Cyclic di-GMP Is Integrated Into a Hierarchal Quorum Sensing Network Regulating Antimicrobial Production and Biofilm Formation in Roseobacter Clade Member Rhodobacterales Strain Y4I
- Department of Microbiology, The University of Tennessee, Knoxville, Knoxville, TN, United States
Microbial biofilms associated with marine particulate organic matter carry out transformations that influence local and regional biogeochemical cycles. Early microbial colonizers are often hypothesized to “set the stage” for biofilm structure, dynamics, and function via N-acyl homoserine lactone (AHL)-mediated quorum sensing (QS). Production of AHLs, as well as antimicrobials, contributes to the colonization success of members of the Roseobacter clade. One member of this group of abundant marine bacteria, Rhodobacterales sp. Y4I, possesses two QS systems, phaRI (QS1) and pgaRI (QS2). Here, we characterize mutants in both QS systems to provide genetic evidence that the two systems work in hierarchical fashion to coordinate production of the antimicrobial indigoidine as well as biofilm formation. A mutation in pgaR (QS2) results in decreased expression of genes encoding both QS systems as well as those governing the biosynthesis of indigoidine. In contrast, mutations in QS1 did not significantly influence gene expression of QS2. Addition of exogenous AHLs to QS1 and QS2 mutants led to partial restoration of indigoidine production (45–60% of WT) for QS1 but not QS2. Mutational disruptions of QS1 had a more pronounced effect on biofilm development than those in QS2. Finally, we demonstrate that c-di-GMP levels are altered in QS and indigoidine biosynthesis Y4I mutants. Together, these results indicate that pgaRI (QS2) is at the top of a regulatory hierarchy governing indigoidine biosynthesis and that the global regulatory metabolite, c-di-GMP, is likely integrated into the QS circuitry of this strain. These findings provide mechanistic understanding of physiological processes that are important in elucidating factors driving competitiveness of Roseobacters in nature.
Introduction
Cell-to-cell signaling known as quorum sensing (QS) allows bacterial populations to coordinate gene expression. This coordination of gene expression typically occurs in a cell density dependent manner, through the production and recognition of small diffusible signaling molecules. A prevalent class of these molecules in Gram-negative Proteobacteria are N-acyl homoserine lactones (AHLs) (Papenfort and Bassler, 2016). AHL-based QS characteristically involves a two-component system consisting of a transcriptional regulator (denoted by -R) and an AHL synthase (designated as -I). When bound to their cognate transcriptional regulator, AHLs can elicit global changes in gene expression (Williams et al., 2007). In model biofilm forming representatives, QS has been shown to genetically regulate surface attachment and biofilm formation (Hammer and Bassler, 2003; Schuster and Peter Greenberg, 2006). Surface attachment and biofilm formation is also often modulated through intracellular signaling of a global secondary messenger of bis-(3′-5′)-cyclic dimeric guanosine monophosphate (c-di-GMP). Concentrations of c-di-GMP are influenced by environmental cues as well as cell-to-cell signaling in bacteria (Römling et al., 2013). Indeed, c-di-GMP signaling is integrated in QS regulatory systems in model biofilm bacteria, such as Pseudomonas aeruginosa (Lin Chua et al., 2017). Integration of this intracellular signaling molecule into QS architectures more tightly links cellular physiology with adaptive behaviors.
Members of the Roseobacter clade are competitive surface colonizers in diverse marine niches. This lineage of heterotrophic Alphaproteobacteria represent one of the most phylogenetically coherent, yet physiologically diverse, groups of marine bacteria. Roseobacters are among the most abundant, metabolically active, and aggressive surface colonizers in marine ecosystems (Brinkhoff et al., 2008). As such, members of this clade inhabit a wide range of niches and readily colonize surfaces both abiotic and biotic in nature, e.g., sinking particulate organic matter (Farnelid et al., 2019), Trichodesmium colonies (Van Mooy et al., 2012), and polymer test surfaces (Dang and Lovell, 2000). Recent genomic evidence suggests up to 87% of sequenced Roseobacter genomes contain at least one QS systems. Of those, half contain multiple QS systems (Zan et al., 2014). Despite the prevalence of QS systems identified in Roseobacter genomes, these systems have been studied in a limited number of strains.
Rhodobacterales sp. Y4I shows promise as an emerging model organism to better understand the roles QS play in Roseobacter physiology. Isolated from a coastal marsh ecosystem in the southeastern United States, Y4I has been shown to be a competitive surface colonizer, prolific biofilm former, and a dominant member in mixed species mesocosm experiments (Cude et al., 2012; Quigley et al., 2019). Specifically, Y4I has been shown to have a competitive advantage against other marine bacteria, including members of its own clade, when grown on a surface. This fitness advantage has been demonstrated to be linked to the production of the blue-pigmented, redox reactive antimicrobial indigoidine (Cude et al., 2012). Indigoidine is regulated by two QS systems, phaRI (QS1) and pgaRI (QS2), in this strain. Previous studies demonstrated a disruption of either QS system influences both indigoidine and AHL production. However, the contributions of each QS system to production of indigoidine were ambiguous due to the expression of a leaky phenotype in a QS1 mutant (Cude et al., 2015). In addition, the regulatory network governing indigoidine biosynthesis is complex, particularly given that surface attachment is a prerequisite for its production, suggesting that cellular mechanisms beyond QS alone may play a role in coordinated cellular responses to environmental conditions. Here, we assess the hierarchical organization of these two QS systems to coordinate production of the antimicrobial indigoidine and biofilm formation. We also present evidence that the secondary global messenger, c-di-GMP, is integrated into the complex regulatory network governing surface attachment and indigoidine production.
Materials and Methods
Strains, Growth Conditions, and Maintenance
Rhodobacterales sp. Y4I was previously isolated from an estuary off the coast of Georgia in the southeastern United States (Buchan et al., 2004; Slightom and Buchan, 2009). Insertional mutants in either the indigoidine synthase gene, igiD::Tn5-KmR (formerly Y401BD4), or QS transcriptional regulators, phaR::Tn5-KmR (formerly Y412AE6) and pgaR::Tn5-KmR (formerly Y411CE4) were previously generated using a mini Tn5 transposon system (Cude et al., 2012). Aliivibrio fischeri ES114 (ATCC 700601) was kindly provided by Eric Stabb (University of Illinois). Unless otherwise noted, all Y4I and A. fischeri strains were routinely grown in YTSS [per liter: 15 g Sea Salts (Sigma) or Instant Ocean (Thermo Fisher Scientific), 4 g tryptone, 2.5 g yeast extract] and incubated at 30°C and 25°C, respectively. Escherichia coli BW20767, which is pir+, was maintained on Luria-Bertani (LB) and routinely grown at 37°C on LB unless otherwise noted (Metcalf et al., 1996).
Construction of phaI Mutant Using Insertional Mutagenesis
Insertional mutagenesis of the phaI (QS1) gene of strain Y4I was achieved using the pKNOCK-KmR plasmid (Alexeyev, 1999). Briefly, an internal fragment of phaI (302 bp) was PCR amplified using primers Y4I_3464_for and Y4I_3464_rev (Supplementary Table 1) and ligated into the PCR cloning vector TA TOPO pCR 2.1 (Invitrogen, Carlsbad, CA, United States) following manufacturer’s guidelines. The phaI-containing plasmid was digested with BamHI and XhoI, liberating the phaI fragment with compatible cohesive ends suitable for ligation into linearized pKNOCK-KmR. The phaI-pKNOCK-KmR plasmid was introduced into Y4I via biparental mating with E. coli strain BW20767. Counterselection for transconjugants was achieved on marine basal medium (MBM) with 2 mM p-hydroxybenzoate (POB) as the sole carbon source and kanamycin (50 μg/mL) [per liter: 1.5% (wt/vol) Sigma Sea Salts, 2.38 μM K2HPO4, 13.35 mM NH4Cl, 71 mM Tris–HCl (pH 7.5), 68 μM Fe-EDTA, trace metals and vitamins] at 30°C (Gonzalez et al., 1997). The resulting mutant was PCR confirmed using primers phaIF and phaIR. Orientation of pKNOCK insert was confirmed using two primer sets: (i) phaIF and pKNOCK_746 and (ii) phaIR and pKNOCK_895 (Supplementary Table 1). Finally, the mutant was verified by sequencing of the phaI gene and designated phaI::pKNOCK.
