MTHFR rs1801133 Polymorphism Is Associated With Liver Fibrosis Progression in Chronic Hepatitis C: A Retrospective Study
- 1Servicio de Laboratorio Clínico, Hospital de Antequera, Málaga, Spain
- 2Servicio de Digestivo, Hospital Virgen de la Salud, Toledo, Spain
- 3Unidad de Infección Viral e Inmunidad, Centro Nacional de Microbiología, Instituto de Salud Carlos III, Majadahonda, Spain
Background: The MTHFR (methylenetetrahydrofolate reductase) rs1801133 polymorphism leads to higher circulating levels of homocysteine, which is related to several liver diseases. We aimed to evaluate the relationship between MTHFR rs1801133 polymorphism and liver fibrosis progression in HCV-infected patients.
Methods: We conducted a preliminary retrospective cohort study in 208 non-cirrhotic HCV-infected patients. These subjects had at least two liver stiffness measurements (LSM), which were assessed using transient elastography, and no patient had cirrhosis at baseline. We analyzed the association between MTHFR rs1801133 and outcome variables using Generalized Linear Models.
Results: HCV-infected patients were 47 years old, around 54% were males, a low frequency of high alcohol intake (13.5%) or prior use of intravenous drugs (10.1%). A total of 26 patients developed cirrhosis (LSM1 ≥ 12.5) during a median follow-up of 46.6 months. The presence of the rs1801133 C allele showed an inverse association with the LSM2/LSM1 ratio (adjusted AMR = 0.90; 95%CI = 0.83–0.98; p = 0.020) and the cirrhosis progression (adjusted OR = 0.43; 95%CI = 0.19–0.95; p = 0.038). Besides, rs1801133 CT/CC genotype had an inverse association with the LSM2/LSM1 ratio (adjusted AMR = 0.80; 95%CI = 0.68–0.95; p = 0.009) and the cirrhosis progression (adjusted OR= 0.21; 95%CI = 0.06–0.74; p = 0.015).
Conclusions: MTHFR rs1801133 C allele carriers presented a diminished risk of liver fibrosis progression and development of cirrhosis than rs1801133 T allele carriers. This statement supports the hypothesis that MTHFR rs1801133 polymorphism appears to play a crucial role in chronic hepatitis C immunopathogenesis.
Introduction
According to the world health organization, viral hepatitis is a significant public health problem that causes 1.34 million deaths per year due to chronic liver disease (720,000 by cirrhosis) and primary liver cancer (47,000 by hepatocellular carcinoma) (1). Globally, around 71 million people suffer chronic hepatitis C (CHC) and the development of the previously described events is frequent (2, 3), even after a sustained virological response (SVR) to treatment with direct-acting antivirals (DAAs) (4, 5). The pathogenic mechanisms involved in the progression of fibrosis and cirrhosis depends, among others, on the genetic background of individuals, including several single nucleotide polymorphisms (SNPs) (6, 7).
The staging of liver fibrosis provides essential clinical information that allows the adequate management and prognosis of CHC patients (8). The liver biopsy has been used to grade the necroinflammatory activity and to stage fibrosis, together with rating scales like METAVIR, which stratify fibrosis as: (i) F0, no fibrosis; (ii) F1, mild fibrosis; (iii) F2, significant fibrosis; (iv) F3, advanced fibrosis; and (v) F4, cirrhosis (9). Nevertheless, non-invasive approaches, as the transient elastography or FibroScan, have been widely used to the liver fibrosis assessment, with excellent accuracy in advanced fibrosis and cirrhosis (10). In this context, the evaluation of liver stiffness measurement (LSM), an intrinsic physical property of liver parenchyma, provides quantitative data that correlates with fibrosis stage in CHC (11).
The methylenetetrahydrofolate reductase (MTHFR) gene encodes an enzyme that plays an important role in the folate metabolism, allowing the conversion from homocysteine to methionine (12). The rs1801133 SNP (also named C677T) is a non-synonymous variant A (Ala) > V (Val) (missense variant) (13). The substitution A > V in the aminoacid 222 (Ala222Val) produces a decrease of the activity of MTHFR protein and an elevation of plasma homocysteine levels (13). Hyper-homocysteinemia was related to several diseases, including hepatocellular carcinoma, steatosis, and cirrhosis (14–16), and also the development of liver fibrosis in CHC (17). There are a few articles published about MTHFR rs1801133 in CHC patients. MTHFR C677T polymorphism has been related to hepatic steatosis (18) and development of liver fibrosis (16, 17), but no association was found in other articles in patients infected with HCV (19–21). Additionally, this polymorphism could also be related to a direct profibrogenic effect, modifying the action of proteins implicated in the degradation of collagen (22).
The main objective in the current study was to investigate the association of MTHFR rs1801133 polymorphism with the progression of liver fibrosis and cirrhosis development, evaluated by LSM, in patients with CHC.
