- 1Molecular Medicine Research Center, Tabriz University of Medical Sciences, Tabriz, Iran
- 2Department of Medical Genetics, Faculty of Medicine, Tabriz University of Medical Sciences, Tabriz, Iran
- 3Department of Molecular Medicine, School of Medicine, Birjand University of Medical Sciences, Birjand, Iran
- 4Department Pharmacognosy, College of Pharmacy, Hawler Medical University, Erbil, Iraq
- 5Skull Base Research Center, Loghman Hakim Hospital, Shahid Beheshti University of Medical Sciences, Tehran, Iran
- 6Institute of Human Genetics, Jena University Hospital, Jena, Germany
- 7Department of Medical Genetics, School of Medicine, Shahid Beheshti University of Medical Sciences, Tehran, Iran
Multiple sclerosis (MS), a chronic inflammatory demyelinating disease of the central nervous system, is one of the most common neurodegenerative diseases worldwide. MS results in serious neurological dysfunctions and disability. Disturbances in coding and non-coding genes are key components leading to neurodegeneration along with environmental factors. Long non-coding RNAs (lncRNAs) are long molecules in cells that take part in the regulation of gene expression. Several studies have confirmed the role of lncRNAs in neurodegenerative diseases such as MS. In the current study, we performed a systematic analysis of the role of lncRNAs in this disorder. In total, 53 studies were recognized as eligible for this systematic review. Of the listed lncRNAs, 52 lncRNAs were upregulated, 37 lncRNAs were downregulated, and 11 lncRNAs had no significant expression difference in MS patients compared with controls. We also summarized some of the mechanisms of lncRNA functions in MS. The emerging role of lncRNAs in neurodegenerative diseases suggests that their dysregulation could trigger neuronal death via still unexplored RNA-based regulatory mechanisms. Evaluation of their diagnostic significance and therapeutic potential could help in the design of novel treatments for MS.
Introduction
Multiple sclerosis (MS) is a chronic inflammatory demyelinating disease of the central nervous system (CNS) and one of the most common neurodegenerative diseases worldwide (1). Pathogenic mechanisms underlying MS development have not been determined up to now. Clinically, different MS subtypes have been identified, including relapsing–remitting (RR), secondary progressive (SP), and primary progressive (PP) subtypes. These subtypes are heterogeneous among affected individuals in terms of clinical course as well as genetic background (2). Complex interactions between genetic susceptibility and environmental factors lead to this neurodegenerative disease. Both innate and adaptive immune-mediated inflammatory mechanisms contribute to the demyelination and neurodegeneration in the context of MS. Previous studies have demonstrated that the inflammatory immune cells such as CD4 T-helper cells (Th1 and Th17) are the main contributors in disease pathogenesis (3, 4). The presence of these cells in the CNS is associated with neuronal demyelination, which can subsequently result in neuroinflammation and neurodegeneration (5, 6). Th17 cells that produce IL-17 are regarded as important inflammatory effectors in this disorder (7). However, the impact of Th17 cells in the pathogenesis of MS is not entirely dependent on the production of this cytokine, and it is supposed that an array of inflammatory factors is responsible in this regard (8). For example, expression of high amounts of the C-C chemokine receptor 6 (CCR6) on the cell surface of Th17 cells (9) facilitates the entry of these cells into the CNS via the choroid plexus (10). Th17 cells also participate in the pathoetiology of MS through production of other proinflammatory cytokines including TNF-α (11).
In recent years, genome-wide association studies (GWAS) and genetic mapping have nominated several candidate loci and variants in autoimmune conditions. However, MS pathogenesis cannot be explained by the genetic susceptibility factors alone. A large amount of evidence has revealed that long non-coding RNAs (lncRNAs) have critical roles in the regulation of cellular immunological pathways and autoimmunity. This new class of non-coding RNA (ncRNAs) contains a large part of the transcriptional output in the human genome but low protein-coding potential (12).
In the current review, we focus on recent reports performed on the roles of lncRNAs in MS pathogenesis. Then, we illustrate the role of some specific lncRNAs and their target genes. Therefore, our manuscript provides new insights into understanding the molecular etiology, diagnosis, and management of MS.
Long Non-Coding RNA Classification and Function
LncRNAs are a class of ncRNAs with sizes more than 200 nt and no protein-coding potential. They are commonly transcribed by RNA Pol II (13). LncRNAs have been detected in a variety of species such as animals, plants, and prokaryotes. The majority of them have a 5′ cap structure, multiple exons, and 3′ polyadenylated tails and are spliced in a way similar to mRNAs (14). Since lncRNAs do not encode proteins, they used to be called as “dark matter.” However, recent studies have demonstrated that they are regulatory molecules and play important roles in several biological processes (14, 15), including gene expression at the epigenetic, transcriptional, and posttranscriptional levels. The vital mechanisms of epigenetic regulation consist of DNA methylation, histone modification, and ncRNA-mediated regulation. Emerging evidence revealed that the normal execution of biological events is controlled by a combination of ncRNAs and transcription factor (TF)-mediated epigenetic modifications (16). Studies on the role of lncRNAs suggest that their dysregulation could trigger neuronal death via still unexplored RNA-based regulatory mechanisms (17). Gene signature in human CNS is precisely regulated by several mechanisms. LncRNAs have a substantial impact on normal neural development, so their abnormal expression affects development and progression of neurodegenerative diseases (18).
According to databases such as the NONCODE (version v5.0) (19), the number of lncRNAs in human has been estimated to be higher than the number of protein-coding genes. The classification of lncRNAs is based on subcellular localization, function, interaction with the protein-coding gene, their size, and their association with protein-encoding genes. Based on their association with protein-encoding genes, they can be categorized to different classes such as sense, intergenic, bidirectional, intronic, antisense, and divergent lncRNAs (20, 21). Long intergenic non-coding RNA (lincRNA) genes are an important group of ncRNAs that participate in many biological processes, such as regulation of gene expression. They also play an essential role in many autoimmune and inflammatory diseases (22). In the current study, we performed a systematic analysis of the role of lncRNAs in MS.
Methods
Review question: Which lncRNAs have been dysregulated in multiple sclerosis?
Inclusion/Exclusion Criteria
The inclusion criteria were as follows: 1) original studies, 2) studies focusing on the expression of lncRNAs in MS patients, 3) studies that confirmed results by RT-PCR, 4) studies with a sample of blood or tissue of human or animal model, and 5) studies that evaluated polymorphisms on lncRNAs. The following documents were excluded from this study: letters, reviews, in vitro studies, or papers with insufficient data.
Search Strategy
The current scoping review was performed according to the PRISMA statement (23). PubMed, Web of Science, ProQuest, and Scopus databases were searched to identify all published studies up to August 10, 2021.
Study Selection
Following the abovementioned search method, all obtained papers were loaded into EndNote version 20. Then, duplicate studies were removed. The title and abstracts of the remaining studies were evaluated, and their full texts were screened using the inclusion criteria. Then, lncRNAs with a role in the pathogenesis of MS were included.
Data Extraction
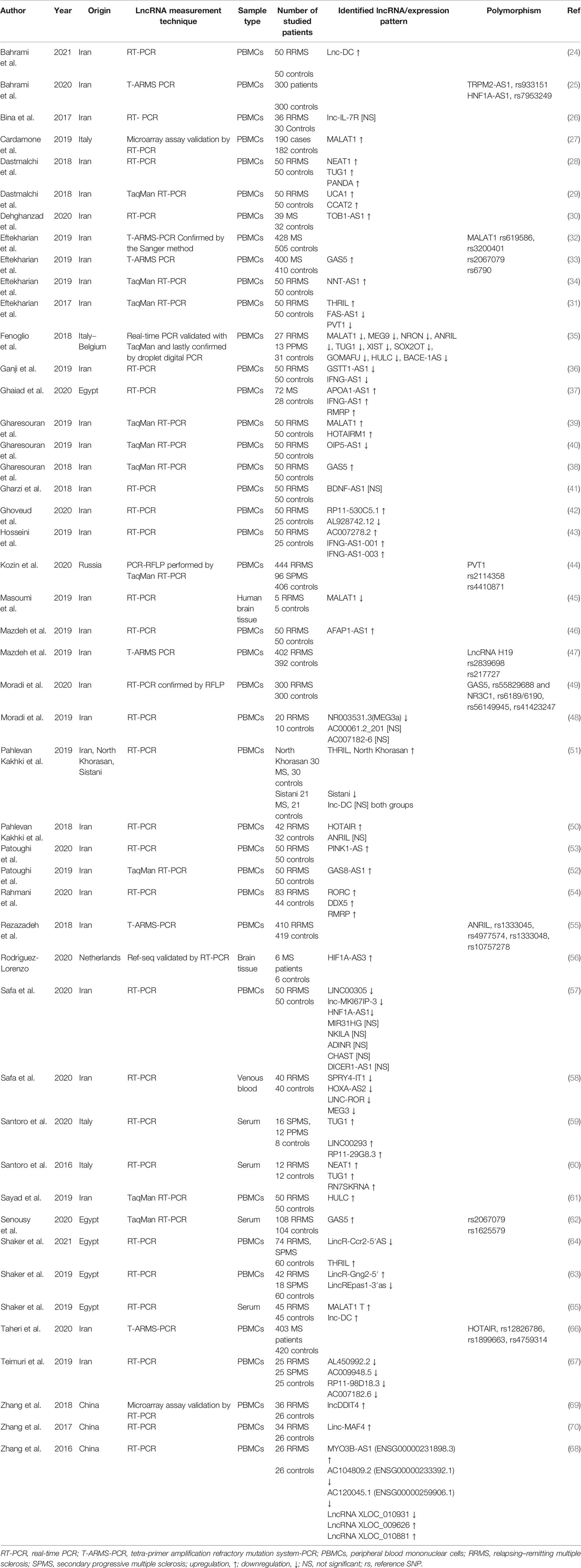
The required data were extracted using a self-constructed data extraction table. Author and year of publication, origin, sample type, studied patients, method for lncRNA analysis, identified lncRNAs and expression pattern, and polymorphisms were extracted from the studies.
Figure 1 shows the flowchart of the study.
