- 1Department of Medical Genetics, School of Medicine, Shahid Beheshti University of Medical Sciences, Tehran, Iran
- 2Men’s Health and Reproductive Health Research Center, Shahid Beheshti University of Medical Sciences, Tehran, Iran
- 3Department of Clinical Analysis, College of Pharmacy, Hawler Medical University, Erbil, Iraq
- 4Institute of Human Genetics, Jena University Hospital, Jena, Germany
- 5Urology and Nephrology Research Center, Shahid Beheshti University of Medical Sciences, Tehran, Iran
- 6Phytochemistry Research Center, Shahid Beheshti University of Medical Sciences, Tehran, Iran
Long Intergenic Non-Protein Coding RNA 511 (LINC00511) is an RNA gene being mostly associated with lung cancer. Further assessments have shown dysregulation of this lncRNA in a variety of cancers. LINC00511 has interactions with hsa-miR-29b-3p, hsa-miR-765, hsa-mir-150, miR-1231, TFAP2A-AS2, hsa-miR-185-3p, hsa-miR-29b-1-5p, hsa-miR-29c-3p, RAD51-AS1 and EZH2. A number of transcription factors have been identified that regulate expression of LINC00511. The current narrative review summarizes the role of LINC00511 in different cancers with an especial focus on its prognostic impact in human cancers.
Introduction
Long non-coding RNAs (lncRNAs) are widely expressed transcripts with essential roles in gene regulation. Based on the results of the Human GENCODE project, the number of lncRNA genes in the human genome is estimated to surpass 16,000 (Fang et al., 2018). These transcripts embrace lncRNAs transcribed by RNA polymerase II, gene-overlapping antisense transcripts as well as lncRNAs from intergenic regions (lincRNAs) (Ghafouri-Fard et al., 2020; Ghafouri-Fard et al., 2021; Statello et al., 2021). Typically, lncRNAs have a 7-methyl guanosine cap at the 5′end and a polyadenylated tail at the 3′ends. Moreover, lncRNAs are spliced in a similar manner to mRNAs (Statello et al., 2021). However, several RNA polymerase II-transcribed lncRNAs are incompetently processed and are held in the nuclear compartment (Statello et al., 2021). LncRNAs interact with DNA, RNA and proteins. Through these interactions, lncRNAs influence chromatin structure and function, affect the assembly of nuclear bodies, change the stability and expression of mRNAs within the cytoplasm and regulate signaling pathways (Statello et al., 2021; Hussen et al., 2022). In comparison with mRNA promoters, lincRNA promoters have been found to be devoid of transcription factor binding sites, but having binding sites for a number of specific factors such as GATA and FOS (Melé et al., 2017).
Long Intergenic Non-Protein Coding RNA 511 (LINC00511) is an RNA gene being mostly associated with lung cancer. This lncRNA is encoded on chr17:72,290,091-72,640,472 (GRCh38/hg38), minus strand. Notably, more than 100 alternatively splice variants have been recognized for LINC00511. Except for two variants with retained introns (LINC00511-279 with 3766 bp and LINC00511-278 with 508 bp), other are affiliated with lncRNA group of transcripts (https://asia.ensembl.org/Homo_sapiens/Gene/Summary?db=core;g=ENSG00000227036; r=17:72290091-72640472). This lncRNA has important roles in the development of cancers and can be used as a possible diagnostic and prognostic marker in cancer. Abnormal expression of LINC00511 in a wide array of malignancies potentiates it as a target for therapeutic interventions. Based on DIANA-LncBase database (https://diana.e-ce.uth.gr/lncbasev3) (Karagkouni et al., 2020), expression of LINC00511 in various tissues and cell types have been investigated. With medium and high TPM levels and in homosapiens, the highest level of expression of LINC00511 is in prostate tissue and PC3 cell type along with cancer/malignant category.
The current narrative review summarizes the role of LINC00511 in different cancers with an especial focus on its prognostic impact in human cancers.
LINC00511 in cancers
Several studies have reported dysregulation (mainly upregulation) of LINC00511 in different cancers. These studies have also identified miRNAs that are sponged by LINC00511.