Materials and Methods
Study Population
A preliminary retrospective study was performed in 208 patients from the Hospital Virgen de la Salud (Toledo, Spain). All patients suffered from chronic hepatitis C and were enrolled between 2008 and 2016, as previously described (see Supplementary Figure 1) (23).
We selected the patients according to these criteria: (i) available DNA sample; (ii) detectable plasma HCV RNA at baseline and during follow-up; and (iii) baseline LSM (LSM1) and final LSM (LSM2) available with a separation of 12 months at least. Regarding the exclusion criteria, we considered: (i) liver cirrhosis at baseline (F4; LSM1 ≥ 12.5 kPa); (ii) coinfection with human immunodeficiency virus or hepatitis B virus; and (iii) autoimmune liver disease.
The study was conducted with the consent of all patients and following the 1975 Declaration of Helsinki. It was approved by the Institutional Review Board of the Instituto de Salud Carlos III (“Comité de Ética de la Investigación y Bienestar Animal” –April 4, 2013).
Clinical Data
As described previously (24), clinical and epidemiological data were collected from medical records. These data included virological, demographic, clinical, and laboratory data. Clinical guidelines available at that time (25, 26) were followed to perform the clinical management of patients during the follow-up.
At baseline, we only included non-responder patients (patients treated for HCV infection before the study), although it was possible to administrate the HCV therapy before or after being included in the study. During the follow-up, the monitoring was stopped when a patient started the HCV therapy and obtained an SVR.
DNA Genotyping
Genomic DNA obtained from 200 microliters of peripheral blood was extracted using the QIAsymphony DNA Mini Kit (Qiagen, Hilden, Germany). MTHFR rs1801133 polymorphism was genotyped at the CeGen (Spanish National Genotyping Center; http://www.cegen.org/). Agena Bioscience's MassARRAY platform (San Diego, CA, USA) and the iPLEX® Gold assay design system were used according to the method described by Gabriel et al. (27).
Evaluation of Liver Fibrosis
The hepatic fibrosis was evaluated using the transient elastography (FibroScan, Echosens, Paris, France), by a trained hepatologist and with a single machine, as we previously described (24). LSM has a range of 2.5 to 75 kilopascals (kPa). When the interquartile-range-to-median ratio for at least 10 successful measurements was <0.30, it was considered reliable. The cut-offs of LSM proposed by Castera et al. were followed for the stratification of patients: (i) <7.1 kPa (F0–F1—absence or mild fibrosis); (ii) 7.1–9.4 kPa (F2—significant fibrosis); (iii) 9.5-12.4 kPa (F3—advanced fibrosis); and (iv) ≥12.5 kPa (F4—cirrhosis) (28).
Outcome Variable
We analyzed how LSM values changed during the follow-up, considering: (i) the date of the first LSM (LSM1) and (ii) the date of the last LSM (LSM2), or the date when the HCV therapy started in responder patients who cleared HCV infection. For this propose, we consider two outcome variables: (1) LSM2/LSM1 ratio; (2) the cirrhosis progression (F4; LSM ≥ 12.5 kPa) measured as +1 [if a patient with LSM <12.5 kPa (F ≤ 3) changed to LSM ≥12.5 kPa (F4)] or 0 (if a patient with F ≤ 3 did not evolve to F4).
Statistical Analysis
To compare independent groups, we used the Mann-Whitney U test for continuous variables and the Chi-square test or Fisher's exact test for categorical variables. In the case of paired measurements, we used the Sign test for categorical variables and the Wilcoxon signed-rank test for continuous variables.
We used Generalized Linear Models (GLM) according to recessive, dominant, and additive inheritance models for the genetic association study with the aim of comparing the outcome variables according to MTHFR rs1801133. First, we used a GLM with a gamma distribution (log-link) to analyze continuous variables (LSM2/LSM1 ratio) and a GLM with a binomial distribution (logit-link) to analyze dichotomous variables (progression to cirrhosis). These tests provide the arithmetic mean ratio (AMR) or difference between groups, and the odds ratio (OR) or probability of occurrence of an event. The most relevant patient characteristics were used to adjust the GLM tests: gender, age, diabetes, high alcohol intake, injection drug use, time since HCV diagnosis, HCV genotype, baseline LSM, time of follow-up, HCV antiviral therapy before baseline and during the follow-up (patients who failed therapy), and other SNPs previously described in this study population (PNPLA3 rs738409 (29), MERTK rs4374383 (30), IL7RA rs6897932 (24), and DARC rs12075 (23). To avoid overfitting the statistical models, we made a previous selection of covariables with the Stepwise algorithm, retaining covariables with a p-value <0.20 at each step.
