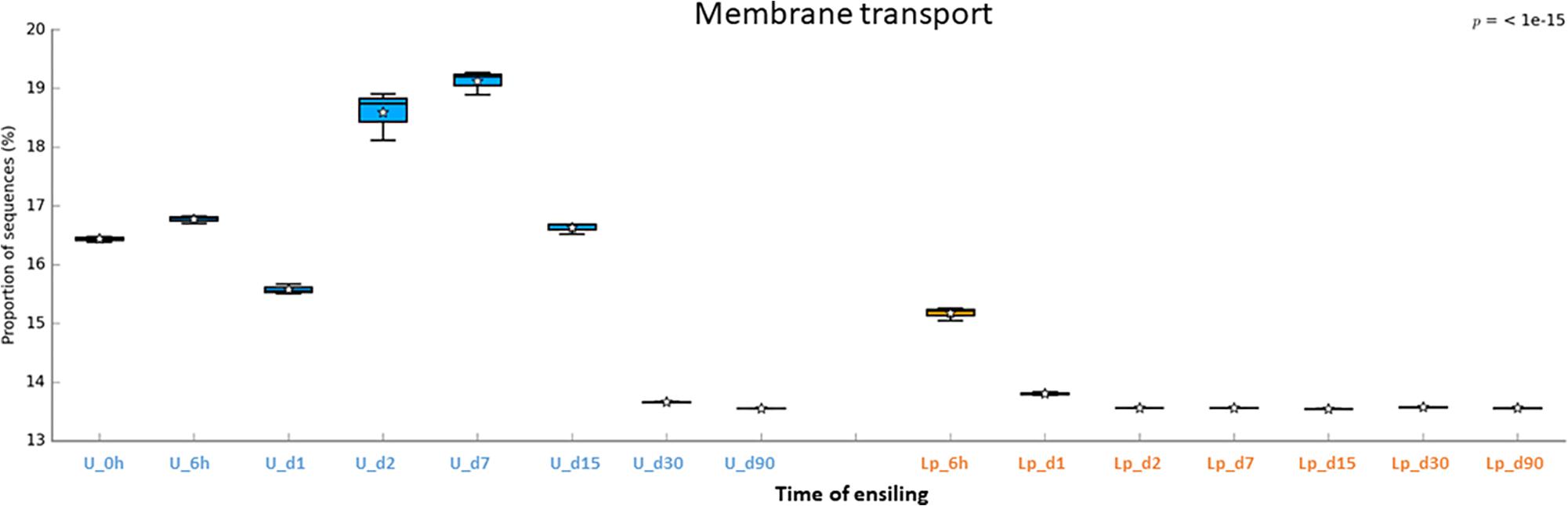
Figure 6. Functional prediction of membrane transporters during untreated and Lp-treated wheat ensiling using PICRUSt.
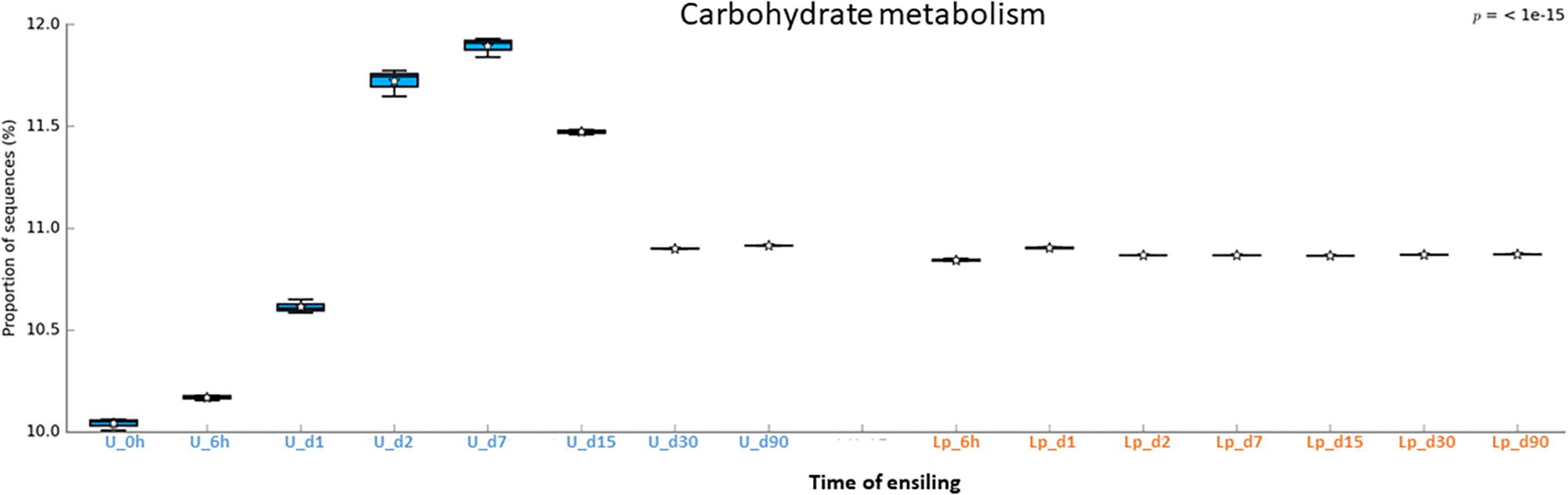
Figure 7. Functional prediction of carbohydrate metabolism during untreated and Lp-treated wheat ensiling using PICRUSt.