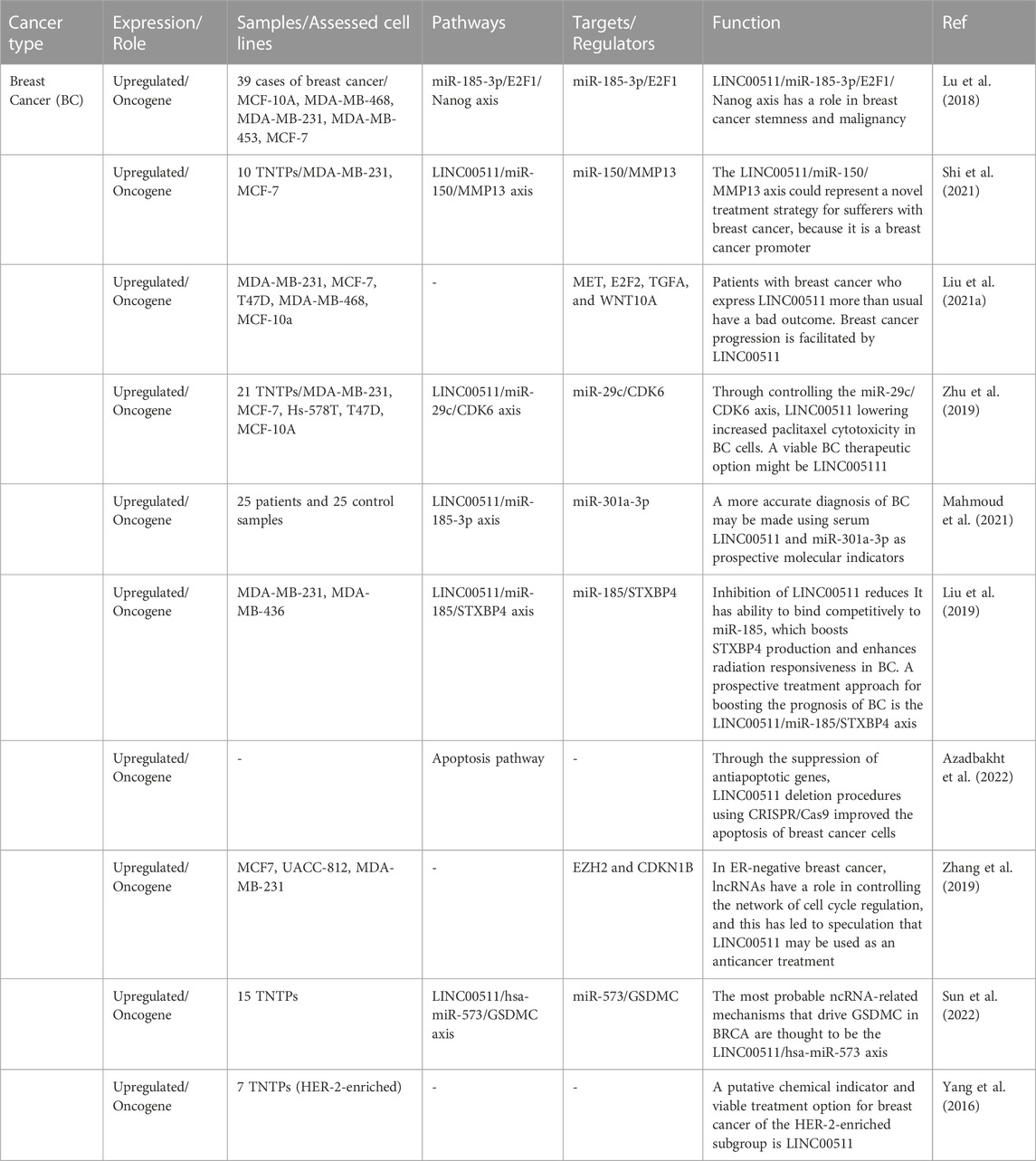
Breast cancer
In breast cancer, LINC00511 has been found to be highly expressed in the clinical samples and its over-expression has been correlated with poor prognosis (Lu et al., 2018). Functional studies have shown that LINC00511 promotes proliferation, sphere-formation capacity, expression of stem factors and growth of breast tumors (Lu et al., 2018). From a mechanical point of view, LINC00511 acts as a molecular sponge for miR-185-3p to enhance expression of E2F1 protein. Besides, E2F1 binds with the promoter of Nanog gene and increases its expression (Figure 1). Therefore, LINC00511/miR-185-3p/E2F1/Nanog axis has been identified as an important route for induction of stemness and tumorigenesis in breast cancer (Lu et al., 2018). Another study in breast cancer has revealed more than 180 potential targets for LINC00511 through siRNA and RNA-seq assays. Bioinformatics analyses have shown relation between differently expressed genes and signaling pathways mediated by p38-α and p38-β. LINC00511 has been found to be mainly located in the cytoplasm regulating expression of MMP13 through sponging miR-150 (Shi et al., 2021). Expression of LINC00511 in breast cancer samples has been closely correlated with the presence of lymph node metastasis, greater tumor size and molecular subtypes of breast cancer. This lncRNA has been found to increase migratory potential and invasive ability of MDA-MB-231 and MCF-7 cells. Moreover, expression of LINC00511 has been shown to be increased by DNA hypomethylation. In turn, LINC00511 could promote expressions of Wnt10A, E2F2, TGFA, and MET and reduce sensitivity of breast cancer cells to Panobinostat (Liu et al., 2021a). LINC00511 can also influence the cytotoxic effects of paclitaxel on breast cancer cells through regulating miR-29c/CDK6 axis (Zhu et al., 2019).