Gene Expression Assays
RNA was isolated from cellular biomass scraped from agar (1.5%) plates and suspended in 1.5 mL of β-mercaptoethanol–RLT buffer (Qiagen, Germantown, MD, United States). The suspension was transferred to a 2.0 mL screw cap tube containing 0.2 g of low-binding 200 μm zirconium beads (OPS Diagnostics, LLC, Lebanon, NJ, United States). The vial was then subjected to vortex mixing at 13,000 rpm for 10 min and incubated in the water bath (70°C) for 5 min. Cell debris was pelleted and lysate was transferred into a clean 1.5 mL tube. Nucleic acids were precipitated using 700 μL volume of 70% ethanol. The lysate was then transferred to a RNeasy Mini spin column and RNA was isolated following the RNeasy minikit protocol (Qiagen, Germantown, MD, United States). Turbo DNase (Invitrogen) was used to remove DNA from samples. Reverse transcriptase (RT) was performed using Moloney murine leukemia virus (M-MLV) reverse transcriptase following manufacturer’s guidelines (Invitrogen). No RT controls were included to identify any residual DNA contamination in RNA samples. Nucleic acid concentration was measured using a Nanodrop spectrophotometer (Nanodrop Technologies, Inc., Wilmington, DE, United States). Total RNA was diluted to 6 ng/μL for use in assays.
Quantitative reverse transcription PCR (qRT-PCR) was used to assess gene expression of igiD, phaR, pgaR, phaI, and pgaI in strains grown on agar plates for 24 h. Position of gene disruptions and location of qPCR primer binding sites are presented in Supplementary Table 2. Amplified products ranged in size from 150 to 240 bp. All assays are normalized to three reference genes (rpoC, alaS, and map; Supplementary Table 1). qRT-PCR data analysis and the normalized relative transcript quantities were calculated using the qBASE method (Hellemans et al., 2007).
Viable Cell Counts and Total Biomass on Surface Attached Cultures
To assess viability of surface attached cells, we inoculated a 96-well polystyrene plate (Costar, Corning Incorporated, Corning, NY, United States) containing a sterilized 4 mm glass bead (Pyrex, Corning Incorporated, Corning, NY, United States) with Y4I variants at ∼1 × 105 cells/mL and incubated at 30°C. Y4I variants were inoculated in biological and technical triplets. A duplicate of each plate was made to assess total biomass described below. Following 20, 44, and 48 h, the glass bead were extracted from wells and cells were dislodged from beads as previously described (Cude et al., 2012). Briefly, beads containing cell biomass were transferred to 2 mL plastic screw cap tubes containing 940 μL of 20% YTSS (per liter: 15 g Instant Ocean, 0.8 g tryptone, 0.5 g yeast extract) and 60 μL of 10% Tween 20, submerged in a sonicating water bath and sonicated for 6 min at 40 kHz. Samples were then vortexed for ∼30 s and diluted to perform viable cell counts.
Total biomass on surface attached cultures was assessed by performing a crystal violet stain on glass beads. Briefly, liquid was removed from wells containing glass beads. The beads were rinsed with 1.5% sea salt solution (Instant Ocean), then extracted and moved to a clean 96-well plate containing 100 μL YTSS. Uninoculated glass beads served as a control. Crystal violet (25 μL) was added to all wells in the 96-well plate containing beads and incubated for 30 min at room temperature. Liquid was aspirated out of wells and stained beads were washed ∼3X with 1.5% sea salt solution. Following wash, 200 μL of 95% EtOH was added to each well and allowed to sit for 1 h to allow crystal violet to solubilize. Following solubilization, liquid was transferred to a clean 96-well plate and diluted 1:10 with 95% EtOH. Absorbance was read at 600 nm using a microplate reader (BioTek Instruments, Inc., Synergy HT Multi-Mode Microplate Reader, SN 270212).
Cyclic-di-GMP Extraction and Quantification
Cyclic-di-GMP was extracted from cell biomass in liquid and growth on agar cultures using procedures adapted from O’Neal et al. (2017). For broth grown cultures, Y4I variants were grown in liquid YTSS at 30°C overnight, shaking in the dark. Cultures were reinoculated in YTSS and allowed to grow for 24 h under the same conditions. Following 24 h, 10 mL of culture was pelleted by centrifugation (3 min at 8,000 rpm). Supernatant was decanted, biomass was resuspended in 1 mL 1.5% sea salt solution and transferred to a 1.5 mL microcentrifuge tube. Biomass was pelleted again, and supernatant was removed by pipetting. For surface attached cultures, Y4I variants were grown in liquid YTSS at 30°C overnight, shaking in the dark. Liquid cultures were spotted (50 μL) onto YTSS agar and incubated for 24 h at 30°C. Biomass was scrapped from agar using cell scrappers and transferred to a 1.5 mL microcentrifuge tube. For Y4I variants in either growth medium technical replicates were combined, if necessary, to achieve an approximate biomass of ∼0.04 g. Biomass was then suspended in 100 μL extraction buffer (40% methanol, 40% acetonitrile, and 0.1 M of N formic acid) per 0.04 g of biomass. Cell slurries were incubated at −20°C for 30 min. Following incubation, cells slurries were centrifuged at 4°C (13,000 rpm for 3 min) to remove insoluble material. Lysate was neutralized with 4 μL of 15% ammonium bicarbonate per 100 μL of sample and transferred to a clean 1.5 mL microcentrifuge tube. Cyclic-di-GMP was extracted from pellet again using 50 μL of extraction buffer per 0.04 g of initial biomass. Incubated for 30 min at −20°C, centrifuged at 4°C at 13,000 rpm for 3 min, and neutralized with 4 μL 15% ammonium bicarbonate and lysates were combined. Following extraction, samples were dried, and shipped for processing to the Mass Spectrometry and Metabolomics Core (Michigan State University). Samples were resuspended in ∼160 μL of mobile phase A [(10 mM tributylamine +15 mM acetic acid) in 97:3/H2O:MeOH]. Fluorinated c-di-GMP (Difluoro 3′3′-c-di-GMP, InvivoGen, San Diego, CA, United States) was added as internal standard prior to being analyzed using mass spectrometry.
Growth Inhibition Assays
Growth inhibition of A. fischeri by Y4I variants was assessed as previously reported (Cude et al., 2012, 2015). Briefly, A. fischeri was grown overnight (23°C) to an optical density (OD) of 1.5–2.0 at 540 nm. Cultures of A. fischeri were diluted 1,000-fold and spread plated on to YTSS. Plates containing A. fischeri lawn were allowed to dry prior to spotting with Y4I variants. Y4I variants were grown to an OD of 0.6 at 540 nm and 10 μl spot plated on A. fischeri lawns and incubated at 27°C as growth rates of the two organisms are comparable at this temperature (Cude et al., 2012). Zones of clearing around Y4I strains was assessed at 48 h post inoculation.
Exogenous AHL Add-Back Assay and Indigoidine Quantification
To test whether addition of exogenous AHLs could restore indigoidine production, we performed AHL add-back assays and measured indigoidine production in each Y4I variant. Y4I and variants were diluted from overnight cultures to an OD540nm of approximately 0.2. This optical density corresponds to early exponential phase in Y4I strains. Cultures were then incubated at 30°C until late exponential phase (OD540nm ∼1.3) was reached. At this time, a final concentration of 13 nM 3OHC12:1-HSL was added to the liquid cultures. Technical replicates containing no added 3OHC12:1-HSL served as controls. All cultures were incubated at 30°C for approximately 16 h until cultures reached stationary phase (OD540nm ∼2.0). Following incubation, cultures were spot plated, in triplicate, onto YTSS agar containing a final concentration of 800 nM C8-HSL. All cultures were also spot plated on YTSS agar containing no C8-HSL. Spot plates were incubated at 30°C for 24 h (Supplementary Figure 1). Following incubation, one set of technical replicates was sacrificed to quantify indigoidine as outlined previously (Cude et al., 2012). Briefly, cells were scrapped from plates and solubilized in dimethyl sulfoxide (DMSO). To obtain purified pigment, the cells were lysed via sonication and cell debris were collect via centrifugation. Pigmented supernatant was passed through a 0.2 μm filter and transferred to a clean tube. Two microliters aliquots of the solubilized pigment were measured on a Nanodrop spectrometer (Thermo Fisher Scientific) at 612 nm. 3OHC12:1-HSL and C8-HSL were synthesized locally as described previously (Cude et al., 2015).
Statistical Analysis
All data was analyzed and visualized using GraphPad Prism8 (GraphPad Software, La Jolla, CA, United States)1. Two-way ANOVA with Dunnett’s test was performed on gene expression assay data. One-way ANOVA with Tukey’s test was used to determine statistical significance in quantitative biofilm assays and cyclic-di-GMP analysis. One-way ANOVA using Dunnett’s test was used to determine statistical significance on AHL add-back assays. Mixed effects analysis was performed on viable cell counts and total biomass assays using Dunnett’s and Tukey’s, respectively, for post-hoc analyses. Outliers were identified with 95% confidence using a robust non-linear repression model (ROUT) (Motulsky and Brown, 2006).