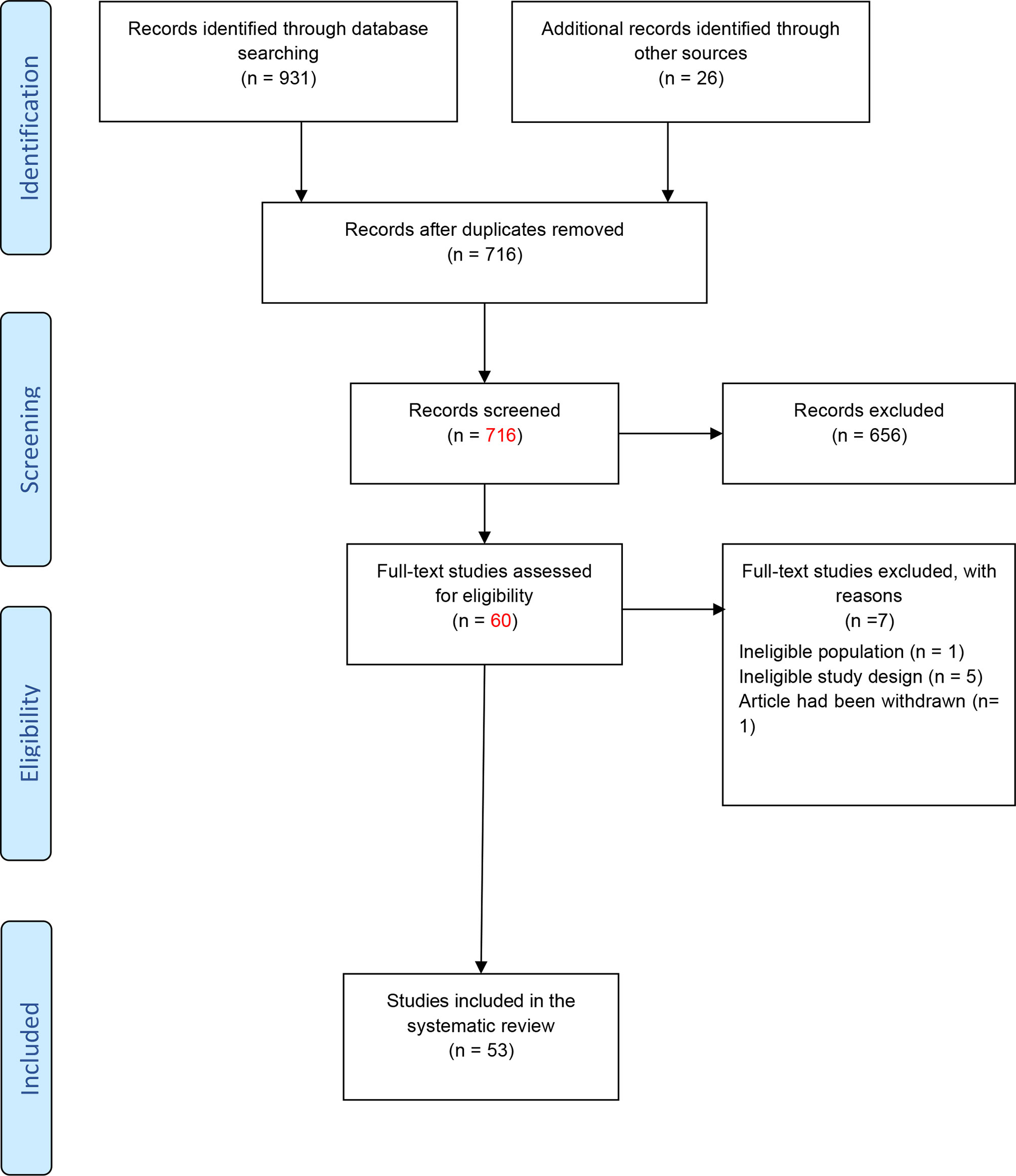
Figure 1 Flowchart of the study (23).
Results
As shown in Figure 1, a total of 931 studies were identified through searching PubMed, Web of Science, ProQuest, and Scopus databases, and 26 studies were identified from other databases. After removing duplicated articles, 716 studies remained. In the next step, based on the evaluation of titles and abstracts, 656 studies were excluded and 60 studies remained. The full text of the articles was evaluated based on our inclusion criteria. After evaluation of the full text, seven studies were removed due to lack of inclusion criteria. At last, 53 studies remained for our systematic review. Among the included studies, 47 studies were conducted on human samples (24–70), 7 studies used animal models (45, 71–76), and only 1 research was conducted on both human samples and animal model (45). Also, 33 studies were conducted in the Iranian population (24–26, 28–34, 36, 38–43, 45–55, 57, 58, 61, 66, 67), 9 studies were in China (68–76), 5 studies were in Egypt (37, 62–65), 4 studies were in Italy (27, 35, 59, 60), 1 study was in Russia (44), and 1 study was in the Netherlands (56). A total of 44 studies evaluated the expression of lncRNAs in MS patients (24, 26–31, 34–43, 45, 46, 48, 50–54, 56–61, 63–65, 67–76), while 9 other studies analyzed polymorphisms of lncRNAs (25, 32, 33, 44, 47, 49, 55, 62, 66). The details of the included studies are summarized in Tables 1, 2.
Recently, several studies revealed the involvement of lncRNAs in the pathogenesis of MS. Figure 2 demonstrates the function of several lncRNAs that are involved in the pathogenesis of MS.
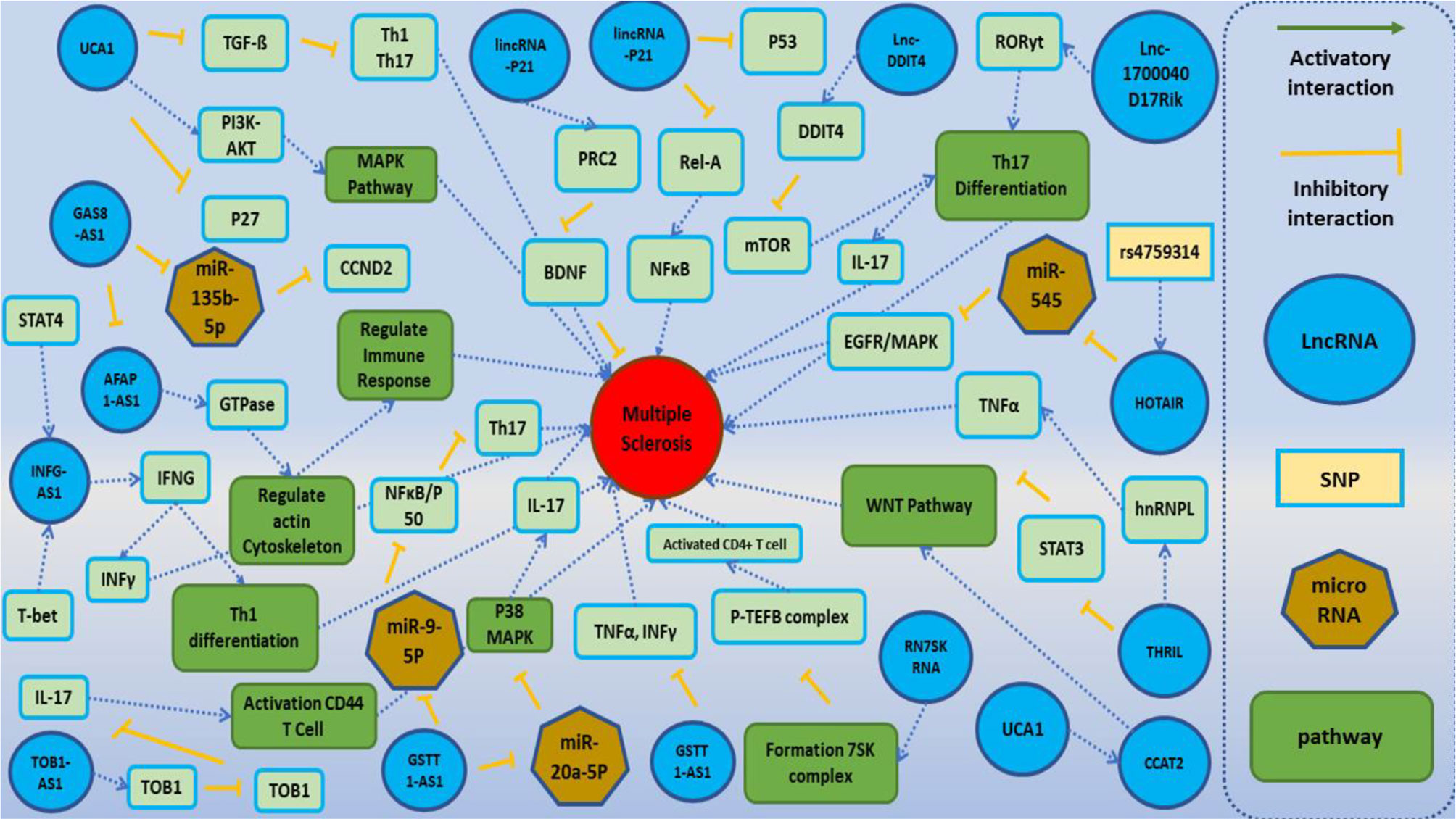
Figure 2 A schematic diagram of the role of several lncRNAs involved in the modulation of the main molecular cascades in multiple sclerosis (MS). One of the main pathophysiological mechanisms associated with the MS involves T cells subsets [regulatory T cells (Treg), Th1, Th2, and Th17 cells]. Dysregulation of these subsets activates inflammatory cascades and cytokine secretion and ultimately leads to demyelination within the brain and spinal cord and neuronal damage. Lnc-DC has been shown to be upregulated in PBMCs of MS patients. Upregulation of this lncRNA activates Toll-like receptor 4 (TLR4) and TLR9. TLR4 has a central role in the secretion of inflammatory cytokines such as IL-1, IL-6, and IL-17 and suppresses Treg cells. Also, TLR4 increases the differentiation of Th17 through inhibition of miR-30a (24, 65). Moreover, lnc-DDIT4 is upregulated in the PBMCs of MS patients. This lncRNA binds to DDIT4 and regulates immune response and differentiation of Th17 (69). BDNF-AS has a role in the recruitment of PRC2 and inhibition of the neuroprotective factor BDNF (41). GSTT1-AS1 inhibits the progression of MS through inhibition of secretion of IFN-γ and TNF-α (36). TUG1 activates p38 MAPK signaling pathway through suppression of miR-20a-5p, so downregulation of TUG1 decreases Th17 differentiation. UCA1 has a role in the regulation of activity of PI3K–AKT, ERK1/2, and MAPK cascades and Th17 differentiation. Also, this lncRNA has interaction with another lncRNA, namely, CCAT2. CCAT2 induces WNT cascade signaling and enhances the production of inflammatory cytokines (28, 59).