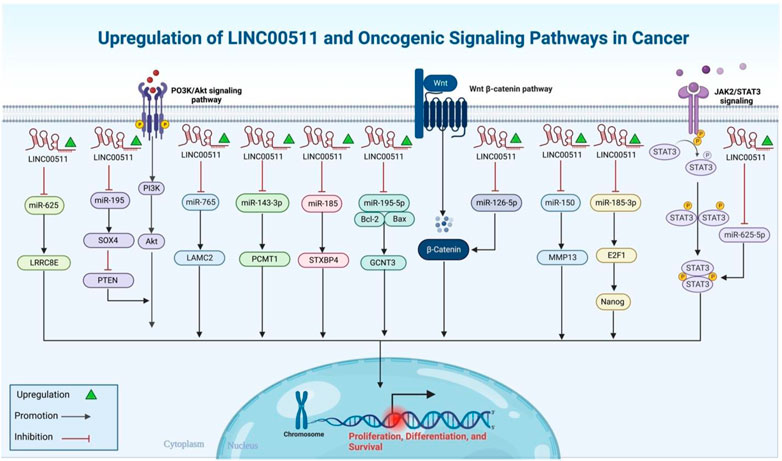
FIGURE 1. A graphical illustration of the oncogenic role that LINC00511 performs in the environment of the many types of cancer.
Another study in patients with breast cancer has reported upregulation of LINC00511 and miR-301a-3p in patients’ blood parallel with downregulation of miR-185-3p (Mahmoud et al., 2021). Notably, LINC00511 expression has been increased in early stages of breast cancer. Area under the reciever operating characteristic curves of LINC00511, miR-185-3p, and miR-301a-3p has been superior to classical tumor markers indicating the diagnostic values of these transcripts as molecular biomarkers in liquid biopsy. Moreover, expression of LINC00511 has been correlated with lymph node metastasis and advanced tumor grades (Mahmoud et al., 2021). Additionally, the sponging effect of LINC00511 on miR-185 has been shown to be involved in breast cancer recurrence and radioresistance via regulation of STXBP4 expression (Liu et al., 2019). In breast cancer cells, LINC00511 expression induced by TFAP-2 expression and directly affected by ER deficiency at the transcriptional level. Through its interaction with EZH2, LINC00511 has been shown to encourage tumor development and suppress apoptosis (Figure 2) (Zhang et al., 2019). Table 1 summarizes the results of studies regarding the role of this lncRNA in breast cancer.