Results
Rhodobacterales sp. Y4I possesses two QS systems: phaRI (QS1) and pgaRI (QS2). We have previously demonstrated that disruption of pgaR (QS2) results in undetectable levels of its cognate AHL, C8-HSL, which is synthesized by PgaI (Cude et al., 2015). This result indicates that the insertional mutation in pgaR leads to downstream effects on pgaI expression. Indeed, this is confirmed by the gene expression data presented in a subsequent section. In contrast, disruption of phaR (QS1) results in detectable, albeit decreased levels, of its cognate AHL, 3OHC12:1-HSL. In addition, indigoidine production is impaired, but not abolished, in this mutant (Cude et al., 2015). Thus, to fully elucidate the relative contributions of each QS system and to define the regulatory architecture governing indigoidine biosynthesis, we first generated a AHL synthase mutant in the QS1 system (phaRI) in the marine isolate Rhodobacterales sp. Y4I. We then assessed various phenotypes of this and previously generated mutants.
The phaI Mutant Does Not Produce Indigoidine and Demonstrates Increased Biofilm Formation
Unlike the phaR::Tn5-KmR (QS1) mutant, phaI::pKNOCK (QS1) does not produce detectable levels of its cognate AHL (data not shown) nor does it produce any visible levels of indigoidine, even after several days of incubation. Consistent with this phenotype, the mutant is unable to inhibit the growth of A. fischeri (Table 1). We also assessed whether insertional mutagenesis of phaI results in growth defects. Growth of phaI::pKNOCK in liquid culture was comparable to that of wildtype (Supplementary Figure 2A). Given that Y4I forms prolific biofilms and mutants in QS1, QS2, and the indigoidine synthase gene (igiD) previously demonstrated increased biofilm formation compared to wildtype, we tested biofilm formation in phaI::pKNOCK. The phaI mutant has significantly increased biofilm formation relative to wildtype as well as the other mutants assayed (Supplementary Figure 2B).
Disruption in pgaRI (QS2) Leads to a System Wide Decrease in QS and igiD Gene Expression
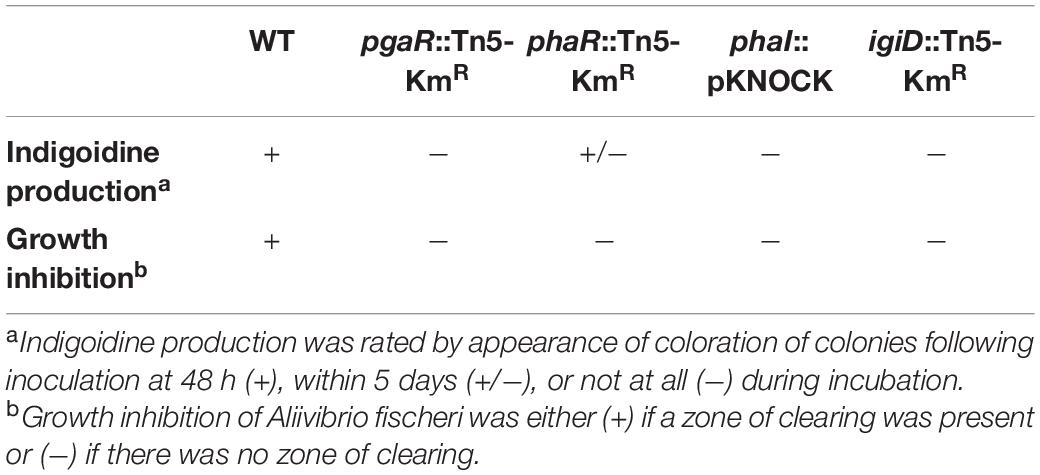
To examine the QS regulatory network governing indigoidine biosynthesis in Y4I, we performed gene expression assays via qRT-PCR on the following five genes: igiD (indigoidine synthase); phaR and -I (QS1); and pgaR and -I (QS2) (Figure 1). A disruption in the luxR homolog of QS2 (pgaRI) resulted in decreased expression in all genes surveyed: a nearly 5-fold decrease in its own gene, more than 10-fold decrease in the luxI homolog of QS2, a 4-fold decrease in phaR, and a 2-fold decrease in phaI. Furthermore, this disruption in pgaR led to more than 1,000-fold decrease in igiD expression (Figure 1A).
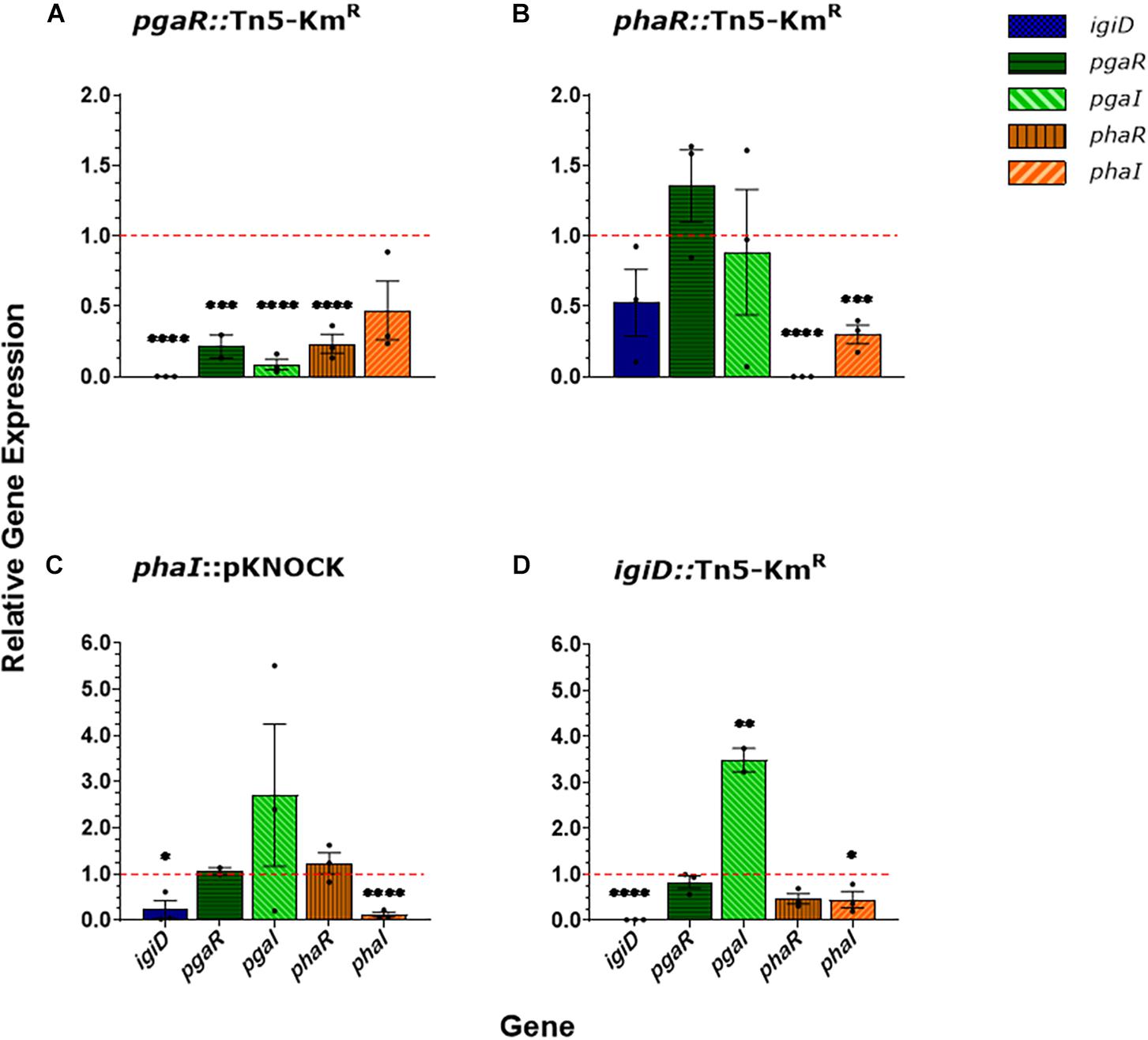
Figure 1. Gene expression of luxRI homologs and igiD synthase from agar grown cultures. Relative gene expression for pgaR::Tn5-KmR (A), phaR::Tn5-KmR (B), phaI::pKNOCK (C), igiD::Tn5-KmR (D). Gene expression was normalized to three housekeeping genes (rpoC, alaS, and map) and expressed relative to WT, set to 1. Each data point represents the average of three technical replicates and errors bars represent standard deviations between three biological replicates. Statistical significance was calculated using a two-way ANOVA [p < 0.05 (∗), 0.01 (∗∗), 0.001 (∗∗∗), 0.0001 (****)].
Insertional disruption of phaR (QS1) resulted in a 2,500-fold decrease of its own expression, but only a ∼3-fold decrease in expression of phaI. Expression of igiD showed a 2-fold decrease compared to wildtype (Figure 1B). Insertional mutagenesis in phaI resulted in an 8-fold decrease in expression of its own gene and a 4-fold decrease in igiD expression (Figure 1C). In the igiD::Tn5-KmR mutant, expression of QS1 lux homologs genes (phaRI) showed a 2-fold reduction for both genes, while expression of the luxR homolog in QS2 (pgaR) remained at wildtype levels. Insertional disruption of igiD resulted in a nearly 4-fold increase in pgaI (QS2) gene expression (Figure 1D).