The Role of LncRNAs in the Pathophysiology of MS
LncRNAs Participating in Adaptive Immune Response or Inflammation
Linc-MAF-4 and lnc-DDIT4 are two upregulated lncRNAs in MS patients which are involved in the regulation of immune responses and inflammation (69, 70). DDIT4 is a cytoplasmic protein that is upregulated during DNA damage. Also, it inhibits the mTORC1 pathway which is a crucial regulator of the immune response (77). Since the mTOR pathway causes differentiation of Th17 and subsequent production of IL-17, it can be a key pathogenic player in MS (78, 79). Lnc-DDIT4 directly binds to and increases DDIT4 expression; thus, it suppresses the differentiation of Th17 (69). Therefore, lnc-DDIT4 might directly regulate Th17 cell differentiation and contribute to the pathogenesis of MS. Linc-MAF-4 is a lincRNA located in the minus strand of 16q23.2, almost 150 kb apart from the gene encoding MAF (19). This lincRNA has an important role in regulating differentiation of Th1/2 cells. MAF is the Th2 lineage-specific TF facilitating Th2 differentiation (70). Linc-MAF-4 is a Th1 lineage-specific factor that recruits chromatin remodeling factors LSD1 and EZH2 to inhibit MAF transcription and elevate Th1 differentiation and IFN-γ production (15). So, linc-MAF-4 can contribute in the pathogenesis of MS. Another study has identified six lncRNAs with abnormal expression in MS. ENSG00000231898.3 (MYO3B-AS1), XLOC_009626, and XLOC_010881 were upregulated, while ENSG00000233392.1 (AC104809.2), ENSG00000259906.1 (AC120045.1), and XLOC_010931 showed downregulation (68).
LincR-Gng2-5′, LincR-Epas1-3′as, and LincR-Ccr2-5′AS
LincR-Gng2-5′ and lincR-Epas1-3 loci were firstly identified by Hu et al. in Th1 and Th2 cells regulated by signal transducer and activator of transcription 4 (STAT4) and (STAT6), respectively (22). According to the data from lncRNAdb (80), LNCipedia (version 5.2), and Ensemble genome browser 99, LincR-Gng2-5′ is located on chromosome 14q22.1 on the plus strand and has a transcript size of 1,233 bp. LincR-Epas1-3′as is located on chromosome 2p.21 on the positive strand and has 758 bp length. They are located in an important place rich in genes with immune regulatory functions. Since they act as enhancers, they might participate in the regulation of neighboring genes, thus modulating immune responses (63). LincR-Gng2-5′ is upregulated in MS patients, while LincR-Epas1-3′as is downregulated in these patients. Dysregulation of these lncRNAs has a role in the pathoetiology of MS through affecting the balance between Th1 and Th2 cells (22, 81). LincR-Ccr2-5′AS is another lncRNA that is expressed in Th2 and has association with GATA-binding protein 3 (GATA3), the “master regulator” of Th2. Shaker et al. have reported the downregulation of lincR-Ccr2-5′AS in MS patients and the subsequent decrease in the production of Th2 cytokines (64).
GSTT1-AS1 and IFNG-AS1
Glutathione S-transferase, Theta1-Anti Sense1 (GSTT1-AS1), also known as lncRNA-CD244, is a novel 284-bp lncRNA, located on the minus strand 22q11.23 with partial overlap with 5′ UTR of the GSTT1 gene (19, 82). This lncRNA was originally discovered as an lncRNA with a crucial role in the pathogenesis of tuberculosis (83). Ganji et al. show downregulation of GSTT1-AS1 in MS patients. Since this lncRNA suppresses the expression of TNF and INFG through recruitment of the epigenetic complex PRC2 and via the EZH2 enzyme complex, it might be involved in the pathogenesis of MS (36).
IFNG-AS1 has been firstly identified as a transcript with a possible role in the regulation of immune system function (84). Also known as Tmevpg1, it is a 1,791-bp intergenic lncRNA located on the plus strand on 12q15 (19), adjacent to the INFG gene (85). It has been shown to be dysregulated in several immune-related disorders (83, 86). This lincRNA acts as an important checkpoint for the expression of IFNG in Th1 cells (87).
AC007278.2 (Expression in T Cells)
Another lncRNA is a 1,200-bp intronic lncRNA, AC007278.2, also known as Lnc-IL18R1-1. This lncRNA is located on the plus strand of the 2q12.1 chromosome and has two exons (19). AC007278.2 has a specific expression in Th1 cells. It is located within the introns of the protein-coding genes IL18RAP and IL18R1, with important roles in Th1 cell differentiation (43). Several studies revealed significant correlations between IL18RAP and IL18R1 and their association with the lncRNA AC007278.2. On the other hand, elevated expression of IL18RAP and IL18R1 is involved in the differentiation of Th1 cells and the pathogenesis MS. During Th1 differentiation, STAT4 and IL-12 recruit chromatin remodeling complexes. Induction of histone acetylases and DNA methylases promotes the expression of IL18RAP and IL18R1 and the release of IL-18 and IL-12 which trigger the differentiation of Th1 and the release of pro-inflammatory cytokines and eventually the progression of MS (43, 88, 89).
TOB1-AS1
TOB1 antisense RNA 1 (TOB1-AS1) is transcribed from the opposite orientation of the TOB1 gene on chromosome 17q21.33, a region with an important role in maintaining immune tolerance (19). Dehghanzad et al. demonstrated the abnormal expression levels of TOB1-AS1 and its targets genes TOB1, TSG, and SKP2 in the blood of MS. Downregulation of TOB1-AS1 might cause dysregulation of the target genes and participate in the progression of MS (30). TOB1-AS1 enhances the expression of the TOB1 gene via suppressing the production of IL-2 (90). An in vitro study revealed the positive feedback between TOB1 and S-phase kinase-associated protein 2 (SKP2). Elevation of TOB1-AS1 levels causes increased TOB1 and thus increased the TSG levels (30).
RMRP
Rahmani et al. demonstrated that RORC, DDX5, and RMRP have been significantly upregulated in patients with MS (54). RORC and DDX5 can affect MS pathogenesis through regulation of Th17 differentiation and the production of inflammatory cytokines such as IL-17A, IL-17F, and IL-22.
LncRNAs With Roles in Innate Immune Response
Lnc-DC and THRIL
TNF and HNRNPL-related immunoregulatory long non-coding RNA (THRIL) is a lincRNA located on the minus strand of the 12q24.31 chromosome. This lncRNA plays an important role in the regulation of the innate immune system (19). This lncRNA has been among the dysregulated lncRNAs in MS (31). THRIL regulates TNF-α expression via its interaction with heterogeneous nuclear ribonucleoprotein L (hnRNPL) and persuades a transcriptional-activating complex, finally connecting to the TNF-α promoter (91). THRIL can suppress STAT3 (51).
Lnc-DC (also known as Wfdc21) is a non-coding RNA gene on the minus strand of chromosome 17q23.1, which was firstly identified by Wang et al. to have an important role in the differentiation of dendritic cells and the regulation of the immune response (92, 93). Lnc-DC positively regulates STAT3 resulting in the differentiation of monocyte cell to dendritic cells (92). This lncRNA is involved in the pathogenesis of sepsis (93), coronary artery disease (94), pre-eclampsia (95), MS (51), and systemic lupus erythematosus (SLE) (96). Xie et al. showed the role of lnc-Dc on the regulation of TLR4 (93). Lnc-DC through the TLR9/STAT3 axis can regulate apoptosis and immune responses, thus can participate in the pathogenesis of MS (97, 98). Bahrami et al. demonstrated the upregulation of lnc-DC level in HLADRB1*15:01-negative MS patients compared with healthy controls (24).
LncRNAs Having a Role in Response to DNA Damage
LincRNA-p21 (Expression in T Cell)
P21-associated ncRNA DNA damage-activated (PANDA) is a lincRNA located on the minus strand 6p21.2. It has a role in response to DNA damage in a p53-dependent pathway (15). Dastmalchi et al. revealed the upregulation of this lncRNA in the peripheral blood of MS patients (28). PANDA controls the cell cycle through suppression of proapoptotic-related genes (15, 99). Dysregulation of the expression of this lncRNA in oligodendrocytes and neurons is associated with the release of free radicals and activation of the apoptosis process (100).
LncRNAs Involved in the Regulation of the Cell Cycle
TUG1, UCA1, and CCAT2
UCA1, CCAT2, and TUG1 are a subgroup of lncRNAs that have a role in the regulation of the cell cycle. UCA1 is located in the plus strand of chromosome 19p13.12 (19). It participates in the pathogenesis of several cancers such as colorectal, breast, and bladder cancer through increasing cell proliferation, apoptosis-resistant cells, invasion, and drug resistance induction (101). UCA1 via modulation of the PI3K–AKT, ERK1/2, and MAPK pathways can regulate the proliferation of cells in various cancers (102). Dastmalchi et al. revealed the upregulation of UCA1 in the blood of MS patients. This lncRNA via inhibiting cell cycle inhibitors such as p27 may cause increased proliferation of T cells (29).
CCAT2 is an intergenic lncRNA on the plus strand of the 8q24.21 chromosome (19). This lncRNA acts as an oncogene and participates in the metastasis, chromosomal instability, and tumor growth in colon cancer (103). Both UCA1 and CCAT2 can regulate the expression of genes participating in WNT pathway (104).
Fenoglio et al. showed the downregulation of TUG1 in MS patients compared with controls (35). TUG1 exerts a repressor function via recruitment of the PRC2 complex. Its promoter has many conserved binding sites for p53, thus after DNA damage, p53 regulates cell cycle and apoptosis via upregulation of TUG1 (35, 105, 106). TUG1 has been found to be upregulated in the serum and PBMCs of RRMS patients (28, 59, 60). TUG1 targets and suppresses different miRNAs such as miR-20a-5p, which has a role in the regulation of p38 MAPK signaling pathway. p38 MAPK promotes the production of proinflammatory cytokines. Downregulation of miR-20a-5p by TUG1 activates p38 MAPK signaling and MS progression (60).
The growth arrest-specific 5 (GAS5) has been recognized as a lncRNA with a possible role in normal growth arrest in T cells. This lncRNA plays a central role in the suppression of glucocorticoid receptor (GR). Gharesouran et al. revealed the correlation between GAS5 and nuclear receptor subfamily 3 group C member 1 (NR3C1) (38). Sun et al. demonstrated that GAS5 can inhibit the transcription factor IRF4, thus suppressing the generation of T cells (75).
LncRNAs With a Role in the CNS
GOMAFU
MIAT or GOMAFU is a lincRNA on the plus strand of 22q12.2 (19), which is highly expressed in the CNS and is suggested to have an important role in regulating the neural stem cell differentiation into oligodendrocytes (107). Fenoglio et al. showed the downregulation of this lncRNA in the blood of MS patients (35). GOMAFU using its repetitive sequence binds to the splicing factor 1 (SF1) protein and prevents the function of the spliceosome complex. Thus, deregulation of GOMAFU causes advent of alternative splicing patterns (108). GOMAFU has a possible role in inflammatory and neurodegenerative processes (35).