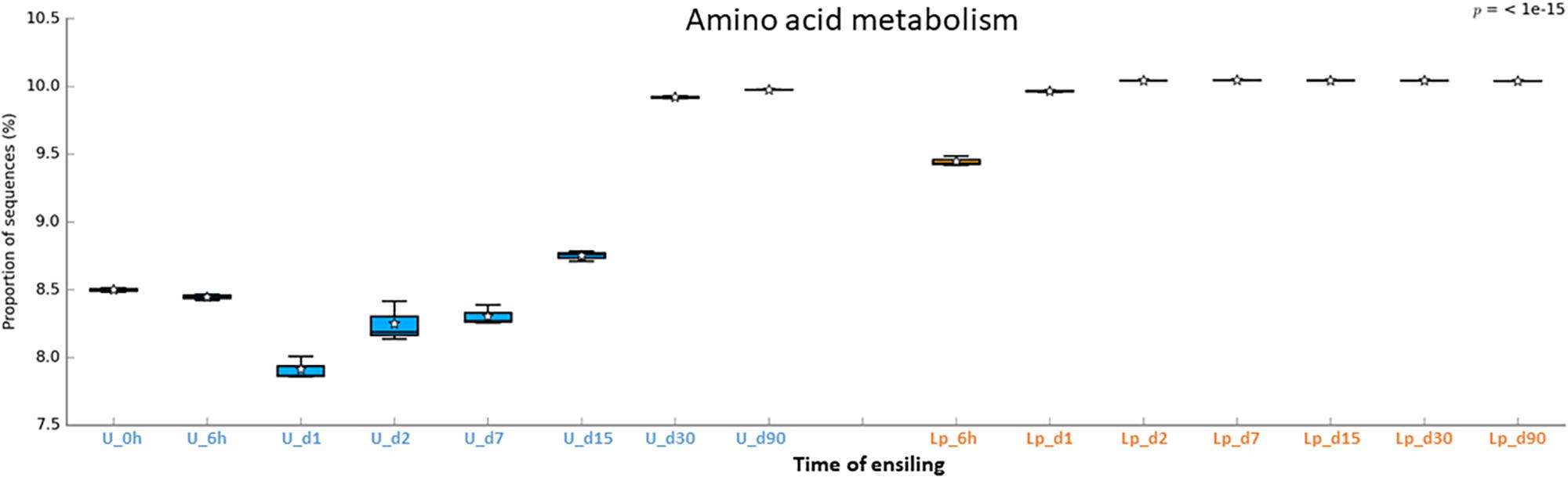
Figure 8. Functional prediction of amino acid metabolism during untreated and Lp-treated wheat ensiling using PICRUSt.
Discussion
Wheat is a major crop used for silage in Israel and China (Weinberg et al., 2009; Ni et al., 2015). While recent studies have analyzed the microbiome dynamics during ensiling of several other crops (Keshri et al., 2018; McAllister et al., 2018), the present study is the first to analyze the dynamics of wheat bacterial community in the course of ensiling, as well as during aerobic stability test. The data presented contribute basic knowledge regarding the bacterial players associated with high-quality wheat ensiling process, as well as regarding the effect of LAB-supplements on bacterial composition.
The DM content of the wheat used for silage preparation was above the recommended DM content 320–380 g/kg for optimal ensiling of wheat (Ashbell et al., 1997). However, since wheat for silages is harvested in Israel in spring, with abrupt changes in weather from cool to hot and dry, sometimes the wheat dries more than planned before the onset of ensiling (Weinberg et al., 2009). The high DM content of the wheat in the current study provided an opportunity to elucidate changes in bacterial community composition in a slow ensiling fermentation, and determine the effects of LAB inoculants under such conditions. The ensiling of the untreated wheat silages was rather slow and the final pH of untreated silage was 4.7, which is similar to the pH of alfalfa silage with a similar DM content (450 g/kg) (Santos and Kung, 2016). Wheat ensiling, with lower DM content (301 g/Kg), also resulted in different pH in untreated (pH 4.4) and Lp-treated (pH 3.8) silages after 56 days of ensiling (Chen and Weinberg, 2014). The pH of the Lp-treated wheat silages, decreased faster and to lower values of the pH. This is in accordance with the previous study performed on wheat ensiling where LAB inoculants have significant effect on decrease of pH and correlated to DM content (Pieper et al., 2011). It is possible that the high DM content of the untreated silage has limited the multiplication of the indigenous LAB of the wheat. In contrast, supplementation of the silage with exogenous L. plantarum inoculant overcame this effect and resulted in significantly faster and improved ensiling. The considerable fermentative activity of the inoculated LAB was also observed in wheat ensiling with high (884 g/kg) DM content (Pieper et al., 2011). DM content is an important parameter in the ensiling process, which may significantly influence the BCC during ensiling with and without inoculants (Pieper et al., 2011), as is evident by BCC of our ensiled wheat silage and, pH dropping that followed different pattern in both cases. Unlike wheat, corn ensiling with rather lower DM (381 ± 7.5 g/kg) resulted in a similar pattern of pH drop in both untreated and Lp-treated silages (Keshri et al., 2018). It is possible that the different patterns of pH change in untreated wheat and corn silages evolved from differences in the moisture and subsequent water activity of the two crops.
The application of NGS to gain insight into the ensiling process is relatively new and has already provided a better understanding regarding the complexity of the microbial ecology of ensiling, role of natural epiphytes and inoculants in high quality silage production (McAllister et al., 2018). The NGS approach is advantageous over culture-dependent and – independent techniques (Bokulich and Mills, 2012; Ni et al., 2017) and to the best of our knowledge, it has been used so far in only five studies to investigate bacterial population dynamics during ensiling. Four studies dealt with corn silages (Zhao et al., 2016; Xu et al., 2017, 2018; Keshri et al., 2018) and one analyzed Manyflower silvergrass silage (Li et al., 2015). In the present study NGS approach was applied for the first time to analyze bacterial population dynamics in wheat silage with and without Lactobacillus inoculants used as additives in commercial ensiling processes in order to facilitate the ensiling process (Weinberg et al., 1999; Filya, 2003; Jatkauskas et al., 2013; Arasu et al., 2014).
Nearly identical bacterial populations were observed in triplicate ensiled laboratory silos, at each time point, supporting the data robustness of the silos model, similar to previous reports regarding corn ensiling (Ni et al., 2017; Keshri et al., 2018). In untreated silage, the richness initially increased and reached its maximum value at 6 h of ensiling. This initial dynamics may reflect the multiplication of aerobic bacteria initially when oxygen was still available. The diversity of Lp-treated silage at 6 h was lower than the untreated silage, probably because of the high concentration of the added L. plantarum. OTUs richness and bacterial diversity were observed to fluctuate during the first weeks of ensiling with different patterns in untreated and Lp-treated silages. In the untreated silage, the reduction in OTU richness might be related to exhausting of available sugars consumed by yeasts, aerobic and facultative aerobic bacteria during the initial aerobic phase of the ensiling process. From d7 to d30, the richness increased, likely due to the development of LAB populations under the increased acidic conditions. In Lp-treated silages, Lactobacillus dominated from d2 of ensiling and imposed a rapid decrease in pH, which apparently inhibited the growth of acid-susceptible bacteria and facilitated the multiplication of selected acid-tolerant LAB population. Indeed, more genera were detected from d7 to d30 in control compared to Lp-treated silage.