For all statistical tests, we used the Stata 15.0 (StataCorp, Texas, USA) and SPSS 24.0 (SPSS INC, Chicago, IL, USA). Statistical significance was defined as p < 0.05 and all p-values were two-tailed.
Results
Characteristics of the Patients
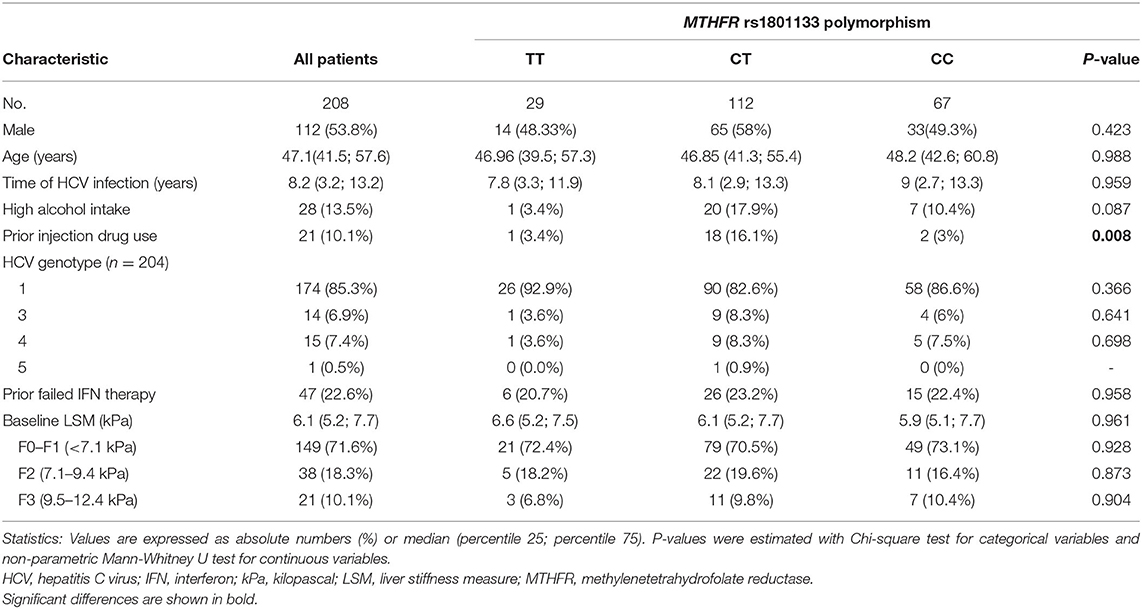
The baseline characteristics of our study population are described in Table 1. HCV-infected patients were 47 years old, around 54% were males, a low frequency of high alcohol intake (13.5%) or prior use of intravenous drugs (10.1%). HCV genotype 1 was the predominant (85.3% of patients), 22.6% of patients previously failed the interferon therapy, and 71.6% of patients had LSM <7.1 kPa. Concerning rs1801133 genotypes, 29 patients were TT genotype, 112 were CT genotype, and 67 were CC genotype. No significant differences in baseline characteristics were found among rs1801133 genotypes, except for prior injection drug use (p = 0.008).
Characteristics of MTHFR rs1801133 Polymorphism
Table 2 describes the allelic and genotypic frequencies of the rs1801133 SNP, which showed <5% of missing values, was in Hardy-Weinberg equilibrium (p = 0.252) and had a minimum allele frequency more than 40%. We compared the genetic frequencies between patients included in this study and the Iberian population in Spain (IBS), a population of healthy subjects published by the 1,000 Genomes Project website (http://www.1000genomes.org/home). No significant differences were found for alleles (p = 0.940) or genotypes (p = 0.665).
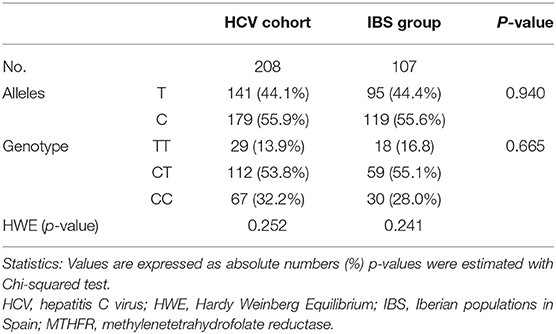
Table 2. Allelic and genotypic frequencies and Hardy Weinberg Equilibrium test for MTHFR rs1801133 polymorphism in HCV-infected patients compared to Iberian population (data from 1,000 Genomes Project Phase 3) (http://grch37.ensembl.org/index.html).
MTHFR rs1801133 SNP and Related Liver Fibrosis Progression
The mean follow-up time (the period between the LSM1 and the LSM2) for all patients was of 46.6 months. In this context, we found a decrease in the proportion of patients with a low stage of fibrosis (F0–F1; p < 0.001), whereas both LSM values and the rate of patients who developed an F4 stage raised (p < 0.001) (Table 3).