OIP5-AS1
OIP5-AS1 (Cyrano) was firstly detected in zebrafish models and it was suggested that it has a role in the development of the CNS (109). Kim et al. revealed that OIP5‐AS1 causes a reduction in the stability a cyclin G‐associated kinase (GAK) mRNA with important roles for mitotic progression (110). It seems that this lncRNA exerts its role in the suppression of cell proliferation through reducing GAK levels by associating with the RNA-binding proteins (RBPs) like HUR1 (ELAV-like protein 1). HuR1 is a protein that in humans is encoded by the ELAVL1 and is regarded as a member of the ELAVL proteins. HUR1 contains three RNA-binding domains and binds to cis-acting AU-rich elements. Since the HuR1 gene is expressed in astrocytes, it might have a role in autoimmune diseases such as encephalomyelitis and MS (111).
BDNF-AS
Brain-derived neurotrophic factor-antisense RNA (BDNF-AS) is a 191-kb-long conserved lncRNA (112), located in the opposite orientation of BDNF on the 11p14.1. It negatively regulates the expression of BDNF at the mRNA and protein levels (113). BDNF is a neuroprotective factor that is synthesized in the brain and is expressed at a high level in the CNS. It has diverse functions such as the promotion of neuronal survival and elevation of growth, maturation, and synaptic plasticity. BDNF is produced and released by neurons and immune cells such as T and B cells under the circumstance of inflammation of the CNS in MS patients (114). BDNF-AS recruits PRC2 and inhibits BDNF expression (113).
Other LncRNAs
NEAT1
This lncRNA has been shown to be upregulated in MS patients compared with healthy individuals (59). NEAT1 plays an important role in the formation of paraspeckle, a nuclear body that comprises numerous protein factors. NEAT1 has been shown to be co-localized with splicing factor proline/glutamine-rich (SFPQ) and NonPOU domain containing, octamer-binding (NONO) (115). Also, NEAT1 is activated by the Toll-like receptor 3 (TLR3)–p38 pathway in antiviral response or endogenous agonists that bind to TLR3 (116, 117). Imamura et al. revealed that upregulation of NEAT1 causes activation and excess IL-8 production via enhancing the relocation of SFPQ proteins from the IL-8 promoter (118).
RN7SK RNA
The lincRNA 7SK small nuclear (RN7SK RNA) is transcribed from the plus strand of the 6p12.2 chromosome. It is involved in the formation of the 7SK snRNP complex with other specific proteins (HEXIM1/2, LARP7, and PIP7S) that can inhibit approximately half of the activity of the cellular kinase P-TEFb complex (119, 120). The P-TEFb complex and its protein component Cdk9/cyclin T1 heterodimer have a role in the activation of CD4+ T cells. So, upregulated RN7SK RNA may cause disturbance in the P-TEFb complex with resulting regulation effects on CD4+ T cells, thus participating in autoimmune diseases such as idiopathic inflammatory myopathy (IIM) and MS (59).
AFAP1-AS1
Actin Filament-Associated Protein 1 Antisense RNA 1 (AFAP1-AS1) is a conserved non-coding RNA transcribed from the plus strand of chromosome 4p16.1 on the opposite strand of the AFAP1 locus. This lncRNA regulates the expression of AFAP1 at the translation level (121). AFAP1-AS1 was found to modulate AFAP1 and act as an adapter molecule that links other proteins such as SRC and PKC with a hypothetical function in blood–brain barrier (BBB) integrity. BBB dysfunction in MS patients allows the enormous influx of immune cells into the brain and, after a series of interactions, leads to demyelination (122). Based on the bioinformatics analyses, AFAP1-AS1 affected the expression of molecules with a vital role in the actin cytoskeleton signaling pathway such as multiple small GTPase family members. As small GTPases are involved in the regulation of immunity and inflammation response, its dysregulation leads to disease progression in many diseases such as autoimmune diseases (123). Upregulation of AFAP1-AS1 promotes metastasis via modulation actin filament integrity (124). Due to its antiapoptotic properties in peripheral immune cells, it might be involved in the pathogenesis of MS (40).
GAS8-AS1
A previous study showed that GAS8-AS1 is a tumor suppressor and regulates the expression of another lncRNA, namely, AFAP1-AS1 (125). GAS8-AS1 has been downregulated, while AFAP-AS1 has been upregulated in MS patients. Regarding the role of AFAP1-AS1 in the pathogenesis and progression of MS, it can be hypothesized that dysregulation of GAS8-AS1 might be involved in the pathogenesis of MS (40, 125). Zha et al. revealed that GAS8-AS1 negatively regulated the expression of UCA1. UCA1 has been shown to regulate various signaling pathways such as FGFR1/ERK and TGF-β (126). TGF-β has a role in the inflammatory condition and acts as an anti-inflammatory factor to inhibit Th1 and Th17 cells (127), so upregulation of GAS8-AS1 resulting in the downregulation of UCA1 and reduced TGF-β might cause progression and aggregate MS.
PINK1-AS
PTEN-induced kinase 1-AS (PINK1-AS) is an intronic non-coding RNA transcribed from the minus strand of chromosome 1p36.12 on the opposite strand of the PINK1 locus. This lncRNA regulates the expression of PINK1. Patoughi et al. (53) revealed the upregulation of the expression level of the PINK1-AS in male MS patients compared with male healthy controls. This might be due to the existence of a gender-based regulatory direction for PINK1-AS expression or variance in the pathogenic process of disease in female and male MS patients. PINK1 is a serine/threonine kinase that preserves the mitochondria and supports its normal function (128). Further studies by Fenoglio et al. have identified 10 lncRNAs with abnormal expression. These lncRNAs consist of MALAT1, MEG9, NRON, ANRIL, TUG1, XIST, SOX2OT, GOMAFU, HULC, and BACE-1AS (35).
The highly upregulated liver cancer (HULC) is another lncRNA found to be upregulated in MS patients in one study (61), whereas Fenoglio et al. have reported an opposite result (35). This lncRNA attaches to miR-200a-3p and also acts as an endogenous sponge for miR-122. Since miR-122 has an anti-inflammatory effect and is significantly downregulated in the blood of MS patients, HULC may be involved in the progression of MS. On the other hand, HULC activates miR-200a-3p/ZEB1 signaling. miR-200a plays an important role in the regulation of the TLR4 pathway and ZEB1 has a neuroprotective protein (129).
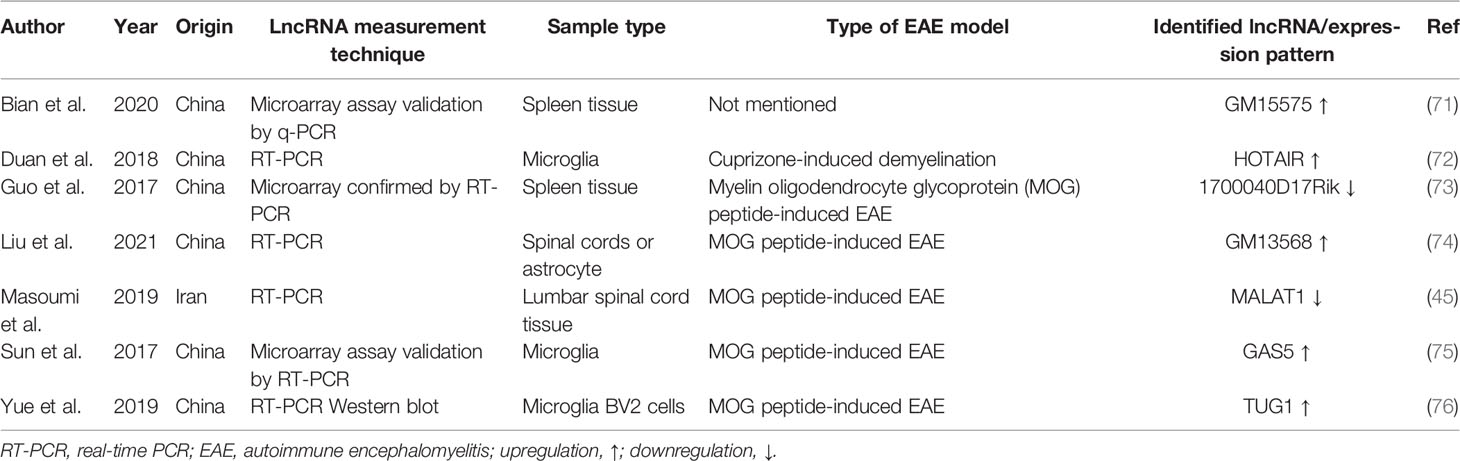
Dysregulated LncRNAs in the Animal Model of MS
One of the useful animal models of MS is EAE mice that share several characteristics with MS. However, there are few studies in this area. Yue et al. (76) demonstrated the abnormal activity of the TUG1/miR-9-5p/NF-κB1/p50 axis in the mouse model of MS. In fact, upregulation of TUG1 causes suppression of miR-9-5p and an increase in the expression of NF-κB1/p50. This transcription factor causes activation of Th17 cell and the production of IL-17 and IL-6. NF-κB also regulates matrix metalloproteinases (MMPs). Downregulation of TUG1 leads to increased levels of miR-9-5p and a decrease in NF-κB1/p50.
Another study by Guo and colleagues showed that lncRNA-1700040D17Rik is a specific mouse lincRNA that is located adjacent to the RORγt gene on chromosome 3 and is downregulated in EAE (73). Then, an in vitro approach revealed that IL23R-CHR is a soluble IL23R that counteracts IL-23 and blocks its signaling pathway, thus inhibiting differentiation of Th17 cell (130). These findings demonstrated that 1700040D17Rik regulates the expression of RORγt, which is an essential transcription factor for Th17 (73).
Liu et al. revealed that IL-9 inducing lncRNA Gm13568 in astrocytes has interaction with CBP/P300. It promotes Notch1 pathway activation and is involved in the construction of inflammatory cytokines in astrocytes in the progression of EAE development (74).
Variants Within LncRNAs and Association With MS
According to the important roles of lncRNAs in the regulation of immune responses, it is expected that functional variants within their coding region or adjacent to them can affect the risk of MS. However, there are few studies on this issue. Bahrami et al. have evaluated the association between rs933151 and rs7953249 polymorphisms in TRPM2-AS and HNF1-AS1, respectively, and MS risk in the Iranian population. They revealed that rs7953249 within HNF1-AS1 has an association with C-reactive protein (CRP) (25).
Taheri et al. assessed the association between three SNPs (rs12826786, rs1899663, and rs4759314) within HOTAIR and MS in 403 Iranian MS patients and 420 controls. Their results showed that the G allele of rs4759314 might be involved in the risk of MS (66).