Lower diversity in LAB treated silages as compared to untreated silages were also reported in rice straw and corn ensiling (Wang et al., 2016; Keshri et al., 2018), which can be attributed to the inhibitory effect of the LAB population upon the multiplication of other microbes, before the development of the indigenous LAB population (Keshri et al., 2018). A decrease in bacterial diversity upon ensiling was also observed in other crop silages and ensiling times using NGS (Eikmeyer et al., 2013; Kasmaei et al., 2016; Ni et al., 2017; Romero et al., 2017; Duniere et al., 2017).
The indigenous bacterial composition present on silage plants is determined by the type of the crop (Kasmaei et al., 2016; Duniere et al., 2017). In freshly chopped corn, Proteobacteria were reported to be the dominant population present at an abundance of 82.6%, and Firmicutes encompassed only 14.7% of the population (Keshri et al., 2018), while in pre-ensiled oat Firmicutes were the most abundant (83.6%), whereas Proteobacteria occupied only 16.2% of the population (Romero et al., 2017). In the case of wheat, the two phyla were represented in comparable abundance (53.5 and 45.7%) in the freshly chopped crop (time 0) and showed antagonistic relationship during ensiling.
Determination of plants bacterial populations may sometimes be biased by the methodology used (Lebeis, 2017). However, Since, the bacterial populations in freshly chopped corn (Keshri et al., 2018) and oat (Romero et al., 2017) plants was analyzed by the same NGS approach using Illumina’s MiSeq methodology, it is likely that the nature of the dominant phyla in each crop is attributed to the type of crop. Still it is possible that the microbial population on the plant might have been influenced by the ecology of the field, too (Berlec, 2012). Regardless of the nature of crop, and the methodology used to determine bacterial community, Firmicutes, and more specifically LAB were reported to dominate silages (McEniry et al., 2008; Parvin and Nishino, 2009; Bao et al., 2016; Duniere et al., 2017; Ni et al., 2017; Romero et al., 2017; Keshri et al., 2018), suggesting that anaerobic and low-pH conditions developing during ensiling, favor their multiplication and persistence in the course of silage maturation.
The Firmicutes genus mostly associated with mature silage is Lactobacillus (Eikmeyer et al., 2013; Li et al., 2015; Keshri et al., 2018; Xu et al., 2018). Species of this genus reached 59.5% relative abundance in untreated, mature wheat-silage, and encompassed a much higher abundance in Lp- and Lb-treated silages (92 and 98%, respectively). Notably, Lactobacillus spp. were comparatively more pronounced in the case of mature corn silage, where they encompassed more 94% of relative abundance in both untreated and Lp-treated silages (Keshri et al., 2018). This difference in response to Lp application between corn and wheat could be attributed to the differences in the DM content between the two crops. Indeed, in the corn ensiling experiment there was no difference in the rate of decrease of pH between untreated and Lp-treated silages, whereas in the dry wheat the pH decrease rate differed significantly between the control and Lp-treated silages. Using NGS approach, a high relative abundance of Lactobacillales was also reported in grass, wilted rye grass (Eikmeyer et al., 2013), and corn silage (Ni et al., 2017). Dominance of LAB (>80% of Lactobacillaceae) was also observed in three out of four good-quality wheat silages taken from commercial bunker silos, whereas less than 10% of Lactobacillus was present in the fourth sample (Kraut-Cohen et al., 2016). Thus, laboratory silos, used in this study, seem to provide a good model for studying the dynamics of bacterial community during commercial ensiling.
It is surprising that at the end of ensiling, fewer genera were detected in untreated mature silage compared to treated silages, since the high lactic acid content and the low pH conditions should have limited bacterial growth to acidophilic or acid-tolerant genera. The reason for this finding is not clear. Notably, in corn ensiling, an opposite trend was observed with fewer genera (22) found in Lp-treated silage as compared to the untreated silages (54 genera) after 3 months of ensiling (Keshri et al., 2018). Future studies using different crop should be employed to verify whether this phenomenon is unique to wheat silage or not.
In order to predict the potential metabolic pathways involved in ensiling of wheat crop, metagenomics functional prediction analysis was performed. The relative abundance of pathways was evaluated using KEGG classification (levels 1–3). Overall, the relative abundance of pathways (at level 3) varied in untreated compared to Lp-treated silages. Low variation of metabolic pathways is observed on days 30 and 90 in the untreated silage, as well as during all time points in the Lp-treated silage. These findings are likely related to the high dominance of distinct Lactobacillus population under these conditions. Notably, the most abundant predicted pathway in both silages was transporters, the 2nd most abundant pathway/function was “General function prediction only, Unclassified,” which mainly consists of “poorly characterized” pathways. This finding underlines the importance of yet unknown pathways/functions in the ensiling process.
The putative abundance of transporters was much higher in untreated silage until d15, compared to Lp-treated silage. To the best of our knowledge, such high abundance of transporters during ensiling was not reported before. These findings may infer a commensal relationship among the microbial populations in the untreated silage. Indeed, the initial ensiling stages are characterized by the presence of heterogeneous aerobic and facultative anaerobic populations, which consume the oxygen trapped in the chopped forage and increase CO2 level. During these stages, metabolic byproducts released by some of the populations may be taken by others, using transporters, in order to obtain essential nutrients for energy and growth. As the pH level decreases up to d15, the silage population becomes more homogeneous and dominated by LAB. Consequently, less transporters are required. In contrast, in the case of Lp-treated silage, the pH level decreases sharply, as early as d2, and Lactobacillus dominated the population, so less transport function was required.
A similar phenomenon was observed regarding carbohydrate metabolism. In the Lp-treated silage, a constant level of carbohydrate metabolisms observed throughout the ensiling stages, which apparently reflects the metabolism of the dominant Lactobacillus population. In contrast, carbohydrate metabolisms increased in untreated silages until d15 to a much higher levels, and then reduced at d30 and remained constant until d90, at a level comparable to that of Lactobacillus treated silages. This is possibly due to the high initial activity of the heterogeneous microbial population at the early ensiling stages. Once oxygen was consumed, and pH level reduced, LAB became the dominant population and anaerobic fermentation took place at d30 through d90.