Disruption of either QS1 gene (phaR and -I) led to greater variance in expression of the other assayed genes. For both mutants, a consistent variation is seen in the pgaI gene expression: in phaR::Tn5-KmR, pgaI transcripts from more than 10-fold decrease to a nearly 2-fold increase in across biological replicates (Figure 1B). In phaI::pKNOCK, pgaI transcripts range from a 5-fold decrease to a 5-fold increase across replicates (Figure 1C). Variability within this mutant is consistent across multiple experimental assays, including biofilm formation, motility, and spontaneous pellicle formation (aka flocking) in liquid cultures (data not shown).
Exogenous Addition of AHLs Partially Restores Indigoidine Production in QS1 Mutants
Given AHL concentrations are decreased in all QS mutants, we next tested to see if we could chemically complement these mutants with the exogenous addition of AHLs. To mimic previously characterized AHL concentrations in Y4I during different phases of growth (Cude et al., 2015), 13 nM 3OHC12:1-HSL was added to liquid cultures during late exponential phase (OD540nm ∼1.3). When cultures reached stationary phase (∼16 h following 3OHC12:1-HSL addition), they were spotted onto agar plates containing a concentration of 800 nM C8-HSL. To assess whether one AHL was driving indigoidine production, we also included 3OHC12:1-HSL and C8-HSL alone controls (Supplementary Figure 1 and Figure 2). To determine whether exogenous addition of AHLs was able to rescue indigoidine production phenotype, we compared the absorbance of our AHL treated mutants to that of the no AHL control in our indigoidine null mutant (igiD::Tn5-KmR) (Figure 2). To assess significance in rescuing indigoidine production, we compared AHL treatments to no AHL treatments within mutant genotypes (Figure 2).
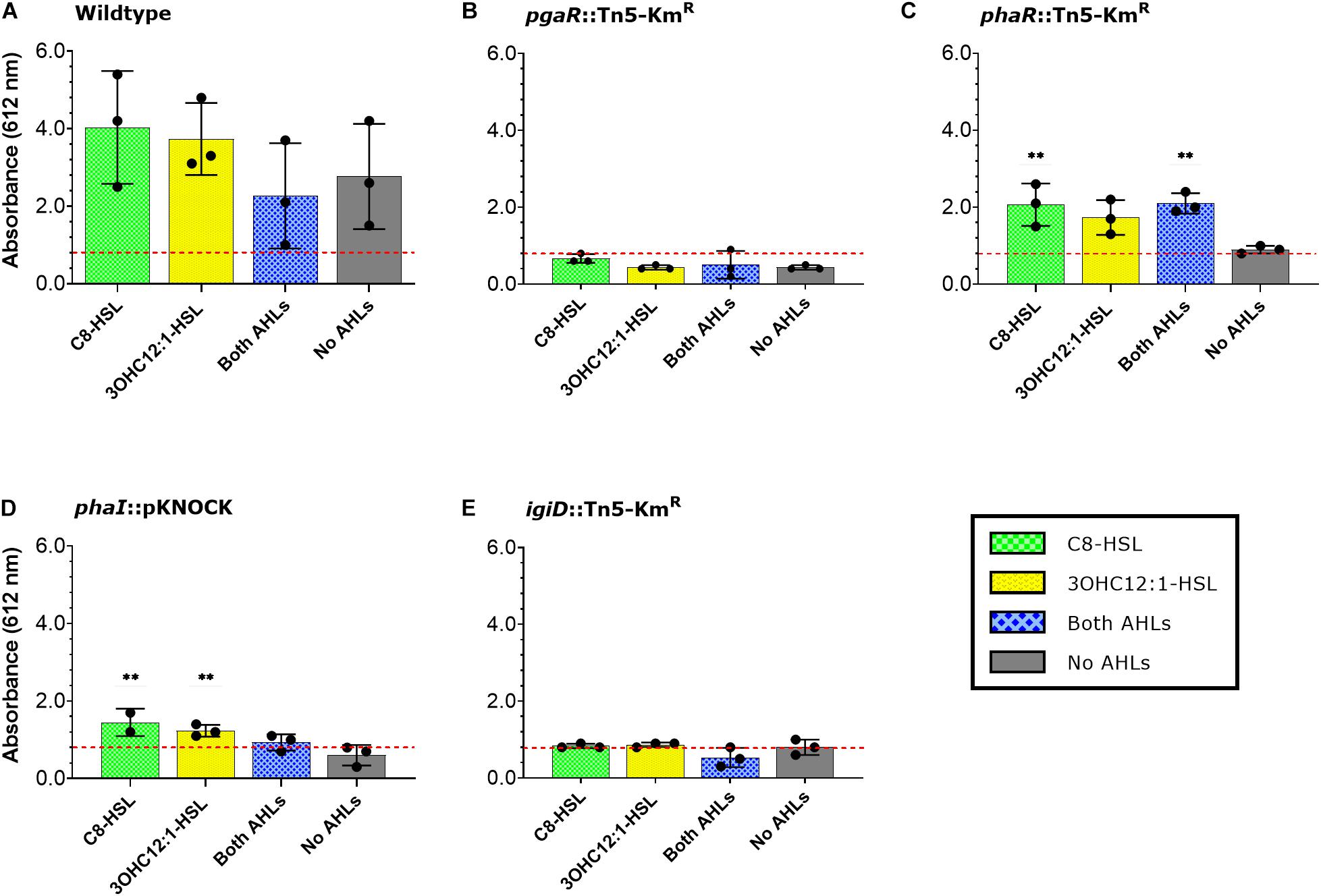
Figure 2. Indigoidine production in response to growth-phase dependent AHL exogenous addition. Indigoidine quantification in Y4I variants: WT (A), pgaR::Tn5-KMR (B), pgaR::Tn5-KMR (C), phaR::pKNOCK (D), and igiD::Tn5-KmR (E) following exogenous addition of AHLs, as indicated in the legend. Indigoidine quantification was normalized to biomass and compared to no a AHL control in the igiD::Tn5-KmR mutant (red dotted line). Error bars represent standard deviations from the mean of three biological replicates. Statistical significance was calculated by comparing AHL treatments to the no AHL control within strain genotypes. Asterisks represent statistical difference (p < 0.01).
Exogenous addition of AHLs did not produce any significant response in indigoidine production in wildtype, the indigoidine null mutant or the pgaR (QS2) mutant (Figures 2A,B,E). Addition of 3OHC12:1-HSL (QS1) alone does not significantly restore pigment production in the phaR mutant, providing additional support for the non-functionality of PhaR in that strain (Figure 2C). Both phaI and -R (QS1) mutants show some restoration of indigoidine production when supplied with exogenous C8-HSL. When supplied with either AHL, but not both, the phaI mutant shows significantly increased indigoidine production. Interestingly, C8-HSL addition restores indigoidine production to 33% that of wildtype in the phaI mutant compared to the chemical complement of its own AHL (3OHC12:1-HSL) which restored production to 22% that of wildtype. Addition of both AHLs in combination resulted in less than 10% wildtype levels of indigoidine production in this mutant background (Figure 2D).
Temporal Analysis Reveals Potential Role for QS in Biofilm Regulation
We next assessed whether alterations in the QS pathways also lead to differences in rates of surface colonization and biofilm formation. The indigoidine biosynthesis mutant was included for reference in these assays. Quantification of viable surface attached cells over a 68-h period revealed all mutants were impaired in their ability to either attach or grow on the surface during the initial growth phase (20 h). However, those differences were insignificant during later sampling efforts (Figure 3A). In contrast, quantification of biofilm biomass showed the opposite trend. All mutants were indistinguishable from wildtype at the 20 h time point, but variation between and among wildtype and the mutants became pronounced with time. For example, both phaR (QS1) and pgaR (QS2) mutants show significantly elevated biofilm biomass relative to wildtype at 44 h, but these biofilms were not significantly different by the final time point (68 h). Conversely, phaI (QS1) mutant was indistinguishable from wildtype until the final time point, at which point it was 32–42% greater than wildtype and all other variants (Figure 3B).
Figure 3. Surface colonization and biofilm production over time in Y4I mutant variants. Viable cell counts of surface attached cultures in Y4I variants (A). Each data point represents the average of three biological and three technical replicates. Data was normalized to cell abundance in liquid culture at the time of inoculation. Error bars represent the standard error of the mean of biological and technical triplicates. The asterisks denote statistical significance from WT at that time point [P < 0.05 (*), 0.01 (**), 0.001 (***)]. Total biofilm of surface attached cultures in Y4I variants as determined by crystal violet assay (B). Data is representative of the mean of three biological and three technical replicates. Letters that are not shared represent significant difference (p < 0.05). Error bars represent the standard deviation from the mean.