Conclusion
In conclusion, the pathogenesis of MS is highly complex including several molecular signaling pathways. Most of the abovementioned studies have assessed the expression of lncRNAs in serum or PBMCs. Although several of these lncRNAs have essential roles in the CNS processes, modulation of peripheral immune responses is the most appreciated route of participation of lncRNAs in the pathogenesis of MS. Few studies have assessed the expressions of lncRNAs in the brain tissues of EAE models. An important study in this field has identified dysregulation of Gm14005, Gm12478, mouselincRNA1117, AK080435, and mouselincRNA0681 in brain tissues of affected animals. Notably, inflammation has been among the mostly enriched pathways among dysregulated genes (131). This observation further emphasized the importance of inflammation-related lncRNAs in the pathoetiology of MS.
In the current review, we highlighted the function of various lncRNAs in the MS pathway. Although few studies have addressed this issue, it is predicted that genomic variation within lncRNAs affecting their function or expression may contribute to the risk of MS or response of subjects to treatments. It has been determined that lncRNAs have roles in the development of the immune system and nerve cells. Further studies are required for understanding the mechanism of lncRNA involvement in the pathogenesis of MS.
Author Contributions
AJ, MT, BH, and SG-F wrote the draft and revised it. MR, HS, JG, and MA collected the data and designed the figures. HD performed the bioinformatics analysis. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
This article is the result of a student dissertation approved by Tabriz University of Medical Sciences code 66211. We also want to thank the Molecular Medicine Research Center of Tabriz University of Medical Sciences for their collaboration.
Glossary
lncRNA: long non-coding RNA
MS: multiple sclerosis
RT-PCR: real-time polymerase chain reaction
AFAP1-AS1: actin filament-associated protein 1 antisense RNA 1
RRMS: relapsing–remitting multiple sclerosis
SPMS: secondary progressive multiple sclerosis
CNS: central nervous system
HOTAIR: Hox transcript antisense intergenic RNA
miRNAs: microRNAs
CD4+ T cells: T helper cells
CD8+ T cells: cytotoxic T cells
GWAS: genome-wide association studies
BDNF: brain-derived neurotrophic factor
BDNF-AS: BDNF antisense RNA
NR3C1: nuclear receptor subfamily 3 group C member 1
PRC2: polycarbonate 2 suppressor complex
DDIT4: DNA-damage-inducible transcript 4
mTORC1: mammalian target of rapamycin complex 1
lncDDIT4: lncRNA DDIT4
Th17: T helper 17 cell
Tregs: regulatory T cells
IFN-γ: interferon gamma
hnRNPs: heterogeneous nuclear ribonucleoproteins
DC: dendritic cells
lnc-DC: lncRNA expressed in DC
PANDA: P21-associated ncRNA DNA damage-activated
FAS-AS1: FAS antisense transcript 1
linc-MAF-4: A lncRNA
THRIL: TNF-α and heterogeneous nuclear ribonucleoprotein L
PVT1: plasmacytoma variant translocation 1
GAK: cyclin G-associated kinase
HuR1: Huantigen R
SIRT1: silent information regulator 1
OIP5-AS1: OIP5 antisense RNA 1
TUG1: taurine-upregulated gene
IL-8: interleukin 8
SFPQ: splicing factor proline- and glutamine-rich
IL-17: interleukin 17
STAT4: signal transducer and activator of transcription 4
EZH2: enhancer of zeste homolog 2
TNF-α: tumor necrosis factor alpha
TLR4: Toll-like receptor 4
NF-κB: nuclear factor kappa-light-chain-enhancer of activated B cells
MAPK: mitogen-activated protein kinase
PI3K: phosphoinositide 3-kinases
ERK1/2: extracellular signal-regulated kinases 1/2
AKT: protein kinase B
WNT: Wnt signaling pathway
SF1: splicing factor 1
GAK: G-associated kinase
NonPOU: non-POU domain-containing octamer-binding protein
P-TEFb: positive transcription elongation factor
BBB: blood–brain barrier
FGFR1: fibroblast growth factor receptor 1
ERK: extracellular signal-regulated kinase
TGF-β: transforming growth factor beta
CRP: C-reactive protein
PINK1-AS: PTEN-induced kinase 1-AS
HIF1-AS3: hypoxia-inducible factor 1-AS3
RMRP: RNA component of the mitochondrial RNA-processing endoribonuclease (RNase MRP)
GATA3: GATA-binding protein 3
GR: glucocorticoid receptor
HULC: highly upregulated liver cancer
ZEB1: zinc finger and homeodomain transcription factor 1
References
1. Yamout BI, Alroughani R. Multiple Sclerosis. Semin Neurol (2018) 38(2):212–25. doi: 10.1055/s-0038-1649502
3. Yadav SK, Mindur JE, Ito K, Dhib-Jalbut S. Advances in the Immunopathogenesis of Multiple Sclerosis. Curr Opin Neurol (2015) 28(3):206–19. doi: 10.1097/WCO.0000000000000205
4. Broux B, Stinissen P, Hellings N. Which Immune Cells Matter? The Immunopathogenesis of Multiple Sclerosis. Crit Rev™ Immunol (2013) 33(4):283–306. doi: 10.1615/CritRevImmunol.2013007453
5. Galli E, Hartmann FJ, Schreiner B, Ingelfinger F, Arvaniti E, Diebold M, et al. GM-CSF and CXCR4 Define a T Helper Cell Signature in Multiple Sclerosis. Nat Med (2019) 25(8):1290–300. doi: 10.1038/s41591-019-0521-4
6. Capone A, Bianco M, Ruocco G, De Bardi M, Battistini L, Ruggieri S, et al. Distinct Expression of Inflammatory Features in T Helper 17 Cells From Multiple Sclerosis Patients. Cells (2019) 8(6):533. doi: 10.3390/cells8060533
7. Brucklacher-Waldert V, Stuerner K, Kolster M, Wolthausen J, Tolosa E. Phenotypical and Functional Characterization of T Helper 17 Cells in Multiple Sclerosis. Brain (2009) 132(12):3329–41. doi: 10.1093/brain/awp289
8. Volpe E, Battistini L, Borsellino G. Advances in T Helper 17 Cell Biology: Pathogenic Role and Potential Therapy in Multiple Sclerosis. Mediators Inflammation (2015) 2015. doi: 10.1155/2015/475158
9. Acosta-Rodriguez EV, Napolitani G, Lanzavecchia A, Sallusto F. Interleukins 1β and 6 But Not Transforming Growth Factor-β Are Essential for the Differentiation of Interleukin 17–Producing Human T Helper Cells. Nat Immunol (2007) 8(9):942–9. doi: 10.1038/ni1496
10. Reboldi A, Coisne C, Baumjohann D, Benvenuto F, Bottinelli D, Lira S, et al. CC Chemokine Receptor 6–Regulated Entry of T H-17 Cells Into the CNS Through the Choroid Plexus Is Required for the Initiation of EAE. Nat Immunol (2009) 10(5):514. doi: 10.1038/ni.1716
11. McGeachy MJ, Bak-Jensen KS, Chen YI, Tato CM, Blumenschein W, McClanahan T, et al. TGF-β and IL-6 Drive the Production of IL-17 and IL-10 by T Cells and Restrain TH-17 Cell–Mediated Pathology. Nat Immunol (2007) 8(12):1390–7. doi: 10.1038/ni1539
12. Wan P, Su W, Zhuo Y. The Role of Long Noncoding RNAs in Neurodegenerative Diseases. Mol Neurobiol (2017) 54(3):2012–21. doi: 10.1007/s12035-016-9793-6
13. Sun M, Kraus WL. From Discovery to Function: The Expanding Roles of Long Noncoding RNAs in Physiology and Disease. Endocrine Rev (2015) 36(1):25–64. doi: 10.1210/er.2014-1034
14. Ma L, Bajic VB, Zhang Z. On the Classification of Long Non-Coding RNAs. RNA Biol (2013) 10(6):924–33. doi: 10.4161/rna.24604
15. Aune TM, Spurlock CF III. Long Non-Coding RNAs in Innate and Adaptive Immunity. Virus Res (2016) 212:146–60. doi: 10.1016/j.virusres.2015.07.003
16. Wu H, Zhao M, Yoshimura A, Chang C, Lu Q. Critical Link Between Epigenetics and Transcription Factors in the Induction of Autoimmunity: A Comprehensive Review. Clin Rev Allergy Immunol (2016) 50(3):333–44. doi: 10.1007/s12016-016-8534-y
17. Riva P, Ratti A, Venturin M. The Long Non-Coding RNAs in Neurodegenerative Diseases: Novel Mechanisms of Pathogenesis. Curr Alzheimer Res (2016) 13(11):1219–31. doi: 10.2174/1567205013666160622112234
18. Quan Z, Zheng D, Qing H. Regulatory Roles of Long Non-Coding RNAs in the Central Nervous System and Associated Neurodegenerative Diseases. Front Cell Neurosci (2017) 11:175. doi: 10.3389/fncel.2017.00175
19. Fang S, Zhang L, Guo J, Niu Y, Wu H, Li H, et al. NONCODEV5: A Comprehensive Annotation Database for Long Non-Coding RNAs. Nucleic Acids Res (2018) 46(D1):D308–14. doi: 10.1093/nar/gkx1107
20. Laurent GS, Wahlestedt C, Kapranov P. The Landscape of Long Noncoding RNA Classification. Trends Genet (2015) 31(5):239–51. doi: 10.1016/j.tig.2015.03.007
21. Quinn JJ, Chang HY. Unique Features of Long Non-Coding RNA Biogenesis and Function. Nat Rev Genet (2016) 17(1):47. doi: 10.1038/nrg.2015.10