Unlike transporters and carbohydrates metabolism, the amino acid metabolism was higher in Lp-treated as compared to untreated silages until d30. The low metabolism might reflect the capacity of the initial microbial populations, in the silage, to synthesize amino acid de novo. In contrast, as LAB do not synthesize all their essential amino acid, they rely on proteolytic systems to provide essential amino acids for their growth (Novik and Savich, 2019). Consequently, the observed dynamic of amino acids metabolism in both Lp-treated and untreated silage, may reflect the metabolism in the dominant population throughout the ensiling process.
Silage are inevitably exposed to air when used for feeding, leading to undesirable microbial multiplication, decreasing nutritive values, and increasing risk of potential pathogens (Driehuis and Oude Elferink, 2000). Aerobic spoilage is primarily initiated by lactic acid assimilating yeasts, which can thrive well in high lactic acid containing silages (Moon and Ely, 1979). Breakdown of lactic acid leads to rise in pH, eventually providing better conditions for multiplication of opportunistic aerobic bacteria (e.g., Bacilli, Listeria monocytogenes) and molds (e.g., Aspergillus, Fusarium, Pencillium), which degrade silages subsequently to severe extent (Woolford, 1990; Pahlow et al., 2003).
Aerobic exposure of mature wheat silage for 5 days (aerobic stability test), resulted in a rapid deterioration of Lp-treated silage compared to untreated and Lb-treated silage. A possible explanation for the lower aerobic stability of Lp- versus Lb-treated silage is that L. plantarum is a homofermentative bacterium, which produce fermentation products high in lactic acid, and cannot produce enough volatile fatty acids to inhibit yeasts and molds (Weinberg et al., 1993). Indeed, the L. plantarum additive used in this study was already shown to enhance spoilage of wheat silage as compared to control (untreated) silage (Chen and Weinberg, 2014) and others have also reported that the usage of homolactic inoculants reduced the aerobic stability of wheat silage ensiled at bunker silos scale (Oliveira et al., 2018).
Unlike Lp, L. buchneri is a heterofermentative species, which produce a mix of products, and is able to ferment lactic acid to acetic acid (Weinberg et al., 1993; Weinberg et al., 1999; Filya, 2003), resulting in a higher content of acetic acid, which is a better inhibitor of yeast and mold (Weinberg et al., 1993; Filya, 2003). Indeed, mature Lb-treated silage contained 14-fold more acetic acid than Lp-treated silage, and no detectable yeasts. In support of the role of L. buchneri is stabilization of the mature silage, most LAB species isolated from untreated, 30 days-old ensiled wheat silage, were heterofermentative (Ni et al., 2015).
At the terminal stage of ensiling (day 90), a significantly higher content of lactic acid was evident in Lp-treated compared to untreated silage. This may set the stage for the emergence of Clostridia, in LP-treated silage, which can utilize lactic acid as substrate (Borreani et al., 2018). Indeed, after aerobic exposure, Clostridium_sensu_stricto_1 has emerged as the second most abundant genus. It is likely that the faster deterioration of this silage was associated with the emergence of this genus. In support of this notion, anaerobic spore-formers were previously linked to spoilage of silage, likely because they are capable of converting lactic acid to acetic- and butyric acids, which further contribute to quality deterioration (Vissers et al., 2007; McEniry et al., 2008; Li and Nishino, 2011). While, aerobic stability test, is considered as an indicative test for predicting the stability of silage in commercial bunkers, data obtained from air-exposed silage in commercial wheat silage bunkers, revealed low abundance of Lactobacillaceae (<2%), unlike the laboratory silos’ model, and were dominated by Bacillaceae, Thermoactinomycetaeae grp I and Enterobacteriaceae (Kraut-Cohen et al., 2016). These differences may be attributed to the more complex environmental factors exist in the field, as well as to the period of air exposure.
Predicted pathways that might be associated with aerobic stability were analyzed by PICRUSt. Pathway analysis using KEGG classification at level 2, did not yield significant differences between untreated and Lp-treated silage, consequently, KEGG pathway analysis at level 3 was employed. A total of seven pathways were significantly different between untreated and Lp-treated silages after aerobic exposure. In untreated silages, pathways for ethylbenzene degradation, D-Alanine metabolisms, terpenoid backbone synthesis and ion channel systems were more abundant, while in Lp-treated silages two component systems, novobiocin biosynthesis were higher. It is possible that production of antibiotics restricted the multiplication of some bacterial communities, which are competing for substrates with other bacterial and fungal communities. Since, the PICRUSt data are based on bacterial 16S sequences, further metagenomics studies should examine if antibiotics biosynthesis pathways are indeed expressed during ensiling in order to support this notion. Future studies based on metagenomics, covering both bacterial and fungal communities will be required for a more comprehensive understanding of the various metabolic networks and interactions present in the silage microbiome.
PICRUSt has been recently applied in pathways prediction also in maize (Gharechahi et al., 2017) and barley silages (Liu et al., 2019). The PICRUSt analysis, using KEGG level 3 classification, confirmed the changes in metabolic pathways after LAB addition during barley ensiling (Liu et al., 2019). It was anticipated that higher abundance of carbohydrate (Glycolysis, Fructose-and-mannose-, Pyruvate-metabolism, Pentose phosphate pathway) and amino acids (Alanine-aspartate-and-glutamate-metabolism) pathways were responsible for the better aerobic stability of LAB-treated barley silages compared to control. Yet, in our study, the Lp-treated wheat silage was less stable than untreated control. The different findings regarding the aerobic stability of wheat and barley silages, may be due to the nature of the LAB inoculants used, as well as to the nature of the ensiled crop. While in our study, a single species (Lp) was used, in the case of barely, three species, L. plantarum, L. casei, and L. buchneri were added at a ratio of 2:1:1 (Liu et al., 2019). In another study, pathway prediction was employed in untreated maize ensiling. Abundance of pathways for nitrotoluene and styrene degradation, cell motility and secretion, and arginine- and proline- metabolism were reduced during ensiling, while pathways for ethylbenzene, naphthalene and xylene degradation, biosynthesis of glycosphingolipid and bile acids were increased (Gharechahi et al., 2017). The changes in pathways abundance of untreated wheat silage during ensiling showed similarity in various pathways/functions (cell secretion and motility, arginine- and proline- metabolisms, ethylbenzene-, naphthalene-degradation, biosynthesis of glycosphingolipid and bile acids) with maize silage (Gharechahi et al., 2017). Nevertheless, the abundance of pathways of nitrotoluene-, styrene-, ethylbezoate- and xylene-degradation, showed an opposite trend until day 30 of ensiling, and showed subsequently similar level of abundance until 90 days.