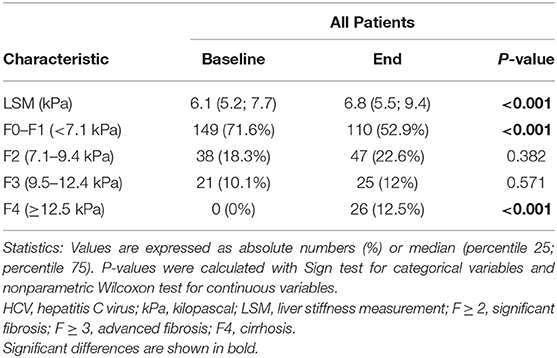
Table 3. Clinical characteristics related to hepatic fibrosis in patients with chronic hepatitis C during the follow-up.
We did not find significant differences among rs1801133 genotypes in the time intervals between the LSM1 and the LSM2 (48.5 months in TT genotype, 46.9 months in CT genotype, and 45.5 months in CC genotype; p = 0.921). We observed decreased values of the LSM2/LSM1 ratio and the rate of cirrhosis progression in rs1801133 T allele carriers (Table 4). Moreover, we analyzed the association between MTHFR rs1801133 polymorphism and liver fibrosis/cirrhosis progression through multivariate GLMs (Table 4, full description in Supplementary Table 1). The presence of the rs1801133 C allele showed an inverse association with the LSM2/LSM1 ratio (adjusted AMR = 0.90; 95%CI = 0.83–0.98; p = 0.020) and the cirrhosis progression (adjusted OR = 0.43; 95%CI = 0.19–0.95; p = 0.038). Besides, rs1801133 CT/CC genotype had an inverse association with the LSM2/LSM1 ratio (adjusted AMR = 0.80; 95%CI = 0.68–0.95; p = 0.009) and the cirrhosis progression (adjusted OR = 0.21; 95%CI = 0.06–0.74; p = 0.015) (Table 4).
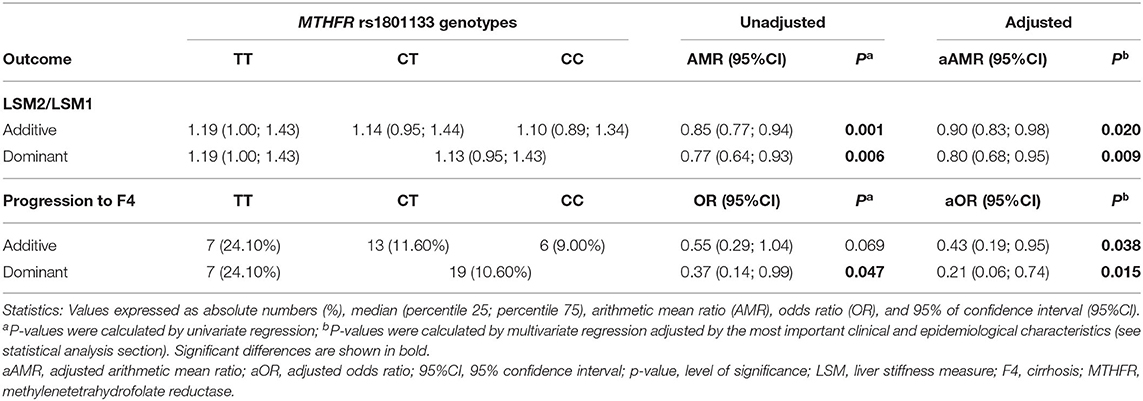
Table 4. Association between MTHFR rs1801133 polymorphism and progression of liver fibrosis in patients with chronic hepatitis C (longitudinal analysis).
Discussion
The present study focused on the potential relationship of MTHFR rs1801133 polymorphism and the development of liver fibrosis and cirrhosis on HCV-infected patients, using two LSM values with an interval of at least 12 months. We observed that CT/CC genotype was related to a decreased risk of progression of liver fibrosis and the occurrence of cirrhosis.
The C677T genetic substitution at the MTHFR gene results in the Ala222Val replacement in the MTHFR protein, resulting in a thermolabile variant associated with lower activity, and therefore, higher circulating levels of homocysteine (13). Moreover, MTHFR rs1801133 SNP is in high linkage disequilibrium (LD) with other MTHFR SNPs, such as A1298C (rs1801131), also related to a decrease in MTHFR activity (31). The LD between the two SNPs is strong in the Spanish population (coefficient of LD = 0.98) (32), so we think that our results may be extrapolated to the A1298C variant and other MTHFR SNPs in LD with rs1801133.