22. Hu G, Tang Q, Sharma S, Yu F, Escobar TM, Muljo SA, et al. Expression and Regulation of Intergenic Long Noncoding RNAs During T Cell Development and Differentiation. Nat Immunol (2013) 14(11):1190–8. doi: 10.1038/ni.2712
23. Moher D, Liberati A, Tetzlaff J, Altman DG, The PRISMA Group. Preferred Reporting Items for Systematic Reviews and Meta-Analyses: The PRISMA Statement. Int J Surg (2010) 8(5):336–41. doi: 10.1016/j.ijsu.2010.02.007
24. Bahrami T, Taheri M, Javadi S, Omrani MD, Karimipour M. Expression Analysis of Long Non-Coding RNA Lnc-DC in HLA-DRB1*15:01-Negative Patients With Multiple Sclerosis: A Probable Cause for Gender Differences in Multiple Sclerosis Susceptibility? J Mol Neurosci (2021) 71(4):821–5. doi: 10.1007/s12031-020-01704-7
25. Bahrami T, Taheri M, Javadi S, Omrani MD, Karimipour M. Associations Between Genomic Variants in lncRNA-TRPM2-AS and lncRNA-HNF1A-AS1 Genes and Risk of Multiple Sclerosis. J Mol Neurosci (2020) 70(7):1050–5. doi: 10.1007/s12031-020-01504-z
26. Bina P, Kakhki MD, Sahraian MA, Behmanesh M. The Expression of lnc-IL-7R Long Non-Coding RNA Dramatically Correlated With Soluble and Membrane-Bound Isoforms of IL-7Ra Gene in Multiple Sclerosis Patients. Neurosci Lett (2017) 642:174–8. doi: 10.1016/j.neulet.2017.01.068
27. Cardamone G, Paraboschi EM, Solda G, Cantoni C, Supino D, Piccio L, et al. Not Only Cancer: The Long Non-Coding RNA MALAT1 Affects the Repertoire of Alternatively Spliced Transcripts and Circular RNAs in Multiple Sclerosis. Hum Mol Genet (2019) 28(9):1414–28. doi: 10.1093/hmg/ddy438
28. Dastmalchi R, Ghafouri-Fard S, Omrani MD, Mazdeh M, Sayad A, Taheri M. Dysregulation of Long Non-Coding RNA Profile in Peripheral Blood of Multiple Sclerosis Patients. Mult Scler Relat Disord (2018) 25:219–26. doi: 10.1016/j.msard.2018.07.044
29. Dastmalchi R, Omrani MD, Mazdeh M, Arsang-Jang S, Movafagh A, Sayad A, et al. Expression of Long Non-Coding RNAs (UCA1 and CCAT2) in the Blood of Multiple Sclerosis Patients: A Case-Control Study. Iranian Red Crescent Med J (2018) 20(8). doi: 10.5812/ircmj.66334
30. Dehghanzad R, Kakhki MP, Alikhah A, Sahraian MA, Behmanesh M. The Putative Association of TOB1-AS1 Long Non-Coding RNA With Immune Tolerance: A Study on Multiple Sclerosis Patients. Neuromol Med (2020) 22(1):100–10. doi: 10.1007/s12017-019-08567-1
31. Eftekharian MM, Ghafouri-Fard S, Soudyab M, Omrani MD, Rahimi M, Sayad A, et al. Expression Analysis of Long Non-Coding RNAs in the Blood of Multiple Sclerosis Patients. J Mol Neurosci (2017) 63(3-4):333–41. doi: 10.1007/s12031-017-0982-1
32. Eftekharian MM, Noroozi R, Komaki A, Mazdeh M, Ghafouri-Fard S, Taheri M. MALAT1 Genomic Variants and Risk of Multiple Sclerosis. Immunol Investig (2019) 48(5):549–54. doi: 10.1080/08820139.2019.1576728
33. Eftekharian MM, et al. GAS5 Genomic Variants and Risk of Multiple Sclerosis. Neurosci Lett (2019) 701:54–7. doi: 10.1016/j.neulet.2019.02.028
34. Eftekharian MM, et al. Nicotinamide Nucleotide Transhydrogenase Expression Analysis in Multiple Sclerosis Patients. Int J Neurosci (2019) 129(12):1256–60. doi: 10.1080/00207454.2019.1660655
35. Fenoglio C, et al. LncRNAs Expression Profile in Peripheral Blood Mononuclear Cells From Multiple Sclerosis Patients. J Neuroimmunol (2018) 324:129–35. doi: 10.1016/j.jneuroim.2018.08.008
36. Ganji M, et al. Expression Analysis of Long Non-Coding RNAs and Their Target Genes in Multiple Sclerosis Patients. Neurol Sci (2019) 40(4):801–11. doi: 10.1007/s10072-019-3720-3
37. Ghaiad HR, et al. Long Noncoding RNAs APOA1-AS, IFNG-AS1, RMRP and Their Related Biomolecules in Egyptian Patients With Relapsing-Remitting Multiple Sclerosis: Relation to Disease Activity and Patient Disability. J Adv Res (2020) 21:141–50. doi: 10.1016/j.jare.2019.10.012
38. Gharesouran J, et al. The Growth Arrest-Specific Transcript 5 (GAS5) and Nuclear Receptor Subfamily 3 Group C Member 1 (NR3C1): Novel Markers Involved in Multiple Sclerosis. Int J Mol Cell Med (2018) 7(2):102–10. doi: 10.22088/IJMCM.BUMS.7.2.102
39. Gharesouran J, et al. A Novel Regulatory Function of Long Non-Coding RNAs at Different Levels of Gene Expression in Multiple Sclerosis. J Mol Neurosci (2019) 67(3):434–40. doi: 10.1007/s12031-018-1248-2
40. Gharesouran J, et al. Integrative Analysis of OIP5-AS1/HUR1 to Discover New Potential Biomarkers and Therapeutic Targets in Multiple Sclerosis. J Cell Physiol (2019) 234(10):17351–60. doi: 10.1002/jcp.28355
41. Gharzi V, et al. Expression Analysis of BDNF Gene and BDNF-AS Long Noncoding RNA in Whole Blood Samples of Multiple Sclerosis Patients: Not Always a Negative Correlation Between Them. Iran J Allergy Asthma Immunol (2018) 17(6):548–56. doi: 10.18502/ijaai.v17i6.619
42. Ghoveud E, et al. Potential Biomarker and Therapeutic LncRNAs in Multiple Sclerosis Through Targeting Memory B Cells. NeuroMol Med (2020) 22(1):111–20. doi: 10.1007/s12017-019-08570-6
43. Hosseini A, et al. LncRNAs Associated With Multiple Sclerosis Expressed in the Th1 Cell Lineage. J Cell Physiol (2019) 234(12):22153–62. doi: 10.1002/jcp.28779
44. Kozin M, et al. Mitonuclear Interactions Influence Multiple Sclerosis Risk. Gene (2020) 758:144962. doi: 10.1016/j.gene.2020.144962
45. Masoumi F, et al. Malat1 Long Noncoding RNA Regulates Inflammation and Leukocyte Differentiation in Experimental Autoimmune Encephalomyelitis. J Neuroimmunol (2019) 328:50–9. doi: 10.1016/j.jneuroim.2018.11.013
46. Mazdeh M, et al. Long Non-Coding RNA AFAP1-AS1 Is Upregulated in a Subset of Multiple Sclerosis Patients. Clin Exp Neuroimmunol (2019) 10(2):105–9. doi: 10.1111/cen3.12513
47. Mazdeh M, et al. Single Nucleotide Polymorphisms of lncRNA H19 Are Not Associated With Risk of Multiple Sclerosis in Iranian Population. Meta Gene (2019) 21. doi: 10.1016/j.mgene.2019.100592
48. Moradi A, et al. Evaluation of the Expression Levels of Three Long Non-Coding RNAs in Multiple Sclerosis. Cell J (2019) 22(2):165–70.
49. Moradi M, et al. Role of NR3C1 and GAS5 Genes Polymorphisms in Multiple Sclerosis. Int J Neurosci (2020) 130(4):407–12. doi: 10.1080/00207454.2019.1694019
50. Pahlevan Kakhki M, et al. HOTAIR But Not ANRIL Long Non-Coding RNA Contributes to the Pathogenesis of Multiple Sclerosis. Immunology (2018) 153(4):479–87. doi: 10.1111/imm.12850
51. Pahlevan Kakhki M, et al. Differential Expression of STAT3 Gene and its Regulatory Long Non-Coding RNAs, Namely lnc-DC and THRIL, in Two Eastern Iranian Ethnicities With Multiple Sclerosis. Neurol Sci (2020) 41(3):561–8. doi: 10.1007/s10072-019-04092-y
52. Patoughi M, et al. GAS8 and its Naturally Occurring Antisense RNA as Biomarkers in Multiple Sclerosis. Immunobiology (2019) 224(4):560–4. doi: 10.1016/j.imbio.2019.04.005
53. Patoughi M, et al. Expression Analysis of PINK1 and PINK1-AS in Multiple Sclerosis Patients Versus Healthy Subjects. Nucleosides Nucleotides Nucleic Acids (2020) 40(2):157–65. doi: 10.1080/15257770.2020.1844229
54. Rahmani S, Noorolyai S, Ayromlou H, Shahgoli VK, Shanehbandi D, Baghbani E, et al. The Expression Analyses of RMRP, DDX5, and RORC in RRMS Patients Treated With Different Drugs Versus Naive Patients and Healthy Controls. Gene (2021) 769:145236. doi: 10.1016/j.gene.2020.145236
55. Rezazadeh M, et al. Association Study of ANRIL Genetic Variants and Multiple Sclerosis. J Mol Neurosci (2018) 65(1):54–9. doi: 10.1007/s12031-018-1069-3
56. Rodríguez-Lorenzo S, Francisco DM, Vos R, van Het Hof B, Rijnsburger M, Schroten H, et al. Altered Secretory and Neuroprotective Function of the Choroid Plexus in Progressive Multiple Sclerosis. Acta Neuropathol Commun (2020) 8(1):1–3. doi: 10.1186/s40478-020-00903-y
57. Safa A, et al. Dysregulation of NF-κb-Associated lncRNAs in Multiple Sclerosis Patients. J Mol Neurosci (2021) 71(1):80–8. doi: 10.1007/s12031-020-01628-2
58. Safa A, et al. Downregulation of Cancer-Associated lncRNAs in Peripheral Blood of Multiple Sclerosis Patients. J Mol Neurosci (2020) 70(10):1533–40. doi: 10.1007/s12031-020-01646-0
59. Santoro M, et al. Expression Profile of Long Non-Coding RNAs in Serum of Patients With Multiple Sclerosis. J Mol Neurosci (2016) 59(1):18–23. doi: 10.1007/s12031-016-0741-8
60. Santoro M, et al. A Pilot Study of lncRNAs Expression Profile in Serum of Progressive Multiple Sclerosis Patients. Eur Rev Med Pharmacol Sci (2020) 24(6):3267–73.