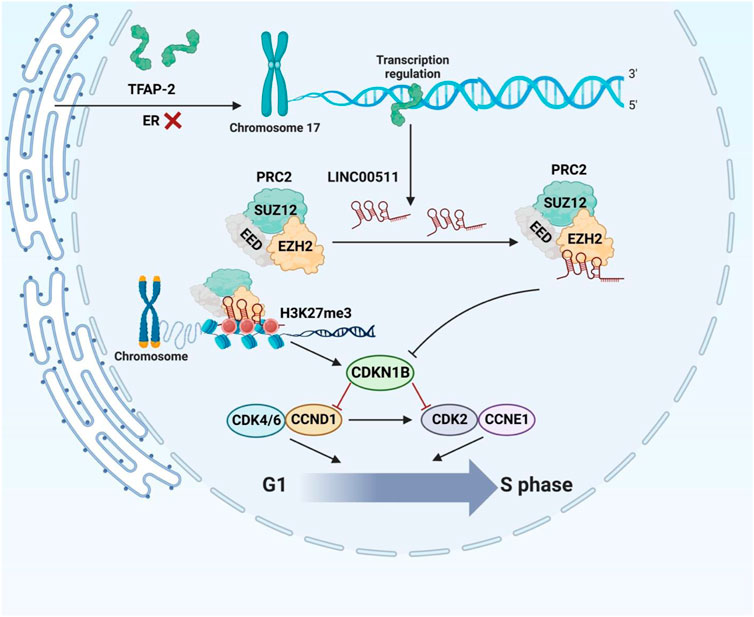
FIGURE 2. The carcinogenic role of LINC00511 in ER-negative tumorigenesis. ER insufficiency enhanced TFAP-2 activity at particular promoter regions, promoting LINC00511 expression. By interacting with EZH2 to attract PRC2 to regulate histone methylation, the ER-negative-associated LINC00511 repressed the expression of CDKN1B, assisting in the G1/S transition to maintain cellular growth.
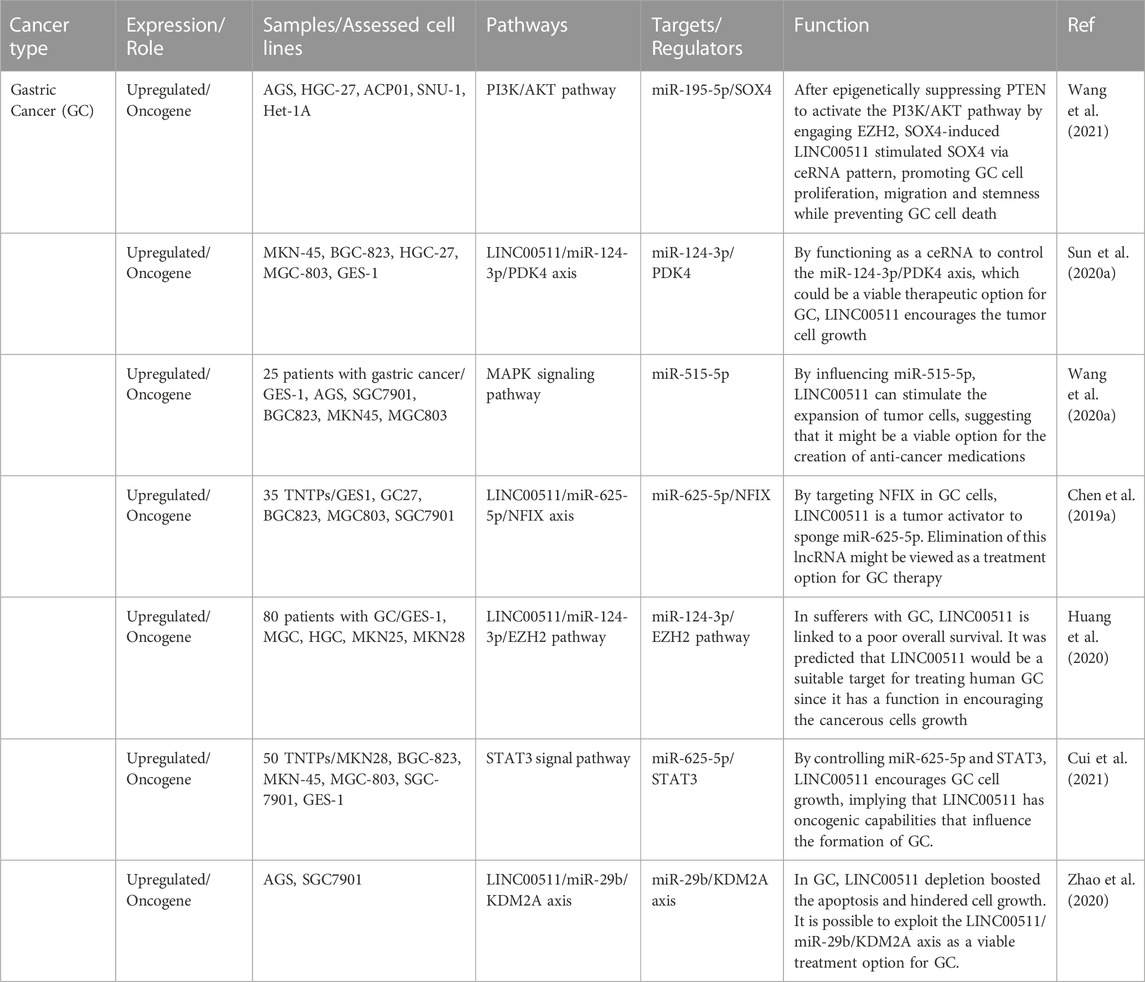
Gastric cancer
LINC00511 has been shown to promote progression of gstaric acnecr through regulating SOX4 expression and epigenetically suppressing PTEN to induce activity of PI3K/AKT pathway (Wang et al., 2021). Moreover, LINC00511 can promote growth of gastric tumors through acting as a molecular sponge for miR-124-3p and regulating expression of PDK4 (Sun et al., 2020a). miR-515-5p is another miRNA that is sponged by LINC00511 in gastric cancer cells leading to enhancemnet of proliferation and invasion of these cells (Wang et al., 2020a). Finally, miR-625-5p/NFIX (Chen et al., 2019a), miR-124-3p/EZH2 (Huang et al., 2020), miR-625-5p/STAT3 (Cui et al., 2021) and miR-29b/KDM2A (Zhao et al., 2020) are other molecular axes regulated by LINC00511 in gastric cancer. Table 2 shows the role of LINC00511 in gastric cancer.
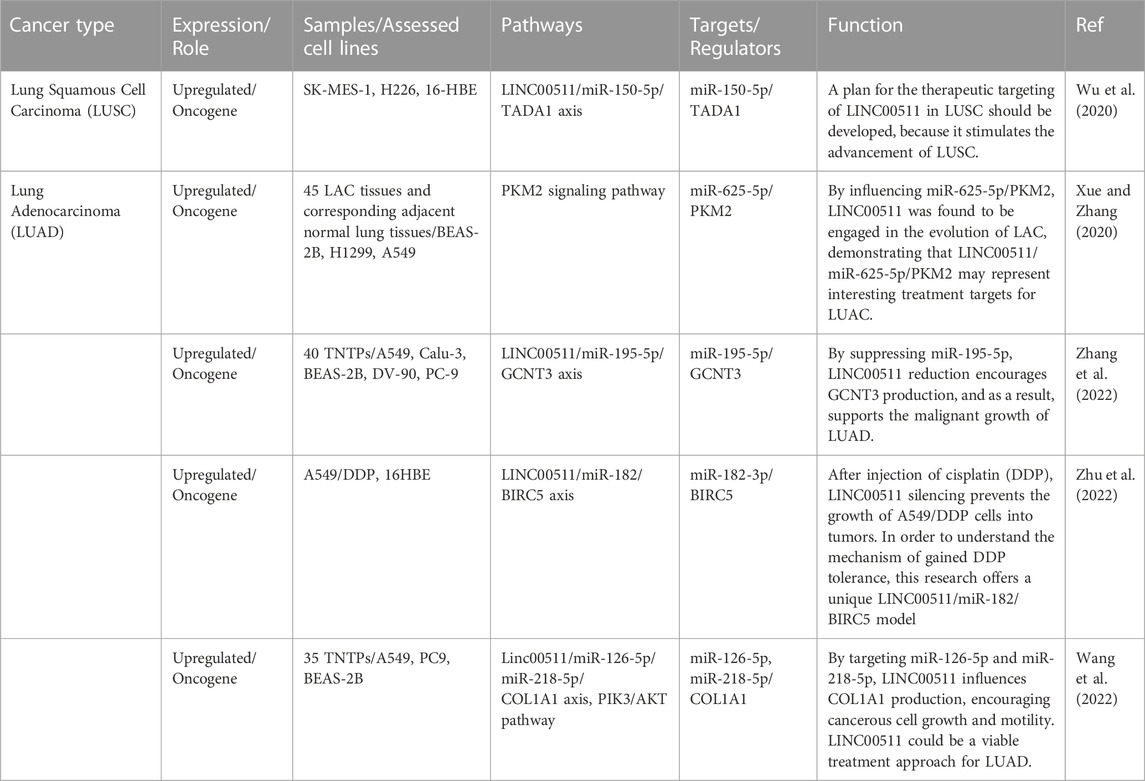
Lung cancer
LINC00511 has ben shown to promote proliferation, invasive capacities, and migration of non-small cell lung cancer cells through regulating miR-625-5p/GSPT1 axis (Cheng et al., 2021). Moreover, LINC00511 can promote progression of this type of cancer through binding to EZH2 and LSD1 and decreasing expression levels of LATS2 and KLF2 (Zhu et al., 2019). Besides, this lncRNA has a role in induction of tumor recurrence in this type of cancer through sponging miR-98-5p and increasing expression of TGFBR1 (Li et al., 2022). LINC00511 can also induce resistance of lung cancer cells to cisplatin through sponging miR-625 and influencing