The MTHFR rs1801133 SNP was previously linked to a long list of conditions and diseases, such as bone disorders, cardiovascular disease, thrombosis, neurological/neuropsychiatric conditions (33), pre-eclampsia, diabetes mellitus (34), longevity (35), and several types of neoplasia (36). Regarding liver diseases, the MTHFR rs1801133 polymorphism is related to altered lipid metabolism (37), which would contribute to the development of steatosis and fibrosis in HCV-infected patients (38), as well as the development of cirrhosis (14, 39–41). Furthermore, in CHC, the MTHFR rs1801133 variant has also been linked to liver fibrosis/cirrhosis (16, 17). Toniutto et al. described that recipients with the presence of MTHFR rs1801133 TT homozygote evolved with more frequency to a significant fibrosis degree during recurrent hepatitis C after liver transplantation (16). Similar to this, Adinolfi et al. described that the T allele is related to a higher prevalence of steatosis, accelerating the fibrosis development and liver disease progression (17). These analyses support that the MTHFR rs1801133 polymorphism and the subsequent hyperhomocysteinemia are associated with liver fibrosis among HCV-infected patients. However, there are also other articles that found no association between the MTHFR rs1801133 variant and liver fibrosis/cirrhosis (19–21). These articles, both those that showed an association and those that did not, had a cross-sectional design, and fibrosis was evaluated by biopsy. Our article, by contrast, had a longitudinal design that provides robustness to our data, and liver fibrosis was evaluated by transient elastography, which has excellent accuracy for cirrhosis diagnosis.
Other issues should be considered for the correct interpretation of the data. Firstly, we performed a retrospective study, which could induce ascertainment and selection biases. Secondly, the low sample size per group could limit the statistical power of the tests performed and increasing the rate of false positives. Therefore, further studies should be conducted to corroborate our preliminary findings on the potential use of MTHFR rs1801133 SNP as a predictive marker of liver fibrosis/cirrhosis progression in HCV-infected patients. Thirdly, the follow-up time (between LSM1 and LSM2) varied between different subjects, but 75% of the patients had more than 28 months of follow-up, and globally, all patients presented more than 12 months of follow-up. Besides, the time of follow-up in patients stratified by MTHFR rs1801133 genotypes was comparable. Fourthly, due to the retrospective design of our investigation, we did not take into account some important clinical variables, including obesity, abdominal ultrasound, metabolic syndrome, and pathological study of the liver (fibrosis, necroinflammation, and steatosis), among others; and biomarkers, such as HCV viral load, transaminases, platelet counts, APRI score, and FIB4 index, among others. We did not have access to these data at the time of the LSM. Moreover, we had not plasma samples available to measure concentrations of homocysteine. Finally, we included in the study more than 20% non-responders to previous interferon therapy, but this HCV therapy does not seem to protect against the progression of CHC in the long term studies (42).
Conclusion
In summary, in this preliminary study, our data suggest an association between MTHFR rs1801133 SNP and the progression of liver fibrosis and the development of cirrhosis in HCV-infected patients. Specifically, MTHFR rs1801133 C allele carriers presented a diminished risk of liver fibrosis progression and development of cirrhosis than rs1801133 T allele carriers. Further studies with higher numbers of patients would be needed to confirm the role of MTHFR in the immune-pathogenesis of CHC.
Data Availability Statement
The datasets used and analyzed during the current study may be made available by the corresponding author upon reasonable request.
Ethics Statement
The studies involving human participants were reviewed and approved by the study was conducted following the 1975 Declaration of Helsinki. The Institutional Review Board of the Instituto de Salud Carlos III (Comité de Ética de la Investigación y Bienestar Animal 04/04/2013) approved the study, and all patients gave their consent for the study. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
MJ-S and SR: funding body, study concept and design. AG-M, MJ-S, JS-R, and TA-V: patients' selection and clinical data acquisition. DP-T, MJ-S, and AG-M: sample preparation, DNA isolation and genotyping. DP-T, MJ-S, AV-B, and SR: statistical analysis and interpretation of data. DP-T, MJ-S, and SR: writing of the manuscript. DP-T, AF-R, and PM: critical revision of the manuscript for relevant intellectual content. SR: supervision and visualization. All authors contributed to the article and approved the submitted version.
Funding
This work has been supported by grants given by Instituto de Salud Carlos III (ISCIII) (grant # PI17CIII/00003 to SR). MJ-S and AF-R are supported by Instituto de Salud Carlos III (grant # CP17CIII/00007 and CP14CIII/00010, respectively).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study would not have been possible without the collaboration of all the patients, medical and nursery staff and data managers who have taken part in the project. The authors also thank the Spanish National Genotyping Center (CEGENPRB2-ISCIII) for providing SNP genotyping services (http://www.cegen.org). CEGEN is supported by grant PT13/0001, ISCIII-SGEFI/FEDER.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmed.2020.582666/full#supplementary-material
Abbreviations
CHC, chronic hepatitis C; HCV, hepatitis C virus; DAAs, direct-acting antivirals; SNPs, single nucleotide polymorphisms; LSM, liver stiffness measurement; MTHFR, methylenetetrahydrofolate reductase; F4; LSM1 ≥ 12.5 kPa, cirrhosis; LSM1, baseline LSM; LSM2, final LSM; ΔLSM, increase of LSM; GLM, generalized linear models; AMR, arithmetic mean ratio; OR, odds ratio; SPSS, statistical package for the social sciences; IBS, Iberian population in Spain.