61. Sayad A, et al. Hepatocellular Carcinoma Up-Regulated Long Non-Coding RNA: A Putative Marker in Multiple Sclerosis. Metab Brain Dis (2019) 34(4):1201–5. doi: 10.1007/s11011-019-00418-z
62. Senousy MA, et al. LncRNA GAS5 and miR-137 Polymorphisms and Expression Are Associated With Multiple Sclerosis Risk: Mechanistic Insights and Potential Clinical Impact. ACS Chem Neurosci (2020) 11(11):1651–60. doi: 10.1021/acschemneuro.0c00150
63. Shaker OG, et al. Correlation Between LincR-Gng2-5’and LincR-Epas1-3’as With the Severity of Multiple Sclerosis in Egyptian Patients. Int J Neurosci (2020) 130(5):515–21. doi: 10.1080/00207454.2019.1695610
64. Shaker OG, et al. lincR-Ccr2-5 ’ AS and THRIL as Potential Biomarkers of Multiple Sclerosis. Egyptian J Med Hum Genet (2021) 22(1). doi: 10.1186/s43042-021-00151-2
65. Shaker OG, et al. LncRNAs, MALAT1 and lnc-DC as Potential Biomarkers for Multiple Sclerosis Diagnosis. Biosci Rep (2019) 39(1). doi: 10.1042/BSR20181335
66. Taheri M, Noroozi R, Sadeghpour S, Omrani MD, Ghafouri-Fard S. The Rs4759314 SNP Within Hotair lncRNA Is Associated With Risk of Multiple Sclerosis. Multiple Sclerosis Relat Disord (2020) 40:101986. doi: 10.1016/j.msard.2020.101986
67. Teimuri S, et al. Integrative Analysis of lncRNAs in Th17 Cell Lineage to Discover New Potential Biomarkers and Therapeutic Targets in Autoimmune Diseases. Mol Ther Nucleic Acids (2018) 12:393–404. doi: 10.1016/j.omtn.2018.05.022
68. Zhang F, et al. Expression Profile of Long Noncoding RNAs in Peripheral Blood Mononuclear Cells From Multiple Sclerosis Patients. CNS Neurosci Ther (2016) 22(4):298–305. doi: 10.1111/cns.12498
69. Zhang F, et al. DDIT4 and Associated Lncddit4 Modulate Th17 Differentiation Through the DDIT4/TSC/mTOR Pathway. J Immunol (2018) 200(5):1618–26. doi: 10.4049/jimmunol.1601689
70. Zhang F, et al. Linc-MAF-4 Regulates Th1/Th2 Differentiation and Is Associated With the Pathogenesis of Multiple Sclerosis by Targeting MAF. FASEB J (2017) 31(2):519–25. doi: 10.1096/fj.201600838R
71. Bian Z, et al. Gm15575 Functions as a ceRNA to Up-Regulate CCL7 Expression Through Sponging miR-686 in Th17 Cells. Mol Immunol (2020) 125:32–42. doi: 10.1016/j.molimm.2020.06.027
72. Duan C, Liu Y, Li Y, Chen H, Liu X, Chen X, et al. Sulfasalazine Alters Microglia Phenotype by Competing Endogenous RNA Effect of miR-136-5p and Long Non-Coding RNA HOTAIR in Cuprizone-Induced Demyelination. Biochem Pharmacol (2018) 155:110–23. doi: 10.1016/j.bcp.2018.06.028
73. Guo W, Lei W, Yu D, Ge Y, Chen Y, Xue W, et al. Involvement of lncRNA-1700040D17Rik in Th17 Cell Differentiation and the Pathogenesis of EAE. Int Immunopharmacol (2017) 47:141–9. doi: 10.1016/j.intimp.2017.03.014
74. Liu X, Zhou F, Wang W, Chen G, Zhang Q, Lv R, et al. IL-9-Triggered lncRNA Gm13568 Regulates Notch1 in Astrocytes Through Interaction With CBP/P300: Contribute to the Pathogenesis of Experimental Autoimmune Encephalomyelitis. J Neuroinflamm (2021) 18(1):1–5. doi: 10.1186/s12974-021-02156-5
75. Sun DY, Yu Z, Fang X, Liu M, Pu Y, Shao Q, et al. LncRNA GAS5 Inhibits Microglial M2 Polarization and Exacerbates Demyelination. EMBO Rep (2017) 18(10):1801–16. doi: 10.15252/embr.201643668
76. Yue P, Jing L, Zhao X, Zhu H, Teng J. Down-Regulation of Taurine-Up-Regulated Gene 1 Attenuates Inflammation by Sponging miR-9-5p via Targeting NF-κb1/P50 in Multiple Sclerosis. Life Sci (2019) 233:116731. doi: 10.1016/j.lfs.2019.116731
77. Powell JD, et al. Regulation of Immune Responses by mTOR. Annu Rev Immunol (2012) 30:39–68. doi: 10.1146/annurev-immunol-020711-075024
78. Lock C, Hermans G, Pedotti R, Brendolan A, Schadt E, Garren H, et al. Gene-Microarray Analysis of Multiple Sclerosis Lesions Yields New Targets Validated in Autoimmune Encephalomyelitis. Nat Med (2002) 8(5):500–8. doi: 10.1038/nm0502-500
79. Matusevicius D, Kivisäkk P, He B, Kostulas N, Özenci V, Fredrikson S, et al. Interleukin-17 mRNA Expression in Blood and CSF Mononuclear Cells Is Augmented in Multiple Sclerosis. Multiple Sclerosis J (1999) 5(2):101–4. doi: 10.1191/135245899678847275
80. Amaral PP, Clark MB, Gascoigne DK, Dinger ME, Mattick JS. LncRNAdb: A Reference Database for Long Noncoding RNAs. Nucleic Acids Res (2011) 39(suppl_1):D146–51. doi: 10.1093/nar/gkq1138
81. Ransohoff JD, Wei Y, Khavari PA. The Functions and Unique Features of Long Intergenic Non-Coding RNA. Nat Rev Mol Cell Biol (2018) 19(3):143. doi: 10.1038/nrm.2017.104
82. Stelzer G, Rosen N, Plaschkes I, Zimmerman S, Twik M, Fishilevich S, et al. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Curr Protoc Bioinf (2016) 54(1):1.30. 1–1.30. 33.
83. Wang Y, Zhong H, Xie X, Chen CY, Huang D, Shen L, et al. Long Noncoding RNA Derived From CD244 Signaling Epigenetically Controls CD8+ T-Cell Immune Responses in Tuberculosis Infection. Proc Natl Acad Sci (2015) 112(29):E3883–92. doi: 10.1073/pnas.1501662112
84. Luo M, Liu X, Meng H, Xu L, Li Y, Li Z, et al. IFNA-AS1 Regulates CD4+ T Cell Activation in Myasthenia Gravis Though HLA-Drb1. Clin Immunol (2017) 183:121–31. doi: 10.1016/j.clim.2017.08.008
85. Aune TM, Crooke PS, Spurlock CF. Long Noncoding RNAs in T Lymphocytes. J Leukocyte Biol (2016) 99(1):31–44. doi: 10.1189/jlb.1RI0815-389R
86. Stein N, Berhani O, Schmiedel D, Duev-Cohen A, Seidel E, Kol I, et al. IFNG-AS1 Enhances Interferon Gamma Production in Human Natural Killer Cells. Iscience (2019) 11:466–73. doi: 10.1016/j.isci.2018.12.034
87. Collier SP, Collins PL, Williams CL, Boothby MR, Aune TM. Cutting Edge: Influence of Tmevpg1, a Long Intergenic Noncoding RNA, on the Expression of Ifng by Th1 Cells. J Immunol (2014) 192(1):533–3. doi: 10.4049/jimmunol.1390068
88. Yu Q, Chang HC, Ahyi AN, Kaplan MH. Transcription Factor-Dependent Chromatin Remodeling of Il18r1 During Th1 and Th2 Differentiation. J Immunol (2008) 181(5):3346–52. doi: 10.4049/jimmunol.181.5.3346
89. Gutcher I, et al. Interleukin 18–Independent Engagement of Interleukin 18 Receptor-α Is Required for Autoimmune Inflammation. Nat Immunol (2006) 7(9):946–53. doi: 10.1038/ni1377
90. Tzachanis D, Freeman GJ, Hirano N, van Puijenbroek AA, Delfs MW, Berezovskaya A, et al. Tob Is a Negative Regulator of Activation That Is Expressed in Anergic and Quiescent T Cells. Nat Immunol (2001) 2(12):1174–82. doi: 10.1038/ni730
91. Li Z, Chao TC, Chang KY, Lin N, Patil VS, Shimizu C, Head SR, et al. The Long Noncoding RNA THRIL Regulates Tnfα Expression Through its Interaction With hnRNPL. Proc Natl Acad Sci (2014) 111(3):1002–7. doi: 10.1073/pnas.1313768111
92. Wang P, Xue Y, Han Y, Lin L, Wu C, Xu S, et al. The STAT3-Binding Long Noncoding RNA lnc-DC Controls Human Dendritic Cell Differentiation. Science (2014) 344(6181):310–3. doi: 10.1126/science.1251456
93. Xie Z, Guo Z, Liu J. Whey Acidic Protein/Four-Disulfide Core Domain 21 Regulate Sepsis Pathogenesis in a Mouse Model and a Macrophage Cell Line via the Stat3/toll-Like Receptor 4 (TLR4) Signaling Pathway. Med Sci Moni: Int Med J Exp Clin Res (2018) 24:4054. doi: 10.12659/MSM.907176
94. Alikhah A, Kakhkia MP, Ahmadi A, Dehghanzad R, Boroumand MA, Behmanesh M. The Role of lnc-DC Long Non-Coding RNA and SOCS1 in the Regulation of STAT3 in Coronary Artery Disease and Type 2 Diabetes Mellitus. J Diabetes Its Complic (2018) 32(3):258–65. doi: 10.1016/j.jdiacomp.2017.12.001
95. Zhang W, Zhou Y, Ding Y. Lnc-DC Mediates the Over-Maturation of Decidual Dendritic Cells and Induces the Increase in Th1 Cells in Preeclampsia. Am J Reprod Immunol (2017) 77(6):e12647. doi: 10.1111/aji.12647
96. Wu G-C, Li J, Leng RX, Li XP, Li XM, Wang DG, et al. Identification of Long Non-Coding RNAs GAS5, Linc0597 and lnc-DC in Plasma as Novel Biomarkers for Systemic Lupus Erythematosus. Oncotarget (2017) 8(14):23650. doi: 10.18632/oncotarget.15569
97. Zhuang L, Tian J, Zhang X, Wang H, Huang C. Lnc-DC Regulates Cellular Turnover and the HBV-Induced Immune Response by TLR9/STAT3 Signaling in Dendritic Cells. Cell Mol Biol Lett (2018) 23(1):1–9. doi: 10.1186/s11658-018-0108-y
98. Zhou Y, Fang L, Peng L, Qiu W. TLR9 and its Signaling Pathway in Multiple Sclerosis. J Neurol Sci (2017) 373:95–9. doi: 10.1016/j.jns.2016.12.027
99. Kim C, Kang D, Lee EK, Lee JS. Long Noncoding RNAs and RNA-Binding Proteins in Oxidative Stress, Cellular Senescence, and Age-Related Diseases. Oxid Med Cell Longevity (2017) 2017. doi: 10.1155/2017/2062384
100. Abedin K, et al. Oxidative DNA Damage in the Spinal Cord in Multiple Sclerosis. Neuropathol Appl Neurobiol (2002) 28(2):168–8. doi: 10.1046/j.1365-2990.2002.39286_53.x
101. Tehrani SS, et al. Multiple Functions of Long Non-Coding RNAs in Oxidative Stress, DNA Damage Response and Cancer Progression. J Cell Biochem (2018) 119(1):223–36. doi: 10.1002/jcb.26217
102. Yang C, et al. Long Non-Coding RNA UCA1 Regulated Cell Cycle Distribution via CREB Through PI3-K Dependent Pathway in Bladder Carcinoma Cells. Gene (2012) 496(1):8–16. doi: 10.1016/j.gene.2012.01.012
103. Xue Y, et al. Long Noncoding RNA CCAT2 as an Independent Prognostic Marker in Various Carcinomas: Evidence Based on Four Studies. Int J Clin Exp Med (2016) 9(2):2567–72.