expression of LRRC8E (Liu et al., 2022). Finally, this lncRNA exerts its oncogenic effects in lung cancer through binding to EZH2 and decaresing expression of p57 (Sun et al., 2016). Table 3 shows the role of LINC00511 in lung cancer.
Other types of cancers
Over-expression of LINC00511 has been reported in a variety of cancers including colorectal, pancreatic, liver, thyroid and other types of cancers (Table 4).
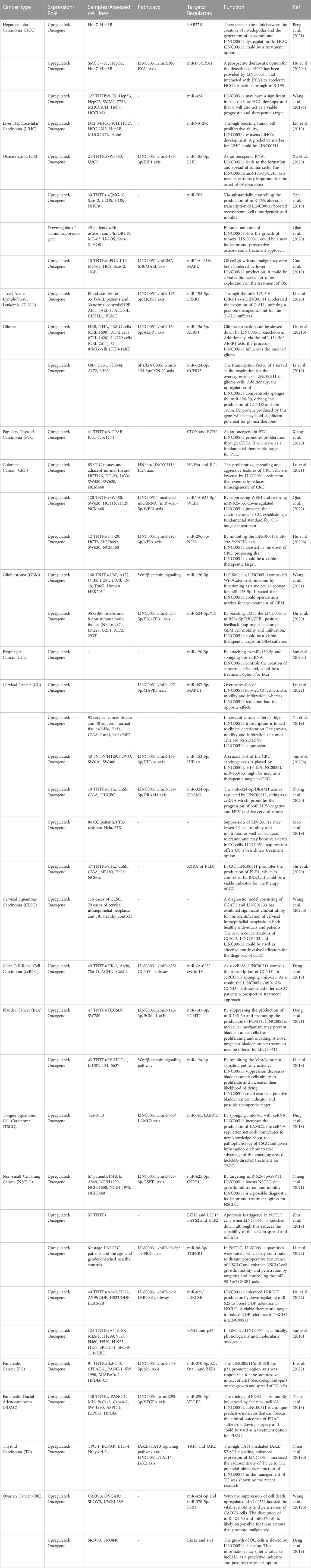
TABLE 4. Dysregulation of LINC00511 in different cancers (TNTP: tumor and non-tumor pairs of tissues).
Transcriptional regulation of LINC00511
Investigations in the Hormonizome database (Rouillard et al., 2016) indicated that 18 transcription factors (CTCF, EP300, ESR1, EZH2, FOXA1, GATA3, H2AFZ, MAX, MYC, NFIC, NR2F2, NR3C1, POLR2A, RAD21, TCF12, TEAD4, YY1, and ZBTB7A) possibly bind to the promoter of LINC00511 gene based on ChIP-seq data from the ENCODE Transcription Factor Target dataset.
LINC00511 related pathways and functions.