Cyclic-di-GMP Is Higher in Surface Attached Cultures and Influenced by QS
In many biofilm forming bacteria the global secondary messenger, c-di-GMP, is integrated into the regulatory network dictating the transition between motile and sessile lifestyles (Römling et al., 2013). Thus, we wanted to investigate intracellular c-di-GMP concentrations in Y4I and the panel of QS and indigoidine mutants. C-di-GMP concentrations are significantly increased in agar grown Y4I cultures relative to broth cultures (Figure 4A). Furthermore, all mutants have significantly reduced c-di-GMP concentrations, ranging from 15 to 33% of wildtype levels when strains are grown in liquid culture and 4–23% of wildtype when grown on agar surfaces (Figures 4B,C).
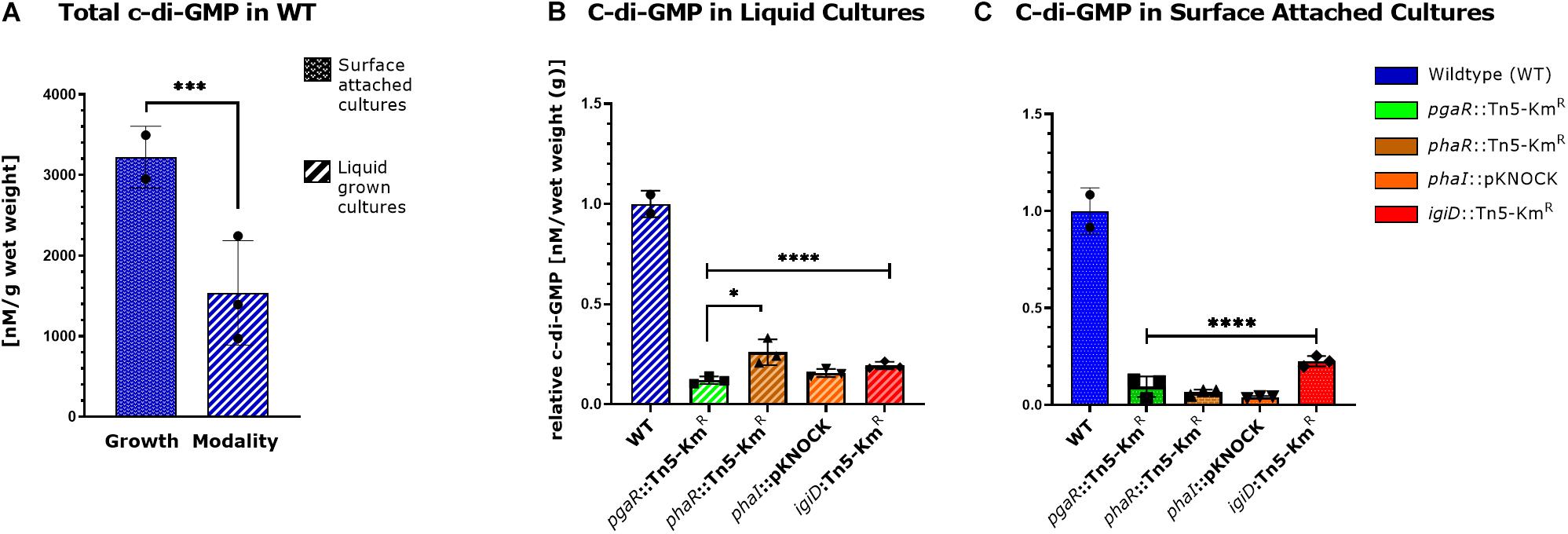
Figure 4. Cyclic-di-GMP concentrations in broth and agar grown cultures. A comparison of intracellular cyclic-di-GMP concentration in WT cultures grown on agar and in broth culture (A). Intracellular c-di-GMP concentration of Y4I variants grown on broth (B) and in agar culture (C) are shown. Each data point represents the average of three technical replicates. Error bars represent the standard deviation of the mean. Asterisks represent statistical significance [p < 0.05 (∗), 0.001 (∗∗∗), 0.0001 (****)].
Discussion
N-acyl homoserine lactone-based QS networks are prevalent in members of the Roseobacter clade where they regulate physiologies predicted to be central to the ecological success of group members (Cude and Buchan, 2013; Zan et al., 2014). The high incidence of multiple QS systems in individual strains presents opportunity for different QS network architectures and signaling circuities to have evolved, the extent of which has not been fully evaluated for Roseobacters. From studies of diverse microbes, three defined QS network architectures have been proposed and provide a valuable comparative framework: (i) “One-to-One” system, in which a single receptor responding to a single signal controls the entire QS response; (ii) “Many-to-One” parallel circuit, in which information contained in multiple autoinducers are integrated together to control the QS response; and (iii) “Many-to-One” hierarchical system, in which many QS receptors are connected in a signaling cascade (Hawver et al., 2016). Of the handful of Roseobacters that have been genetically characterized with regards to QS, two appear to employ a hierarchal regulatory circuit: Ruegeria mobilis KLH11 (Zan et al., 2012) and Dinoroseobacter shibae (Koppenhöfer et al., 2019). A third, Phaeobacter inhibens, appears to employ a parallel regulatory circuit to regulate the production of the antimicrobial trophodithietic acid (Beyersmann et al., 2017). Here, we propose that Y4I uses a “Many-to-One” hierarchal QS circuit to mediate indigoidine production and biofilm formation. We also provide evidence for integration of a global regulatory metabolite (c-di-GMP) into this QS circuitry.
Several lines of evidence suggest that QS2 (pgaRI) is at the top of the hierarchical regulatory network that integrates input from QS1 to govern indigoidine biosynthesis. Expression of both QS systems and the indigoidine synthase gene, igiD, is negatively influenced when the QS2 transcriptional regulator (pgaR) is disrupted. This mutant is also null for its cognate AHL. In contrast, disruption of either QS1 component (phaR or -I) has no significant effect on gene expression of QS2 components, though pgaI expression shows considerable and consistent variation across biological replicates in both QS1 mutants. We postulate that the variation in pgaI expression is a manifestation of global scale misregulation resulting in some degree of stochasticity within these mutants. This suggests a role for the QS1 system in regulatory homeostasis in this strain. Generation and study of a pgaI mutant would further support this finding.
The phenotypic response of the mutants to exogenous AHL additions provide further evidence for the predominant role of QS2 as well as cross-signaling between the two QS systems. With regards to indigoidine production, the QS2 mutant (pgaR) is unresponsive to any of the AHL additions. However, both QS1 mutants can be partially restored for this phenotype. When supplied with exogenous C8-HSL (QS2), indigoidine production is evident in both QS1 (phaRI) mutants. Addition of its own cognate AHL (3OHC12:1-HSL) results in partial restoration of indigoidine in the phaI mutant. However, addition of both AHLs does not restore pigment production. That both AHLs in combination are unable to chemically complement indigoidine production is intriguing. One explanation for this result could lie in the promiscuous nature of AHLs and their receptors. Alternations in the length and degree of saturation in acyl chain structures impact binding affinity of the transcriptional regulator, influencing the genetic response (Churchill and Chen, 2011; Rajput et al., 2016). Simultaneous addition of both AHLs may oversaturate the system, resulting in promiscuous binding of the transcriptional regulators and disruption of the regulatory network. Indeed, addition of AHL structural analogs is a proposed mechanism for combating bacterial pathogens reliant on QS for virulence (LaSarre and Federle, 2013). Genetic complementation of these mutants would enhance confidence in these results and help elucidate the mechanism underlying the results presented here.
The two QS mutants exhibit temporal differences regarding biofilm formation, indicating a role for this regulatory network in surface colonization and biofilm dynamics in Y4I. This finding is consistent with studies in other bacteria where QS can influence all stages of biofilm progression: from attachment and recruitment to maturation and dispersal (McDougald et al., 2012; Dang and Lovell, 2016). By monitoring Y4I biofilm formation over the course ∼70 h, the cyclical nature of this growth modality is evident. Comparison between and amongst mutant and wildtype strains reveals a degree of asynchronicity in the biofilm mass of QS mutants, supporting a role for QS in mediating periodic biofilm dispersal. QS-regulated biofilm dispersion has been proposed to prevent overcrowding in the sponge symbiont R. mobilis (Zan et al., 2014). Furthermore, induction of biofilm dispersal via QS in P. inhibens is thought to contribute to the switch between mutualism (attachment) and pathogenesis (dispersion) with its host (Beyersmann et al., 2017). The production of the Y4I QS1 signal (3OHC12:1-HSL) is biphasic across the growth cycle, highest during lag and late stationary phases of growth. A previous interpretation of these data was that this AHL represents an important signal when cell growth is relatively static and, thus, may impart information on metabolic status (Cude et al., 2015). This interpretation combined with data presented here suggests a role for cellular metabolic status in biofilm dynamics, particularly dispersion. Indeed, this topic has recently gained attention for pathogenic microbes (Rumbaugh and Sauer, 2020).