104. Ong MS, et al. ‘Lnc’-Ing Wnt in Female Reproductive Cancers: Therapeutic Potential of Long Non-Coding RNAs in Wnt Signalling. Br J Pharmacol (2017) 174(24):4684–700. doi: 10.1111/bph.13958
105. Khalil AM, et al. Many Human Large Intergenic Noncoding RNAs Associate With Chromatin-Modifying Complexes and Affect Gene Expression. Proc Natl Acad Sci (2009) 106(28):11667–72. doi: 10.1073/pnas.0904715106
106. Li J, et al. LncRNA TUG1 Acts as a Tumor Suppressor in Human Glioma by Promoting Cell Apoptosis. Exp Biol Med (2016) 241(6):644–9. doi: 10.1177/1535370215622708
107. Mercer TR, et al. Long Noncoding RNAs in Neuronal-Glial Fate Specification and Oligodendrocyte Lineage Maturation. BMC Neurosci (2010) 11(1):14. doi: 10.1186/1471-2202-11-14
108. Barry G, et al. The Long Non-Coding RNA Gomafu Is Acutely Regulated in Response to Neuronal Activation and Involved in Schizophrenia-Associated Alternative Splicing. Mol Psychiatry (2014) 19(4):486–94. doi: 10.1038/mp.2013.45
109. Arunkumar G, et al. LncRNA OIP5-AS1 Is Overexpressed in Undifferentiated Oral Tumors and Integrated Analysis Identifies as a Downstream Effector of Stemness-Associated Transcription Factors. Sci Rep (2018) 8(1):1–13. doi: 10.1038/s41598-018-25451-3
110. Kim J, et al. LncRNA OIP5-AS1/cyrano Suppresses GAK Expression to Control Mitosis. Oncotarget (2017) 8(30):49409. doi: 10.18632/oncotarget.17219
111. Chen J, et al. Posttranscriptional Gene Regulation of IL-17 by the RNA-Binding Protein HuR Is Required for Initiation of Experimental Autoimmune Encephalomyelitis. J Immunol (2013) 191(11):5441–50. doi: 10.4049/jimmunol.1301188
112. Pruunsild P, et al. Dissecting the Human BDNF Locus: Bidirectional Transcription, Complex Splicing, and Multiple Promoters. Genomics (2007) 90(3):397–406. doi: 10.1016/j.ygeno.2007.05.004
113. Modarresi F, et al. Inhibition of Natural Antisense Transcripts In Vivo Results in Gene-Specific Transcriptional Upregulation. Nat Biotechnol (2012) 30(5):453–9. doi: 10.1038/nbt.2158
114. Yoshimura S, Ochi H, Isobe N, Matsushita T, Motomura K, Matsuoka T, et al. Altered Production of Brain-Derived Neurotrophic Factor by Peripheral Blood Immune Cells in Multiple Sclerosis. Multiple Sclerosis J (2010) 16(10):1178–88. doi: 10.1177/1352458510375706
115. Sasaki YT, Ideue T, Sano M, Mituyama T, Hirose T. Menϵ/β Noncoding RNAs Are Essential for Structural Integrity of Nuclear Paraspeckles. Proc Natl Acad Sci (2009) 106(8):2525–30. doi: 10.1073/pnas.0807899106
116. Bsibsi M, Bajramovic JJ, Vogt MH, van Duijvenvoorden E, Baghat A, Persoon-Deen C, et al. The Microtubule Regulator Stathmin Is an Endogenous Protein Agonist for TLR3. J Immunol (2010) 184(12):6929–37. doi: 10.4049/jimmunol.0902419
117. Wu G-C, Pan HF, Leng RX, Wang DG, Li XP, Li XM, et al. Emerging Role of Long Noncoding RNAs in Autoimmune Diseases. Autoimmun Rev (2015) 14(9):798–805. doi: 10.1016/j.autrev.2015.05.004
118. Imamura K, Imamachi N, Akizuki G, Kumakura M, Kawaguchi A, Nagata K, et al. Long Noncoding RNA NEAT1-Dependent SFPQ Relocation From Promoter Region to Paraspeckle Mediates IL8 Expression Upon Immune Stimuli. Mol Cell (2014) 53(3):393–406. doi: 10.1016/j.molcel.2014.01.009
119. Markert A, Grimm M, Martinez J, Wiesner J, Meyerhans A, Meyuhas O, et al. The La-Related Protein LARP7 Is a Component of the 7SK Ribonucleoprotein and Affects Transcription of Cellular and Viral Polymerase II Genes. EMBO Rep (2008) 9(6):569–75. doi: 10.1038/embor.2008.72
120. He N, Jahchan NS, Hong E, Li Q, Bayfield MA, Maraia RJ, et al. A La-Related Protein Modulates 7SK snRNP Integrity to Suppress P-TEFb-Dependent Transcriptional Elongation and Tumorigenesis. Mol Cell (2008) 29(5):588–99. doi: 10.1016/j.molcel.2008.01.003
121. Bo H, Gong Z, Zhang W, Li X, Zeng Y, Liao Q, et al. Upregulated Long Non-Coding RNA AFAP1-AS1 Expression Is Associated With Progression and Poor Prognosis of Nasopharyngeal Carcinoma. Oncotarget (2015) 6(24):20404. doi: 10.18632/oncotarget.4057
122. Kamphuis W, Derada Troletti C, Reijerkerk A, Romero I, de Vries EH. The Blood-Brain Barrier in Multiple Sclerosis: microRNAs as Key Regulators. CNS Neurol Disorders-Drug Targets (Formerly Curr Drug Targets-CNS Neurol Disorders) (2015) 14(2):157–67. doi: 10.2174/1871527314666150116125246
123. Reedquist KA, Tak PP. Suppl 2: Signal Transduction Pathways in Chronic Inflammatory Autoimmune Disease: Small GTPases. Open Rheumatol J (2012) 6:259. doi: 10.2174/1874312901206010259
124. Zhang J-Y, Weng M-Z, Song F-B, Xu Y-G, Liu Q, Wu J-Y, et al. Long Noncoding RNA AFAP1-AS1 Indicates a Poor Prognosis of Hepatocellular Carcinoma and Promotes Cell Proliferation and Invasion via Upregulation of the RhoA/Rac2 Signaling. Int J Oncol (2016) 48(4):1590–8. doi: 10.3892/ijo.2016.3385
125. Zhao Y, Chu Y, Sun J, Song R, Li Y, Xu F. LncRNA Gas8-AS Inhibits Colorectal Cancer (CRC) Cell Proliferation by Downregulating lncRNA AFAP1-As1. Gene (2019) 710:140–4. doi: 10.1016/j.gene.2019.05.040
126. Zha Z, Han Q, Liu W, Huo S. lncRNA GAS8-AS1 Downregulates lncRNA UCA1 to Inhibit Osteosarcoma Cell Migration and Invasion. J Orthopaedic Surg Res (2020) 15(1):1–6. doi: 10.1186/s13018-020-1550-x
127. Tuosto L. Targeting Inflammatory T Cells in Multiple Sclerosis: Current Therapies and Future Challenges. Austin J Mult Scler Neuroimmunol (2015) 2(1):1009. doi: 10.26420/austinjmultsclerneuroimmunol.2015.1009
128. Scheele C, Petrovic N, Faghihi MA, Lassmann T, Fredriksson K, Rooyackers O, et al. The Human PINK1 Locus Is Regulated In Vivo by a Non-Coding Natural Antisense RNA During Modulation of Mitochondrial Function. BMC Genomics (2007) 8(1):1–13. doi: 10.1186/1471-2164-8-74
129. Wendlandt EB, Graff JW, Gioannini TL, McCaffrey AP, Wilson ME. The Role of microRNAs miR-200b and miR-200c in TLR4 Signaling and NF-κb Activation. Innate Immun (2012) 18(6):846–55. doi: 10.1177/1753425912443903
130. Guo W, Luo C, Wang C, Wang Yh, Wang X, Gao Xd, et al. Suppression of Human and Mouse Th17 Differentiation and Autoimmunity by an Endogenous Interleukin 23 Receptor Cytokine-Binding Homology Region. Int J Biochem Cell Biol (2014) 55:304–10. doi: 10.1016/j.biocel.2014.09.019
Keywords: lncRNAs, multiple sclerosis, neurodegenerative disease, polymorphism, expression
Citation: Jalaiei A, Asadi MR, Sabaie H, Dehghani H, Gharesouran J, Hussen BM, Taheri M, Ghafouri-Fard S and Rezazadeh M (2021) Long Non-Coding RNAs, Novel Offenders or Guardians in Multiple Sclerosis: A Scoping Review. Front. Immunol. 12:774002. doi: 10.3389/fimmu.2021.774002
Received: 10 September 2021; Accepted: 08 November 2021;
Published: 07 December 2021.
Edited by:
Luisa María Villar, Ramón y Cajal University Hospital, SpainReviewed by:
Francesca Gilli, Dartmouth College, United StatesPaul Roy Heath, The University of Sheffield, United Kingdom
Copyright © 2021 Jalaiei, Asadi, Sabaie, Dehghani, Gharesouran, Hussen, Taheri, Ghafouri-Fard and Rezazadeh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Soudeh Ghafouri-Fard, s.ghafourifard@sbmu.ac.ir; Maryam Rezazadeh, Rezazadehm@tbzmed.ac.ir
 Abbas Jalaiei
Abbas Jalaiei Mohammad Reza Asadi
Mohammad Reza Asadi Hani Sabaie
Hani Sabaie Hossein Dehghani3
Hossein Dehghani3 Jalal Gharesouran
Jalal Gharesouran Mohammad Taheri
Mohammad Taheri Soudeh Ghafouri-Fard
Soudeh Ghafouri-Fard