References
2. Lagging LM, Westin J, Svensson E, Aires N, Dhillon AP, Lindh M, et al. Progression of fibrosis in untreated patients with hepatitis C virus infection. Liver. (2002) 22:136–44. doi: 10.1034/j.1600-0676.2002.01623.x
3. Hoofnagle JH. Course and outcome of hepatitis C. Hepatology. (2002) 36(S1):S21–9. doi: 10.1053/jhep.2002.36227
4. Conti F, Buonfiglioli F, Scuteri A, Crespi C, Bolondi L, Caraceni P, et al. Early occurrence and recurrence of hepatocellular carcinoma in HCV-related cirrhosis treated with direct-acting antivirals. J Hepatol. (2016) 65:727–33. doi: 10.1016/j.jhep.2016.06.015
5. FornerA R. BruixJ. Hepatocellular carcinoma. Lancet. (2018) 391:1301–14. doi: 10.1016/S0140-6736(18)30010-2
6. Rüeger S, Bochud P, Dufour J-F, Müllhaupt B, Semela D, Heim M, et al. Impact of common risk factors of fibrosis progression in chronic hepatitis C. Gut. (2015) 64:1605–15. doi: 10.1136/gutjnl-2014-306997
7. Heim MH, Bochud P-Y, George J. Host–hepatitis C viral interactions: the role of genetics. J Hepatol. (2016) 65:S22–32. doi: 10.1016/j.jhep.2016.07.037
8. Pawlotsky J-M, Negro F, Aghemo A, Berenguer M, Dalgard O, Dusheiko G, et al. EASL recommendations on treatment of hepatitis C 2018. J Hepatol. (2018) 69:461–511. doi: 10.1016/j.jhep.2018.03.026
9. Bedossa P, Poynard T. An algorithm for the grading of activity in chronic hepatitis C. Hepatology. (1996) 24:289–93. doi: 10.1002/hep.510240201
10. Resino S, Sánchez-Conde M, Berenguer J. Coinfection by human immunodeficiency virus and hepatitis C virus: noninvasive assessment and staging of fibrosis. Curr Opin Infect Dis. (2012) 255:564–9. doi: 10.1097/QCO.0b013e32835635df
11. Castera L. Invasive and non-invasive methods for the assessment of fibrosis and disease progression in chronic liver disease. Best Pract Res Clin Gastroenterol. (2011) 25:291–303. doi: 10.1016/j.bpg.2011.02.003
12. Kopp M, Morisset R, Rychlik M. Characterization and interrelations of one-carbon metabolites in tissues, erythrocytes, and plasma in mice with dietary induced folate deficiency. Nutrients. (2017) 9:462. doi: 10.3390/nu9050462
13. Frosst P, Blom HJ, Milos R, Goyette P, Sheppard CA, Matthews RG, et al. A candidate genetic risk factor for vascular disease: a common mutation in methylenetetrahydrofolate reductase. Nat Genet. (1995) 10:111–3. doi: 10.1038/ng0595-111
14. Bosy-Westphal A, Petersen S, Hinrichsen H, Czech N, M JM. Increased plasma homocysteine in liver cirrhosis. Hepatol Res. (2001) 20:28–38. doi: 10.1016/s1386-6346(00)00119-4
15. Ventura P, Rosa MC, Abbati G, Marchini S, Grandone E, Vergura P, et al. Hyperhomocysteinaemia in chronic liver diseases: role of disease stage, vitamin status and methylenetetrahydrofolate reductase genetics. Liver Int. (2005) 25:49–56. doi: 10.1111/j.1478-3231.2005.01042.x
16. Toniutto P, Fabris C, Falleti E, Cussigh A, Fontanini E, Bitetto D, et al. Methylenetetrahydrofolate reductase C677T polymorphism and liver fibrosis progression in patients with recurrent hepatitis C. Liver Int. (2008) 28:257–63. doi: 10.1111/j.1478-3231.2007.01591.x
17. Adinolfi LE, Ingrosso D, Cesaro G, Cimmino A, D'Antò M, Capasso R, et al. Hyperhomocysteinemia and the MTHFR C677T polymorphism promote steatosis and fibrosis in chronic hepatitis C patients. Hepatology. (2005) 41:995–1003. doi: 10.1002/hep.20664
18. Dawood RM, Mahmoud EM, Ibrahim MK, Din NG, Aboul-Enein A, Zayed N, et al. Methylene tetrahydrofolate reductase gene polymorphism is associated with severity of liver steatosis in chronically infected patients with HCV genotype 4. Clin Lab. (2017) 63:419–26. doi: 10.7754/Clin.Lab.2016.160624
19. Fernández-Miranda C, Manzano ML, Fernández I, López-Alonso G, Gómez P, Ayala R, et al. Asociación entre hiperhomocisteinemia y esteatosis hepática en pacientes con hepatitis crónica C. Medicina Clínica. (2011) 136:45–9. doi: 10.1016/j.medcli.2010.05.024