C-di-GMP Is Integrated Into QS Network
The global secondary messenger, c-di-GMP, relays information in response to metabolic state and environmental cues. This metabolite has been shown to modulate biofilm dispersal in many bacteria (Valentini and Filloux, 2016; Rumbaugh and Sauer, 2020). Consistent with the general paradigm (Römling et al., 2013), intracellular c-di-GMP concentrations in Y4I correlate with growth modality: they are elevated in surface-grown cultures relative to liquid cultures. C-di-GMP concentrations are not only altered in Y4I strains with disruptions in either QS system but also the indigoidine biosynthesis mutant, suggestive of a direct link between these secondary metabolites. In fact, glutamate has been shown to play a key role in biofilm formation by regulating intercellular pools of c-di-GMP in P. aeruginosa (Roy et al., 2012; Roy and Sauer, 2014). In Y4I, glutamate is an essential precursor for indigoidine (Kuhn et al., 1965; Cude et al., 2012; Day et al., 2017). Thus, the integration of c-di-GMP and QS signaling pathways may provide an avenue for convergence of information on cellular status from different central metabolic pathways in Y4I.
Integration of c-di-GMP with bacterial regulatory signaling pathways is not uncommon. For example, in several Alphaproteobacteria, c-di-GMP contributes to the regulation of the biphasic swim-stick switch controlled by the CtrA phosphorelay “master regulator” system (Paul et al., 2004; Boutte et al., 2011; D’Alvise et al., 2014; Koppenhöfer et al., 2019; Su et al., 2019). It is plausible that c-di-GMP and the CtrA phosphorelay system are also integrated into the QS regulatory network architecture governing indigoidine biosynthesis in Y4I. Indeed, we have found a putative DNA binding half-site for the master regulator, CtrA, located in the QS2 (pgaRI) operon (Supplementary Figure 3). Furthermore, integration of the CtrA and QS has been demonstrated in other Roseobacters. In R. mobilis, QS regulates the lifestyle switch from motile (swim) to sessile (stick) by controlling the expression of CtrA (Zan et al., 2013). Previous studies have shown that c-di-GMP is also integrated into the QS of D. shibae (Wang et al., 2014) and regulates the expression of ctrA in a related alphaproteobacterium, Rhodobacter capsulatus (Leung et al., 2013). Thus, integration of these central signaling pathways may be a mechanism utilized by members of the Roseobacter clade to aid in their colonization success.
In an effort to better contextualize the regulatory network architecture that may govern these key Roseobacter physiologies in marine ecosystems, we present a working model that integrates the key players discussed here (Figure 5). Implicit in this model is a connection between metabolism and physiology. Quorum sensing molecules are dependent on metabolic precursors (Goo et al., 2015). Additionally, c-di-GMP, a key intermediate linked to metabolic state has been demonstrated to regulate QS, either directly or indirectly, in Alphaproteobacteria (Hengge, 2009; McDougald et al., 2012; D’Alvise et al., 2014). We postulate this mechanism of regulation applies to the QS networks in Y4I. We also hypothesize a role for the CtrA phosphorelay system in regulating surface growth associated physiologies, including attachment, mechanosensing, flagella and biofilm formation in Y4I (Poncin et al., 2018). We anticipate future studies will more fully elucidate the extent and nature of c-di-GMP and CtrA to antimicrobial production and biofilm in this strain.
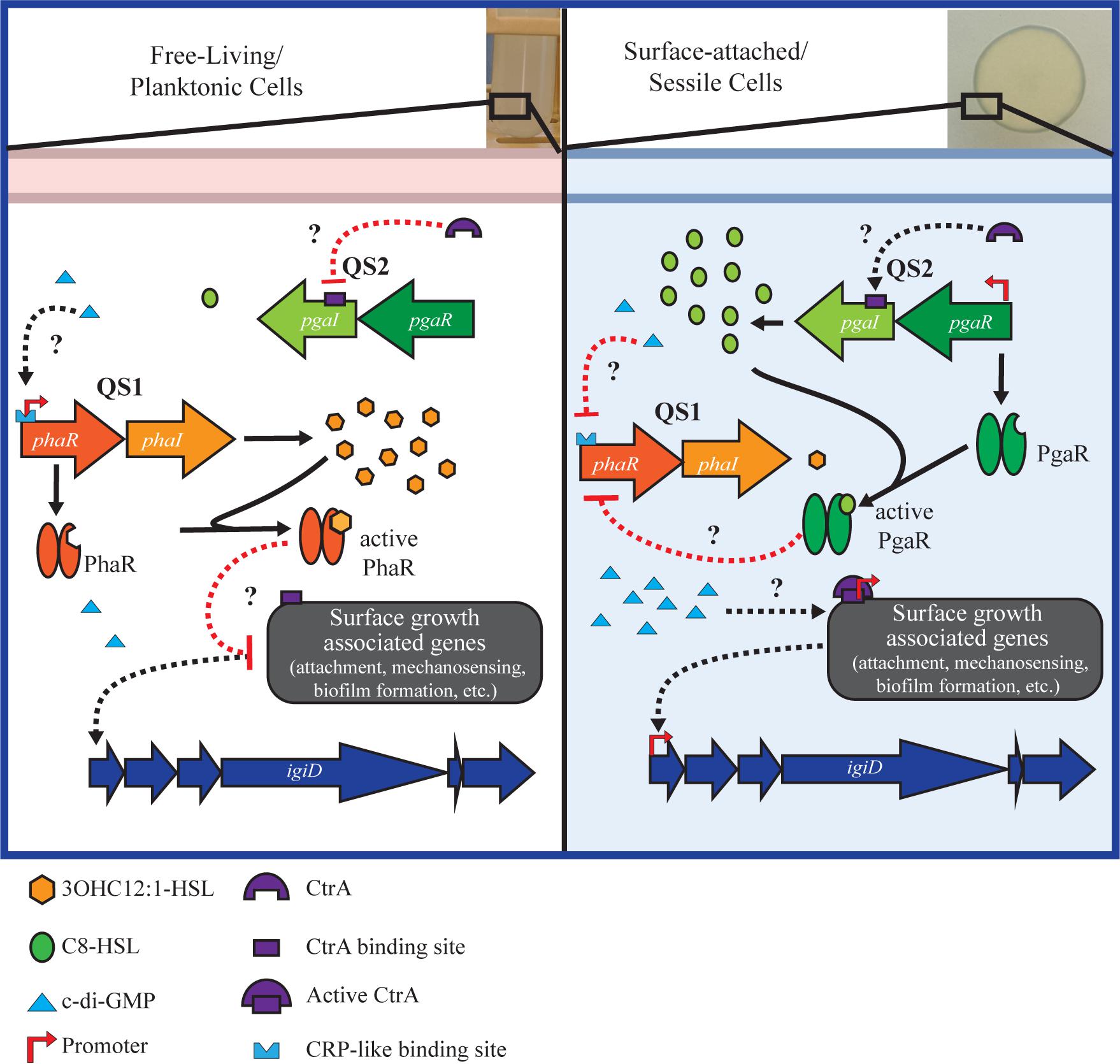
Figure 5. Working model of the regulatory network governing key physiologies in strain Y4I. Left panel shows genetic regulation during growth at low cell densities when Y4I is growing planktonically. During this growth modality, 3OHC12:1-HSL (QS1) concentrations are highest and we hypothesize that surface growth associated genes (e.g., mechanosensing, attachment, biofilm formation, flagella biosynthesis, and/or ctrA phosphorelay) are repressed, either directly or indirectly through QS1. Right panel shows genetic regulation during growth on a surface. It is predicted that when concentrations of C8-HSL are highest, QS2 represses expression of QS1, either directly or indirectly, allowing for the transcription of surface growth associated genes. Both c-di-GMP and CtrA are expected to play a role in activating surface-growth associated genes, though the specific mechanism of this regulation is currently unknown. Black lines are indicative of activation while red represents inhibition. Solid lines represent known pathways, while dotted lines are hypothesized.
Ecological Context of QS Signaling Dynamics
Quorum sensing has been shown to contribute to competitive physiologies, such as biofilm formation and antimicrobial production, in members of the Roseobacter clade (e.g., Rao et al., 2006; Berger et al., 2011; Beyersmann et al., 2017 and reviewed in Zan et al., 2014). Here, we provide evidence that Y4I employs a hierarchical QS circuit to mediate biofilm formation and indigoidine production. At the top of the hierarchical network in Y4I is one of the most common marine signaling molecules: C8-HSL (Wagner-Döbler et al., 2005; Mohamed et al., 2008; Cude et al., 2012; Gromek et al., 2016; Krupke et al., 2016; Ziesche et al., 2018). It is intriguing that this dominant AHL, which is produced by a number of aquatic microorganisms, including A. fischeri and Aliivibrio anguillarum, is also required for indigoidine biosynthesis in Y4I, which inhibits the growth of both of these species (García-Aljaro et al., 2012; Verma and Miyashiro, 2013; Cude et al., 2015). Given the universality of C8-HSL, the exploitation of this ubiquitous signaling molecule by Y4I may be central to its successful competitive strategies.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors upon request, without undue reservation.