The Lb-treated silage, which was rather stable following aerobic exposure, differed from non-treated silage at day 90 in three pathways that belonged to base excision repair, pyruvate metabolism and transcription machinery. Notably, a higher abundance of pyruvate metabolism, among few other pathways, was also observed in LAB-treated barley silage, and was suggested as a possible reason for enhancing of silage stability (Liu et al., 2019).
While taxonomic analyses were based on the more updated SILVA database, the PICRUSt profiling was based on the taxonomic IDs obtained from Greengenes database. Consequently, changes in the bacterial composition based on the two databases might affect the obtained PICRUSt results. However, comparison of the main OTUs, obtained according to both databases, showed high similarity in the taxonomic assignments (data not shown).
Finally, it should be noted the data obtained from functional metagenome prediction must be analyzed cautiously, since the prediction relies on genomic data and 16SrRNA amplification, rather than on direct mRNA measurements (Gharechahi et al., 2017). Therefore, further studies should employ metagenomics and transcriptomics in order to decipher the important metabolic pathways of both bacteria and fungi involved in the ensiling process of various crops.
Conclusion
The bacterial community of fresh whole crop wheat was found to be dominated by Pantoea, Weissella, Pseudomonas, Exiguobacterium, and Paenibacillus. During wheat ensiling with and without LAB-inoculant, the bacterial community shifted dramatically and the terminal silage was dominated mainly by Lactobacillus. There was a substantial difference in the abundance of Lactobacillus, between untreated and LAB-supplemented silages. Exogenous L. plantarum significantly affect the bacterial composition and dynamics during ensiling and resulted in a higher dominance of LAB in the mature silage and a higher content of lactic acid. Notably, addition of L. plantarum, but not L. buchneri reduced aerobic stability of the wheat silage. The lower aerobic stability of the L. plantarum-treated silage may be attributed to the different fermentation pathways in the two LAB inoculants. PICRUSt analysis provided knowledge regarding the role of transporters and carbohydrate metabolism in the initial stages of ensiling until stabilization of the bacterial populations. Since, ensiling is an uncontrolled fermentation process, depending on multiple parameters, the ability to monitor the bacterial dynamics, through the process, holds promise for a better understanding and improvement of the silage quality.
Author Contributions
SSS conceived and coordinated the study and edited the manuscript. JK, ZW, and SSS designed the experiments. JK, YC, RP, and ZW ensiled the wheat crop and collected the samples. JK carried out the molecular biology and bioinformatics analyses. YC and YK carried out the microbiological and biochemical analyses. JK and ZW drafted the manuscript with input from all co-authors. All authors read and approved the final manuscript.
Funding
This study was partially supported by the BARD (United States-Israel Binational Agricultural Research and Development Fund), Project IS-4704-14. SSS was a member of the EU COST Action FA1202 (CGAFA1202): An European Network for Mitigating Bacterial Colonization and Persistence on Foods and Food Processing Environments (http://www.bacfoodnet.org/) and is a current member of the EU COST Action 16110: “Control of Human Pathogenic Micro-organisms in Plant Production Systems” (http://huplantcontrol.igzev.de/), and acknowledges these actions. JK was partially supported by an ARO Postdoctoral Fellowship Fund under the Indo-Israeli Research and Development Program.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.01532/full#supplementary-material
FIGURE S1 | Heatmap of microbial function pathways dynamics in untreated and Lp-treated wheat silages. Colors reflect relative abundance from low (blue) to high (yellow).
FIGURE S2 | PICRUSt prediction of functional profiling of the microbial communities based on the 16S rRNA gene sequences. Extended error bar plot indicating differences (0.001%) in the abundance of functional profiles at day 90, in untreated and L. plantarum treated silages. The colored circles denote the difference in mean proportion of a particular pathway that is higher in Lp-treated silage (orange), or in untreated silage (blue).
FIGURE S3 | PICRUSt prediction of functional profiling of the microbial communities based on the 16S rRNA gene sequences. Extended error bar plot indicating differences in functional profiles of the day 90, untreated and L. buchneri-treated silage. The difference higher than 0.001% are presented; the red dots on difference in mean denotes that particular pathway abundance is higher in Lb-treated silage while blue dots denotes it is higher in untreated silage.
TABLE S1 | Weighted UniFrac analysis of bacterial community composition natural, L. plantarum (Lp)- and L. buchneri (Lb)-supplemented (treated) silages during ensiling.
TABLE S2 | Relative abundance (%) of bacterial genera found in terminal wheat silages.
TABLE S3 | Relative abundance (%) of bacterial genera found after 5 days of aerobic exposure of mature wheat silages.
TABLE S4 | Pathways predictions during ensiling of wheat silage with and without L. plantarum supplement.
Footnotes
References
Arasu, M. V., Jung, M. W., Kim, D. H., Ilavenil, S., Jane, M., Park, H. S., et al. (2014). Enhancing nutritional quality of silage by fermentation with Lactobacillus plantarum. Indian J. Microbiol. 54, 396–402. doi: 10.1007/s12088-014-0473-9
Ashbell, G., Weinberg, Z. G., Bruckental, I., Tabori, K., and Sharet, N. (1997). Wheat silage: the effect of cultivar and stage of maturity on yield and degradability in situ. J. Agric. Food Chem. 45, 709–712. doi: 10.1021/jf960336l
Bal, E. B. B., and Bal, M. A. (2012). Effects of chemical additives and ensiling time on whole plant wheat silage microbial pro?les inferred by phenotypic and 16S ribosomal DNA analyses. World J. Microbiol. Biotechnol. 28, 767–776. doi: 10.1007/s11274-011-0864-6
Bao, W., Mi, Z., Xu, H., Zheng, Y., Kwok, L. Y., Zhang, H., et al. (2016). Assessing quality of Medicago sativa silage by monitoring bacterial composition with single molecule, real-time sequencing technology and various physiological parameters. Sci. Rep. 6:28358. doi: 10.1038/srep28358
Barker, B. S., and Summerson, W. H. (1941). The colorimetric determination of lactic acid in biological material. J. Biol. Chem. 138, 535–554.