20. Petta S, Bellia C, Mazzola A, Cabibi D, Camma C, Caruso A, et al. Methylenetetrahydrofolate reductase homozygosis and low-density lipoproteins in patients with genotype 1 chronic hepatitis C. J Viral Hepat. (2012) 19:465–72. doi: 10.1111/j.1365-2893.2011.01557.x
21. Samokhodskaya LM, Starostina EE, Sulimov AV, Krasnova capital Te C, Rosina TP, Avdeev VG, et al. Prediction of features of the course of chronic hepatitis C using Bayesian networks. Ter Arkh. (2019) 91:32–9. doi: 10.26442/00403660.2019.02.000076
22. Garcia-Tevijano ER, Berasain C, Rodriguez JA, Corrales FJ, Arias R, Martin-Duce A, et al. Hyperhomocysteinemia in liver cirrhosis: mechanisms and role in vascular and hepatic fibrosis. Hypertension. (2001) 38:1217–21. doi: 10.1161/hy1101.099499
23. Jiménez-Sousa MÁ, Gómez-Moreno AZ, Pineda-Tenor D, Sánchez-Ruano JJ, Artaza-Varasa T, Martin-Vicente M, et al. Impact of DARC rs12075 variants on liver fibrosis progression in patients with chronic hepatitis C: a retrospective study. Biomolecules. (2019) 9:143. doi: 10.3390/biom9040143
24. Jiménez-Sousa MÁ, Gómez-Moreno AZ, Pineda-Tenor D, Medrano LM, Sánchez-Ruano JJ, Fernández-Rodríguez A, et al. The IL7RA rs6897932 polymorphism is associated with progression of liver fibrosis in patients with chronic hepatitis C: repeated measurements design. PloS ONE. (2018) 13:e0197115. doi: 10.1371/journal.pone.0197115
25. Calvaruso V, Craxì A. 2011 European association of the study of the liver hepatitis C virus clinical practice guidelines. Liver Int. (2012) 32(Suppl. 1):2–8. doi: 10.1111/j.1478-3231.2011.02703.x
26. EASL Clinical Practice Guidelines: management of hepatitis C virus infection. J Hepatol. (2014) 60:392–420. doi: 10.1016/j.jhep.2013.11.003
27. Gabriel S, Ziaugra L, Tabbaa D. SNP genotyping using the Sequenom MassARRAY iPLEX platform. Curr Protoc Hum Genet. (2009) 60:2.12. 1–2. 8. doi: 10.1002/0471142905.hg0212s60
28. Castéra L, Vergniol J, Foucher J, Le Bail B, Chanteloup E, Haaser M, et al. Prospective comparison of transient elastography, Fibrotest, APRI, and liver biopsy for the assessment of fibrosis in chronic hepatitis C. Gastroenterology. (2005) 128:343–50. doi: 10.1053/j.gastro.2004.11.018
29. Jiménez-Sousa MÁ, Gómez-Moreno AZ, Pineda-Tenor D, Sánchez-Ruano JJ, Fernández-Rodríguez A, Artaza-Varasa T, et al. PNPLA3 rs738409 polymorphism is associated with liver fibrosis progression in patients with chronic hepatitis C: a repeated measures study. J Clin Virol. (2018) 103:71–4. doi: 10.1016/j.jcv.2018.04.008
30. Jiménez-Sousa MÁ, Gómez-Moreno AZ, Pineda-Tenor D, Brochado-Kith O, Sánchez-Ruano JJ, Artaza-Varasa T, et al. The myeloid-epithelial-reproductive tyrosine kinase (MERTK) rs4374383 polymorphism predicts progression of liver fibrosis in hepatitis C virus-infected patients: a longitudinal study. J Clin Med. (2018) 7:473. doi: 10.3390/jcm7120473
31. Weisberg I, Tran P, Christensen B, Sibani S, Rozen R. A second genetic polymorphism in methylenetetrahydrofolate reductase (MTHFR) associated with decreased enzyme activity. Mol Genet Metab. (1998) 64:169–72. doi: 10.1006/mgme.1998.2714
32. Fernandez-Vega B, Alvarez L, Garcia M, Artime E, Dineiro Soto M, Nicieza J, et al. Association study of mthfr polymorphisms with nonarteritic anterior ischemic optic neuropathy in a spanish population. Biomed Hub. (2020) 5:34–46. doi: 10.1159/000505431
33. Levin BL, Varga E. MTHFR: addressing genetic counseling dilemmas using evidence-based literature. J Genet Couns. (2016) 25:901–11. doi: 10.1007/s10897-016-9956-7
34. Arai K, Yamasaki Y, Kajimoto Y, Watada H, Umayahara Y, Kodama M, et al. Association of methylenetetrahydrofolate reductase gene polymorphism with carotid arterial wall thickening and myocardial infarction risk in NIDDM. Diabetes. (1997) 46:2102–4. doi: 10.2337/diab.46.12.2102
35. Brattström L, Zhang Y, Hurtig M, Refsum H, Östensson S, Fransson L, et al. A common methylenetetrahydrofolate reductase gene mutation and longevity. Atherosclerosis. (1998) 141:315–9.