Author Contributions
AA and AB conceived and designed the experiments and wrote the manuscript. AA conducted the experiments and carried out the data analysis. Both authors contributed to the article and approved the submitted version.
Funding
This work was funded by the National Science Foundation (OCE-1357242 to AB).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank the Campagna Lab (UTK), specifically Amanda May and Ashley Lato, for their help with AHL detection and analysis. For assistance with experimentation, the authors acknowledge Jillian Walton. For their contributions to the discussion and scope of this manuscript, the authors acknowledge Sarah Lebeis, Karen Lloyd, Gladys Alexandre, and Nathan Cude. The authors would also like to thank Katherine M. Fullerton for editorial comments.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.681551/full#supplementary-material
Footnotes
References
Alexeyev, M. F. (1999). The pKNOCK Series of broad-host-range mobilizable suicide vectors for gene knockout and targeted DNA Insertion into the chromosome of gram-negative bacteria. Biotechniques 26, 824–828. doi: 10.2144/99265bm05
Berger, M., Neumann, A., Schulz, S., Simon, M., and Brinkhoff, T. (2011). Tropodithietic acid production in Phaeobacter gallaeciensis is regulated by N-acyl homoserine lactone-mediated quorum sensing. J. Bacteriol. 193, 6576–6585. doi: 10.1128/JB.05818-11
Beyersmann, P. G., Tomasch, J. J., Son, K., Stocker, R., Göker, M., Wagner-Döbler, I., et al. (2017). Dual function of tropodithietic acid as antibiotic and signaling molecule in global gene regulation of the probiotic bacterium Phaeobacter inhibens. Sci. Rep. 7:730. doi: 10.1038/s41598-017-00784-7
Boutte, C. C., Henry, J. T., and Crosson, S. (2011). ppGpp and polyphosphate modulate cell cycle progression in Caulobacter crescentus. J. Bacteriol. 194, 28–35. doi: 10.1128/JB.05932-11
Brinkhoff, T., Giebel, H.-A., and Simon, M. (2008). Diversity, ecology, and genomics of the Roseobacter clade: a short overview. Arch. Microbiol. 189, 531–539. doi: 10.1007/s00203-008-0353-y
Buchan, A., Neidle, E. L., and Moran, M. A. (2004). Diverse organization of genes of the B-ketoadipate pathway in members of the marine roseobacter lineage. Appl. Environ. Microbiol. 70, 1658–1668. doi: 10.1128/aem.70.3.1658-1668.2004
Churchill, M. E. A., and Chen, L. (2011). Structural basis of acyl-homoserine lactone-dependent signaling. Chem. Rev. 111, 68–85. doi: 10.1021/cr1000817
Cude, W. N., and Buchan, A. (2013). Acyl-homoserine lactone-based quorum sensing in the Roseobacter clade: complex cell-to-cell communication controls multiple physiologies. Front. Microbiol. 4:336. doi: 10.3389/fmicb.2013.00336
Cude, W. N., Mooney, J., Tavanaei, A. A., Hadden, M. K., Frank, A. M., Gulvik, C. A., et al. (2012). Production of the antimicrobial secondary metabolite indigoidine contributes to competitive surface colonization by the marine roseobacter Phaeobacter sp. strain Y4I. Appl. Environ. Microbiol. 78, 4771–4780. doi: 10.1128/AEM.00297-12
Cude, W. N., Prevatte, C. W., Hadden, M. K., May, A. L., Smith, R. T., Swain, C. L., et al. (2015). Phaeobacter sp. strain Y4I utilizes two separate cell-to-cell communication systems to regulate production of the antimicrobial indigoidine. Appl. Environ. Microbiol. 81, 1417–1425. doi: 10.1128/AEM.02551-14
D’Alvise, P. W., Magdenoska, O., Melchiorsen, J., Nielsen, K. F., and Gram, L. (2014). Biofilm formation and antibiotic production in Ruegeria mobilis are influenced by intracellular concentrations of cyclic dimeric guanosinmonophosphate. Environ. Microbiol. 16, 1252–1266. doi: 10.1111/1462-2920.12265
Dang, H., and Lovell, C. R. (2000). Bacterial primary colonization and early succession on surfaces in marine waters as determined by amplified rRNA gene restriction analysis and sequence analysis of 16S rRNA genes. Appl. Environ. Microbiol. 66, 467–475. doi: 10.1128/aem.66.2.467-475.2000
Dang, H., and Lovell, C. R. (2016). Microbial surface colonization and biofilm development in marine environments. Microbiol. Mol. Biol. Rev. 80, 91–138. doi: 10.1128/MMBR.00037-15
Day, P. A., Villalba, M. S., Herrero, O. M., Arancibia, L. A., and Alvarez, H. M. (2017). Formation of indigoidine derived-pigments contributes to the adaptation of Vogesella sp. strain EB to cold aquatic iron-oxidizing environments. Antonie Van Leeuwenhoek Int. J. Gen. Mol. Microbiol. 110, 415–428. doi: 10.1007/s10482-016-0814-2
Farnelid, H., Turk-Kubo, K., Ploug, H., Ossolinski, J. E., Collins, J. R., Van Mooy, B. A. S., et al. (2019). Diverse diazotrophs are present on sinking particles in the North Pacific Subtropical Gyre. ISME J. 13, 170–182. doi: 10.1038/s41396-018-0259-x
García-Aljaro, C., Vargas-Cespedes, G. J., and Blanch, A. R. (2012). Detection of acylated homoserine lactones produced by Vibrio spp. and related species isolated from water and aquatic organisms. J. Appl. Microbiol. 112, 383–389. doi: 10.1111/j.1365-2672.2011.05199.x
Gonzalez, J. M., Mayer, F., Moran, M. A., Hodson, R. E., and Whitman, W. B. (1997). Sagittula stellata gen. nov., sp. nov., a lignin-transforming bacterium from a coastal environment. Int. J. Syst. Bacteriol. 47, 773–780. doi: 10.1099/00207713-47-3-773
Goo, E., An, J. H., Kang, Y., and Hwang, I. (2015). Control of bacterial metabolism by quorum sensing. Trends Microbiol. 23, 567–576. doi: 10.1016/J.TIM.2015.05.007
Gromek, S. M., Suria, A. M., Fullmer, M. S., Garcia, J. L., Gogarten, J. P., Nyholm, S. V., et al. (2016). Leisingera sp. JC1, a bacterial isolate from hawaiian bobtail squid eggs, produces indigoidine and differentially inhibits vibrios. Front. Microbiol. 7:1342. doi: 10.3389/fmicb.2016.01342
Hammer, B. K., and Bassler, B. L. (2003). Quorum sensing controls biofilm formation in Vibrio cholerae. Mol. Microbiol. 50, 101–104. doi: 10.1046/j.1365-2958.2003.03688.x
Hawver, L. A., Jung, S. A., and Ng, W.-L. (2016). Specificity and complexity in bacterial quorum-sensing systems. FEMS Microbiol. Rev. 40, 738–752. doi: 10.1093/femsre/fuw014
Hellemans, J., Mortier, G., De Paepe, A., Speleman, F., and Vandesompele, J. (2007). qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 8:19. doi: 10.1186/gb-2007-8-2-r19
Hengge, R. (2009). Principles of c-di-GMP signalling in bacteria. Nat. Rev. Microbiol. 7, 263–273. doi: 10.1038/nrmicro2109
Koppenhöfer, S., Wang, H., Scharfe, M., Kaever, V., Wagner-Döbler, I., and Tomasch, J. (2019). Integrated transcriptional regulatory network of quorum sensing, replication control, and SOS response in Dinoroseobacter shibae. Front. Microbiol. 10:803. doi: 10.3389/fmicb.2019.00803
Krupke, A., Hmelo, L. R., Ossolinski, J. E., Mincer, T. J., and Van Mooy, B. A. S. (2016). Quorum sensing plays a complex role in regulating the enzyme hydrolysis activity of microbes associated with sinking particles in the ocean. Front. Mar. Sci. 3:55. doi: 10.3389/fmars.2016.00055
Kuhn, R., Starr, M. P., Kuhn, D. A., Bauer, H., and Knackmuss, H.-J. J. (1965). Indigoidine and other bacterial pigments related to 3,3’-bipyridyl. Arch. Mikrobiol. 51, 71–84. doi: 10.1007/BF00406851
LaSarre, B., and Federle, M. J. (2013). Exploiting quorum sensing to confuse bacterial pathogens. Microbiol. Mol. Biol. Rev. 77, 73–111. doi: 10.1128/mmbr.00046-12