Berlec, A. (2012). Novel techniques and findings in the study of plant microbiota: search for plant probiotics. Plant Sci. 193, 96–102. doi: 10.1016/j.plantsci.2012.05.010
Bokulich, N. A., and Mills, D. A. (2012). Next-generation approaches to the microbial ecology of food fermentations. BMB Rep. 45, 377–389. doi: 10.5483/BMBRep.2012.45.7.148
Borreani, G., Tabacco, E., Schmidt, R. J., Holmes, B. J., and Muck, R. E. (2018). Silage review: factors affecting dry matter and quality losses in silages. J. Dairy Sci. 101, 3952–3997. doi: 10.3168/jds.2017-13837
Cai, Y., Benno, Y., Ogawa, M., Ohmomo, S., Kumai, S., and Nakase, T. (1998). Influence of Lactobacillus spp. from an inoculant and of Weissella and Leuconostoc spp. from forage crops on silage fermentation. Appl. Environ. Microbiol. 64, 2982–2987.
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Lozupone, C. A., Turnbaugh, P. J., et al. (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. U.S.A. 108, 4516–4522. doi: 10.1073/pnas.1000080107
Chen, Y., and Weinberg, Z. G. (2014). The effect of relocation of whole-crop wheat and corn silages on their quality. J. Dairy Sci. 97, 406–410. doi: 10.3168/jds.2013-7098
Driehuis, F., and Oude Elferink, S. J. (2000). The impact of the quality of silage on animal health and food safety: a review. Vet. Q. 22, 212–216. doi: 10.1080/01652176.2000.9695061
Duniere, L., Xu, S., Long, J., Elekwachi, C., Wang, Y., Turkington, K., et al. (2017). Bacterial and fungal core microbiomes associated with small grain silages during ensiling and aerobic spoilage. BMC Microbiol. 17:50. doi: 10.1186/s12866-017-0947-0
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C., and Knight, R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. doi: 10.1093/bioinformatics/btr381
Eikmeyer, F. G., Kofinger, P., Poschenel, A., Junemann, S., Zakrzewski, M., Heinl, S., et al. (2013). Metagenome analyses reveal the influence of the inoculant Lactobacillus buchneri CD034 on the microbial community involved in grass ensiling. J. Biotechnol. 167, 334–343. doi: 10.1016/j.jbiotec.2013.07.021
Filya, I. (2003). The effect of Lactobacillus buchneri, with or without homofermentative lactic acid bacteria, on the fermentation, aerobic stability and ruminal degradability of wheat, sorghum and maize silages. J. Appl. Microbiol. 95, 1080–1086. doi: 10.1046/j.1365-2672.2003.02081.x
Gharechahi, J., Kharazian, Z. A., Sarikhan, S., Jouzani, G. S., Aghdasi, M., and Hosseini Salekdeh, G. (2017). The dynamics of the bacterial communities developed in maize silage. Microb. Biotechnol. 10, 1663–1676. doi: 10.1111/1751-7915.12751
Green, S. J., Venkatramanan, R., and Naqib, A. (2015). Deconstructing the polymerase chain reaction: understanding and correcting bias associated with primer degeneracies and primer-template mismatches. PLoS One 10:e0128122. doi: 10.1371/journal.pone.0128122
Jatkauskas, J., Vrotniakiene, V., Ohlsson, C., and Lund, B. (2013). The effects of three silage inoculants on aerobic stability in grass, clover-grass, lucerne and maize silages. Agric. Food Sci. 22, 137–144. doi: 10.23986/afsci.6698
Kanehisa, M., Araki, M., Goto, S., Hattori, M., Hirakawa, M., Itoh, M., et al. (2008). KEGG for linking genomes to life and the environment. Nucleic Acids Res. 36, D480–D484. doi: 10.1093/nar/gkm882
Kasmaei, K. M., Dicksved, J., Sporndly, R., and Uden, P. (2016). Separating the effects of forage source and field microbiota on silage fermentation quality and aerobic stability. Grass For. Sci. 72, 281–289. doi: 10.1111/gfs.12238
Keshri, J., Chen, Y., Pinto, R., Kroupitski, Y., Weinberg, Z. G., and Sela Saldinger, S. (2018). Microbiome dynamics during ensiling of corn with and without Lactobacillus plantarum inoculant. Appl. Microbiol. Biotechnol. 102, 4025–4037. doi: 10.1007/s00253-018-8903-y
Kozich, J. J., Westcott, S. L., Baxter, N. T., Highlander, S. K., and Schloss, P. D. (2013). Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq illumina sequencing platform. Appl. Environ. Microbiol. 79, 5112–5120. doi: 10.1128/AEM.01043-13
Kraut-Cohen, J., Tripathi, V., Chen, Y., Gatica, J., Volchinski, V., Sela, S., et al. (2016). Temporal and spatial assessment of microbial communities in commercial silages from bunker silos. Appl. Microbiol. Biotechnol. 100, 6827–6835. doi: 10.1007/s00253-016-7512-x
Langille, M. G., Zaneveld, J., Caporaso, J. G., McDonald, D., Knights, D., Reyes, J. A., et al. (2013). Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 31, 814–821. doi: 10.1038/nbt.2676
Lebeis, S. L. (2017). Plant microbiome identification and characterization. Curr. Protoc. Plant Biol. 2, 135–146. doi: 10.1002/cppb.20048
Li, L., Sun, Y., Yuan, Z., Kong, X., Wao, Y., Yang, L., et al. (2015). Effect of microalgae supplementation on the silage quality and anaerobic digestion performance of manyflower silvergrass. Bioresour. Technol. 189, 334–340. doi: 10.1016/j.biortech.2015.04.029
Li, Y., and Nishino, N. (2011). Effects of inoculation of Lactobacillus rhamnosus and Lactobacillus buchneri on fermentation, aerobic stability and microbial communities in whole crop corn silage. Grassl. Sci. 57, 184–191. doi: 10.1111/j.1744-697X.2011.00226.x
Liu, B., Huan, H., Gu, H., Xu, N., Shen, Q., and Ding, C. (2019). Dynamics of a microbial community during ensiling and upon aerobic exposure in lactic acid bacteria inoculation-treated and untreated barley silages. Bioresour. Technol. 273, 212–219. doi: 10.1016/j.biortech.2018.10.041
Lozupone, C., Hamady, M., and Knight, R. (2006). UniFrac-an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinform. 7:371. doi: 10.1186/1471-2105-7-371