36. Zhang S, Jiang J, Tang W, Liu L. Methylenetetrahydrofolate reductase C677T (Ala>Val, rs1801133 C>T) polymorphism decreases the susceptibility of hepatocellular carcinoma: a meta-analysis involving 12,628 subjects. Biosci Rep. (2020) 40:BSR20194229. doi: 10.1042/BSR20194229
37. Luo Z, Lu Z, Muhammad I, Chen Y, Chen Q, Zhang J, et al. Associations of the MTHFR rs1801133 polymorphism with coronary artery disease and lipid levels: a systematic review and updated meta-analysis. Lipids Health Dis. (2018) 17:1–15. doi: 10.1186/s12944-018-0837-y
38. Lonardo A, Adinolfi LE, Loria P, Carulli N, Ruggiero G, Day CP. Steatosis and hepatitis C virus: mechanisms and significance for hepatic and extrahepatic disease. Gastroenterology. (2004) 126:586–97. doi: 10.1053/j.gastro.2003.11.020
39. Fabris C, Toniutto P, Falleti E, Fontanini E, Cussigh A, Bitetto D, et al. MTHFR C677T polymorphism and risk of HCC in patients with liver cirrhosis: role of male gender and alcohol consumption. Alcohol Clin Exp Res. (2009) 33:102–7. doi: 10.1111/j.1530-0277.2008.00816.x
40. Avila MA, Berasain C, Torres L, Martin-Duce A, Corrales FJ, Yang H, et al. Reduced mRNA abundance of the main enzymes involved in methionine metabolism in human liver cirrhosis and hepatocellular carcinoma. J Hepatol. (2000) 33:907–14. doi: 10.1016/s0168-8278(00)80122-1
41. Peres NP, Galbiatti-Dias AL, Castanhole-Nunes MM, da Silva RF, Pavarino EC, Goloni-Bertollo EM, et al. Polymorphisms of folate metabolism genes in patients with cirrhosis and hepatocellular carcinoma. World J Hepatol. (2016) 8:1234–43. doi: 10.4254/wjh.v8.i29.1234
Keywords: chronic hepatitis C, liver stiffness measure, hepatic fibrosis, cirrhosis, MTHFR (C677T), SNPs (single nucleotide polymorphism)
Citation: Pineda-Tenor D, Gómez-Moreno AZ, Sánchez-Ruano JJ, Artaza-Varasa T, Virseda-Berdices A, Fernández-Rodríguez A, Mendoza PM, Jiménez-Sousa MÁ and Resino S (2020) MTHFR rs1801133 Polymorphism Is Associated With Liver Fibrosis Progression in Chronic Hepatitis C: A Retrospective Study. Front. Med. 7:582666. doi: 10.3389/fmed.2020.582666
Received: 13 July 2020; Accepted: 12 October 2020;
Published: 13 November 2020.
Edited by:
Rikke Norregaard, Aarhus University, DenmarkReviewed by:
Ekaterina Kolesanova, Russian Academy of Medical Sciences (RAMS), RussiaJing He, Guangzhou Medical University, China
Copyright © 2020 Pineda-Tenor, Gómez-Moreno, Sánchez-Ruano, Artaza-Varasa, Virseda-Berdices, Fernández-Rodríguez, Mendoza, Jiménez-Sousa and Resino. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: María Ángeles Jiménez-Sousa, majimenezsousa@yahoo.es; Salvador Resino, sresino@isciii.es
†These authors have contributed equally to this work
 Daniel Pineda-Tenor
Daniel Pineda-Tenor Ana Zaida Gómez-Moreno2
Ana Zaida Gómez-Moreno2  Amanda Fernández-Rodríguez
Amanda Fernández-Rodríguez Salvador Resino
Salvador Resino