Leung, M. M., Brimacombe, C. A., and Beatty, J. T. (2013). Transcriptional regulation of the Rhodobacter capsulatus response regulator CtrA. Microbiology 159, 96–106. doi: 10.1099/mic.0.062349-0
Lin Chua, S., Liu, Y., Li, Y., Jun Ting, H., Kohli, G. S., Cai, Z., et al. (2017). Reduced intracellular c-di-GMP content increases expression of quorum sensing-regulated genes in Pseudomonas aeruginosa. Front. Cell. Infect. Microbiol. 7:451. doi: 10.3389/fcimb.2017.00451
McDougald, D., Rice, S. A., Barraud, N., Steinberg, P. D., and Kjelleberg, S. (2012). Should we stay or should we go: mechanisms and ecological consequences for biofilm dispersal. Nat. Rev. Microbiol. 10, 39–50. doi: 10.1038/nrmicro2695
Metcalf, W. W., Jiang, W., Daniels, L. L., Kim, S.-K., Haldimann, A., and Wanner, B. L. (1996). Conditionally replicative and conjugative plasmids carrying lacZα for cloning, mutagenesis, and allele replacement in bacteria. Plasmid 35, 1–13. doi: 10.1006/plas.1996.0001
Mohamed, N. M., Cicirelli, E. M., Kan, J., Chen, F., Fuqua, C., and Hill, R. T. (2008). Diversity and quorum-sensing signal production of Proteobacteria associated with marine sponges. Environ. Microbiol. 10, 75–86. doi: 10.1111/j.1462-2920.2007.01431.x
Motulsky, H. J., and Brown, R. E. (2006). Detecting outliers when fitting data with nonlinear regressiona new method based on robust nonlinear regression and the false discovery rate. BMC Bioinform. 7:123. doi: 10.1186/1471-2105-7-123
O’Neal, L., Ryu, M., Gomelsky, M., and Alexandre, G. (2017). Optogenetic manipulation of cyclic di-GMP (c-di-GMP) levels reveals the role of c-di-GMP in regulating aerotaxis receptor activity in Azospirillum brasilense. J. Bacteriol. 199, 1–18. doi: 10.1128/JB.00020-17
Papenfort, K., and Bassler, B. L. (2016). Quorum sensing signal-response systems in gram-negative bacteria. Nat. Rev. Microbiol. 14, 576–588. doi: 10.1038/nrmicro.2016.89
Paul, R., Weiser, S., Amiot, N. C., Chan, C., Schirmer, T., Giese, B., et al. (2004). Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes Dev. 18, 715–727. doi: 10.1101/gad.289504
Poncin, K., Gillet, S., and De Bolle, X. (2018). Learning from the master: targets and functions of the CtrA response regulator in Brucella abortus and other alpha-proteobacteria. FEMS Microbiol. Rev. 42, 500–513. doi: 10.1093/femsre/fuy019
Quigley, L. N. M., Edwards, A., Steen, A. D., Buchan, A., and Fagervold, S. K. (2019). Characterization of the interactive effects of labile and recalcitrant organic matter on microbial growth and metabolism. Front. Microbiol. 10:493. doi: 10.3389/fmicb.2019.00493
Rajput, A., Kaur, K., and Kumar, M. (2016). SigMol: repertoire of quorum sensing signaling molecules in prokaryotes. Nucleic Acids Res. 44, 634–639. doi: 10.1093/nar/gkv1076
Rao, D., Webb, J. S., and Kjelleberg, S. (2006). Microbial colonization and competition on the marine alga Ulva australis. Appl. Environ. Microbiol. 72, 5547–5555. doi: 10.1128/AEM.00449-06
Römling, U., Galperin, M. Y., and Gomelsky, M. (2013). Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol. Mol. Biol. Rev. 77, 1–52. doi: 10.1128/MMBR.00043-12
Roy, A. B., Petrova, O. E., and Sauer, K. (2012). The Phosphodiesterase DipA (PA5017) is essential for Pseudomonas aeruginosa biofilm dispersion. J. Bacteriol. 194, 2904–2915. doi: 10.1128/JB.05346-11
Roy, A. B., and Sauer, K. (2014). Diguanylate cyclase NicD-based signalling mechanism of nutrient-induced dispersion by Pseudomonas aeruginosa. Mol. Microbiol. 94, 771–793. doi: 10.1111/mmi.12802
Rumbaugh, K. P., and Sauer, K. (2020). Biofilm dispersion. Nat. Rev. Microbiol. 18, 571–586. doi: 10.1038/s41579-020-0385-0
Schuster, M., and Peter Greenberg, E. (2006). A network of networks: Quorum-sensing gene regulation in Pseudomonas aeruginosa. Int. J. Med. Microbiol. 296, 73–81. doi: 10.1016/J.IJMM.2006.01.036
Slightom, R. N., and Buchan, A. (2009). Surface colonization by marine roseobacters: integrating genotype and phenotype. Appl. Environ. Microbiol. 75, 6027–6037. doi: 10.1128/AEM.01508-09
Su, Y., Tang, K., Liu, J., Wang, Y., Zheng, Y., and Zhang, X.-H. (2019). Quorum sensing system of Ruegeria mobilis Rm01 controls lipase and biofilm formation. Front. Microbiol. 9:3304. doi: 10.3389/fmicb.2018.03304
Valentini, M., and Filloux, A. (2016). Biofilms and cyclic di-GMP (c-di-GMP) signaling: lessons from Pseudomonas aeruginosa and other bacteria. J. Biol. Chem. 291, 12547–12555. doi: 10.1074/jbc.R115.711507
Van Mooy, B. A. S., Hmelo, L. R., Sofen, L. E., Campagna, S. R., May, A. L., Dyhrman, S. T., et al. (2012). Quorum sensing control of phosphorus acquisition in Trichodesmium consortia. ISME J. 6, 422–429. doi: 10.1038/ismej.2011.115
Verma, S. C., and Miyashiro, T. (2013). Quorum sensing in the squid-Vibrio symbiosis. Int. J. Mol. Sci. 14, 16386–16401. doi: 10.3390/ijms140816386
Wagner-Döbler, I., Thiel, V., Eberl, L., Allgaier, M., Bodor, A., Meyer, S., et al. (2005). Discovery of complex mixtures of novel long-chain quorum sensing signals in free-living and host-associated marine Alphaproteobacteria. Chem. Bio. Chem. 6, 2195–2206. doi: 10.1002/cbic.200500189
Wang, H., Ziesche, L., Frank, O., Michael, V., Martin, M., Petersen, J., et al. (2014). The CtrA phosphorelay integrates differentiation and communication in the marine alphaproteobacterium Dinoroseobacter shibae. BMC Genom. 15:130. doi: 10.1186/1471-2164-15-130
Williams, P., Winzer, K., Chan, W. C., and Cámara, M. (2007). Look who’s talking: communication and quorum sensing in the bacterial world. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 362, 1119–1134. doi: 10.1098/rstb.2007.2039
Zan, J., Cicirelli, E. M., Mohamed, N. M., Sibhatu, H., Kroll, S., Choi, O. O., et al. (2012). A complex LuxR-LuxI type quorum sensing network in a roseobacterial marine sponge symbiont activates flagellar motility and inhibits biofilm formation. Mol. Microbiol. 85, 916–933. doi: 10.1111/j.1365-2958.2012.08149.x
Zan, J., Heindl, J. E., Liu, Y., Fuqua, C., and Hill, R. T. (2013). The CckA-ChpT-CtrA phosphorelay system is regulated by quorum sensing and controls flagellar motility in the marine sponge symbiont Ruegeria sp. KLH11. PLoS One 8:e66346. doi: 10.1371/journal.pone.0066346
Zan, J., Liu, Y., Fuqua, C., and Hill, R. (2014). Acyl-homoserine lactone quorum sensing in the roseobacter clade. Int. J. Mol. Sci. 15, 654–669. doi: 10.3390/ijms15010654
Keywords: quorum sensing, AHLs, Roseobacter clade bacteria, biofilm, cyclic-di-GMP
Citation: Armes AC and Buchan A (2021) Cyclic di-GMP Is Integrated Into a Hierarchal Quorum Sensing Network Regulating Antimicrobial Production and Biofilm Formation in Roseobacter Clade Member Rhodobacterales Strain Y4I. Front. Mar. Sci. 8:681551. doi: 10.3389/fmars.2021.681551
Received: 16 March 2021; Accepted: 10 June 2021;
Published: 05 July 2021.
Edited by:
Pia H. Moisander, University of Massachusetts Dartmouth, United StatesReviewed by:
Brett Mellbye, Oregon State University, United StatesMargot Doberva, Polytechnique Montréal, Canada
Julie Stoudenmire, Georgia State University, United States
Copyright © 2021 Armes and Buchan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alison Buchan, abuchan@utk.edu
 April C. Armes
April C. Armes Alison Buchan
Alison Buchan