McAllister, T. A., Duniere, L., Drouin, P., Xu, S., Wang, Y., Munns, K., et al. (2018). Silage review: using molecular approaches to define the microbial ecology of silage. J. Dairy Sci. 101, 4060–4074. doi: 10.3168/jds.2017-13704
McEniry, J., O’kiely, P., Clipson, N. J., Forristal, P. D., and Doyle, E. M. (2008). Bacterial community dynamics during the ensilage of wilted grass. J. Appl. Microbiol. 105, 359–371. doi: 10.1111/j.1365-2672.2008.03802.x
McGarvey, J. A., Franco, R. B., Palumbo, J. D., Hnasko, R., Stanker, L., and Mitloehner, F. M. (2013). Bacterial population dynamics during the ensiling of Medicago sativa (alfalfa) and subsequent exposure to air. J. Appl. Microbiol. 114, 1661–1670. doi: 10.1111/jam.12179
Moon, N. J., and Ely, L. O. (1979). Identification and properties of yeasts associated with the aerobic deterioration of wheat and alfalfa silages. Mycopathologia 69, 153–156. doi: 10.1007/bf00452827
Ni, K., Minh, T. T., Tu, T. T., Tsuruta, T., Pang, H., and Nishino, N. (2017). Comparative microbiota assessment of wilted Italian ryegrass, whole crop corn, and wilted alfalfa silage using denaturing gradient gel electrophoresis and next-generation sequencing. Appl. Microbiol. Biotechnol. 101, 1385–1394. doi: 10.1007/s00253-016-7900-2
Ni, K., Wang, Y., Cai, Y., and Pang, H. (2015). Natural lactic acid bacteria population and silage fermentation of whole-crop wheat. Asian Austr. J. Anim. Sci. 28, 1123–1132. doi: 10.5713/ajas.14.0955
Novik, G., and Savich, V. (2019). “Beneficial lactic acid bacteria Use of lactic acid bacteria in probiotics,” in Pocket Guide to Bacterial Infections, eds K. Balamurugan and U. Prithika (Boca Raton, FL: CRC Press).
Ogunade, I. M., Jiang, Y., Cervantes, A. A. P., Kim, D. H., Oliveira, A. S., Vyas, D., et al. (2018). Bacterial diversity and composition of alfalfa silage as analyzed by illumina MiSeq sequencing: effects of Escherichia coli O157:H7 and silage additives. J. Dairy Sci. 101, 2048–2059. doi: 10.3168/jds.2017-12876
Oliveira, M. R., Jobim, C. C., Neumann, M., Bueno, A. V. I., Leão, G. F. M., and Daniel, J. L. P. (2018). Effects of inoculation with homolactic bacteria on the conservation of wheat silage stored in bunker-silos. Ital. J. Anim. Sci. 17, 81–86. doi: 10.1080/1828051X.2017.1345664
Pahlow, G., Muck, R. E., Driehuis, F., Oude Elferink, S. J. W. H., and Spoelstra, S. F. (2003). “Microbiology of ensiling,” in Silage Science and Technology, eds D. R. Buxton, R. E. Muck, and J. H. Harrison (Madison: American Society of Agronomy), 31–93.
Pang, H., Qin, G., Tan, Z., Li, Z., Wang, Y., and Cai, Y. (2011). Natural populations of lactic acid bacteria associated with silage fermentation as determined by phenotype, 16S ribosomal RNA and recA gene analysis. Syst. Appl. Microbiol. 34, 235–241. doi: 10.1016/j.syapm.2010.10.003
Parks, D. H., Tyson, G. W., Hugenholtz, P., and Beiko, R. G. (2014). STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics 30, 3123–3124. doi: 10.1093/bioinformatics/btu494
Parvin, S., and Nishino, N. (2009). Bacterial community associated with ensilage process of wilted guinea grass. J. Appl. Microbiol. 107, 2029–2036. doi: 10.1111/j.1365-2672.2009.04391.x
Parvin, S., and Nishino, N. (2010). Succession of lactic acid bacteria in wilted rhodesgrass silage assessed by plate culture and denaturing gradient gel electrophoresis. Grassl. Sci. 56, 51–55. doi: 10.1111/j.1744-697X.2009.00173.x
Pieper, R., Hackl, W., Korn, U., Zeyner, A., Souffrant, W. B., and Pieper, B. (2011). Effect of ensiling triticale, barley and wheat grains at different moisture content and addition of Lactobacillus plantarum (DSMZ 8866 and 8862) on fermentation characteristics and nutrient digestibility in pigs. Anim. Feed. Sci. Technol. 164, 96–105. doi: 10.1016/j.anifeedsci.2010.11.013
Pruesse, E., Quast, C., Knittel, K., Fuchs, B. M., Ludwig, W., Peplies, J., et al. (2007). SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35, 7188–7196. doi: 10.1093/nar/gkm864
Romero, J. J., Zhao, Y., Balseca-Paredes, M. A., Tiezzi, F., Gutierrez-Rodriguez, E., and Castillo, M. S. (2017). Laboratory silo type and inoculation effects on nutritional composition, fermentation, and bacterial and fungal communities of oat silage. J. Dairy Sci. 100, 1812–1828. doi: 10.3168/jds.2016-11642
Santos, M. C., and Kung, L. (2016). The effects of dry matter and length of storage on the composition and nutritive value of alfalfa silage. J. Dairy Sci. 99, 5466–5469. doi: 10.3168/jds.2016-10866
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541. doi: 10.1128/AEM.01541-09
Stevenson, D. M., Muck, R. E., Shinners, K. J., and Weimer, P. J. (2006). Use of real time PCR to determine population profiles of individual species of lactic acid bacteria in alfalfa silage and stored corn stover. Appl. Microbiol. Biotechnol. 71, 329–338. doi: 10.1007/s00253-005-0170-z
Strauber, H., Lucas, R., and Kleinsteuber, S. (2016). Metabolic and microbial community dynamics during the anaerobic digestion of maize silage in a two-phase process. Appl. Microbiol. Biotechnol. 100, 479–491. doi: 10.1007/s00253-015-6996-0
Vissers, M. M., Driehuis, F., Te Giffel, M. C., De Jong, P., and Lankveld, J. M. (2007). Concentrations of butyric acid bacteria spores in silage and relationships with aerobic deterioration. J. Dairy Sci. 90, 928–936. doi: 10.3168/jds.S0022-0302(07)71576-X
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/aem.00062-07
Wang, Q., Tang, B., and Han, Z. (2013). The effects of the different silage on milk yield and composition in dairy cows. China Dairy Cattle 9, 52–54.
Wang, Y., Wang, C., Zhou, W., Yang, F. Y., Chen, X. Y., and Zhang, Q. (2018). Effects of wilting and Lactobacillus plantarum addition on the fermentation quality and microbial community of Moringa oleifera leaf silage. Front. Microbiol. 9:1817. doi: 10.3389/fmicb.2018.01817
Wang, Y. S., Shi, W., Huang, L. T., Ding, C. L., and Dai, C. C. (2016). The effect of lactic acid bacterial starter culture and chemical additives on wilted rice straw silage. Anim. Sci. J. 87, 525–535. doi: 10.1111/asj.12449
Weinberg, Z. G., Ashbell, G., Hen, Y., and Azrieli, A. (1993). The effect of applying lactic acid bacteria at ensiling on the aerobic stability of silages. J. Appl. Bacteriol. 75, 512–518. doi: 10.1111/j.1365-2672.1993.tb01588.x
Weinberg, Z. G., and Chen, Y. (2013). Effects of storage period on the composition of whole crop wheat and corn silages. Anim. Feed Sci. Technol. 185, 196–200. doi: 10.1016/j.anifeedsci.2013.08.009
Weinberg, Z. G., Chen, Y., and Solomon, R. (2009). The quality of commercial wheat silages in Israel. J. Dairy Sci. 92, 638–644. doi: 10.3168/jds.2008-1120
Weinberg, Z. G., and Muck, R. E. (1996). New trends and opportunities in the development and use of inoculants for silage. FEMS Microbiol. Rev. 19, 53–68. doi: 10.1016/0168-6445(96)00025-3
Weinberg, Z. G., Szakacs, G., Ashbell, G., and Hen, Y. (1999). The effect of Lactobacillus buchneri and L. plantarum, applied at ensiling, on the ensiling fermentation and aerobic stability of wheat and sorghum silages. J. Ind. Microbiol. Biotech. 23, 218–222. doi: 10.1038/sj.jim.2900726
Wilkins, R., and Wilkinson, M. (2015). “Major contributions in 45 years of International Silage Conferences,” in Proceedings of XVII International Silage Conference, (Piracicaba: University of São Paulo), 26–51.
Wilkinson, J. M., Bolsen, K. K., and Lin, C. J. (2003). “History of silage,” in Silage Science and Technology, eds D. R. Buxton, R. E. Muck, and J. H. Harrison (Madison: American Society of Agronomy), 1–30.
Woolford, M. K. (1990). The detrimental effects of air on silage. J. Appl. Bacteriol. 68, 101–116. doi: 10.1111/j.1365-2672.1990.tb02554.x
Xie, Z. L., Zhang, T. F., Chen, X. Z., Li, G. D., and Zhang, J. G. (2012). Effects of maturity stages on the nutritive composition and silage quality of whole crop wheat. Asian Austr. J. Anim. Sci. 25, 1374–1380. doi: 10.5713/ajas.2012.12084
Xu, Z., He, H., Zhang, S., and Kong, J. (2017). Effects of inoculants Lactobacillus brevis and Lactobacillus parafarraginis on the fermentation characteristics and microbial communities of corn stover silage. Sci. Rep. 7:13614. doi: 10.1038/s41598-017-14052-1
Xu, Z., Zhang, S., Zhang, R., Li, S., and Kong, J. (2018). The changes in dominant lactic acid bacteria and their metabolites during corn stover ensiling. J. Appl. Microbiol. 15, 675–685. doi: 10.1111/jam.13914
Zhao, X., Liu, J., Liu, J., Yang, F., Zhu, W., Yuan, X., et al. (2017). Effect of ensiling and silage additives on biogas production and microbial community dynamics during anaerobic digestion of switchgrass. Bioresour. Technol. 241, 349–359. doi: 10.1016/j.biortech.2017.03.183
Keywords: bacterial diversity, bacterial community, 16S rRNA, silage, aerobic-stability
Citation: Keshri J, Chen Y, Pinto R, Kroupitski Y, Weinberg ZG and Sela Saldinger S (2019) Bacterial Dynamics of Wheat Silage. Front. Microbiol. 10:1532. doi: 10.3389/fmicb.2019.01532
Received: 29 October 2018; Accepted: 19 June 2019;
Published: 09 July 2019.
Edited by:
Annamaria Bevivino, Italian National Agency for New Technologies, Energy and Sustainable Economic Development (ENEA), ItalyReviewed by:
Lysiane Duniere, Lallemand, CanadaAna Griselda Binetti, Instituto de Lactología Industrial (INLAIN, CONICET), Argentina
Copyright © 2019 Keshri, Chen, Pinto, Kroupitski, Weinberg and Sela Saldinger. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zwi G. Weinberg, zgw@volcani.agri.gov.il; Shlomo Sela Saldinger, shlomos@volcani.agri.gov.il
†Present address: Jitendra Keshri, College of Veterinary Medicine, Western University of Health Sciences, Pomona, CA, United States
 Jitendra Keshri†
Jitendra Keshri† Zwi G. Weinberg
Zwi G. Weinberg Shlomo Sela Saldinger
Shlomo Sela Saldinger