Based on lncHUB database (https://maayanlab.cloud/lnchub/), 10 KEGG pathways with the highest Z-score in which LINC00511 is predicted to be involved include glycosphingolipid biosynthesis, bacterial invasion of epithelial cells, basal cell carcinoma, central carbon metabolism in cancer, notch signaling pathway, RNA polymerase, DNA replication, cell cycle, bladder cancer and mismatch repair. Additionally, 10 gene ontology (GO) terms with the highest Z-score that are associated with LINC00511 include regulation of hydrogen peroxide-induced cell death (GO:1903205), negative regulation of response to reactive oxygen species (GO:1901032), protein heterotetramerization (GO:0051290), viral release from host cell (GO:0019076), exit from host cell (GO:0035891), signal complex assembly (GO:0007172), regulation of striated muscle tissue development (GO:0016202), regulation of myelination (GO:0031641), positive regulation of kidney development (GO:0090184) and regulation of proteolysis (GO:0030162). Also, IGSF11, PHLPP1, CPOX, SOX2, PACC1, SOX21, TMPRSS5, HEY1, MARCKS, KCTD5, OLIG2, ATAT1, CDK5R1, BCAN and BAALC are 15 genes with the highest Z-score predicted to be co-expressed with LINC00511.
LINC00511 interactions with miRNAs and other molecules
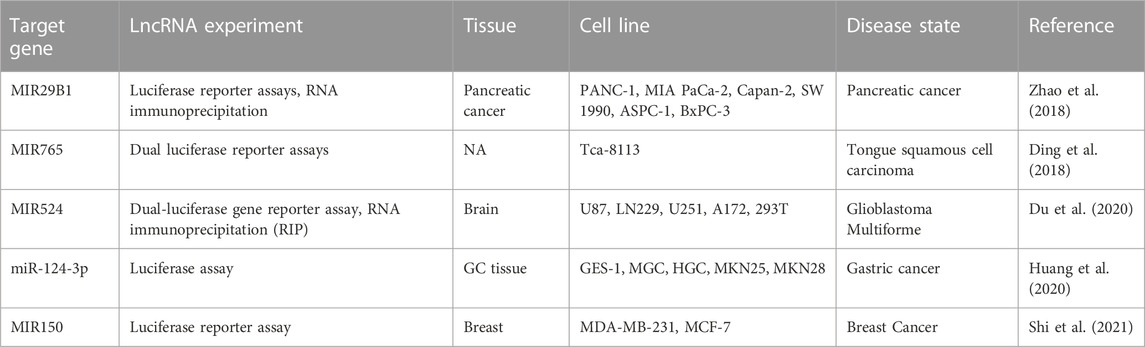
Based on RNAInter (RNA Interactome Database) (Kang et al., 2022), LINC00511 has interactions with hsa-miR-29b-3p, hsa-miR-765, hsa-mir-150, miR-1231, TFAP2A-AS2, hsa-miR-185-3p, hsa-miR-29b-1-5p, hsa-miR-29c-3p, RAD51-AS1 and EZH2 with score ≥0.1. Also, based on LncRNA2Target v3.0 (Cheng et al., 2019), LINC00511 interactions with miRNAs has been showed in Table 5.
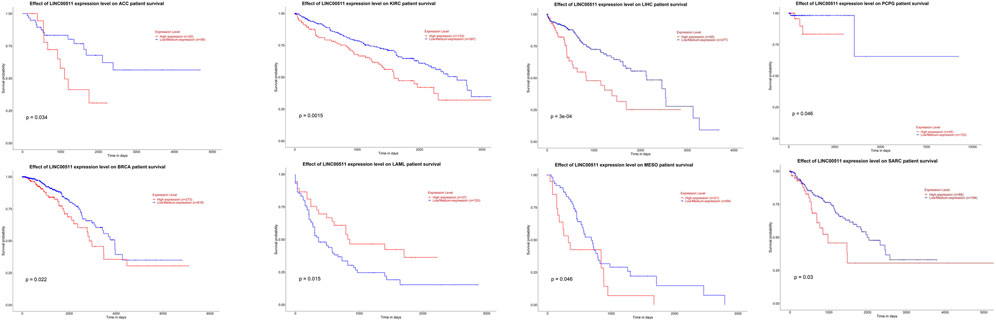
Impact of LINC00511 dysregulation on clinical outcome of patients with cancers
We investigated the survival rate caused by LINC00511 in different cancers using ualcan database (Chandrashekar et al., 2017). This database performs survival analysis using TCGA data. The difference was statistically significant with a log-rank p-value less than 0.05. As a result, LINC00511 has an effect on the survival rate of patients with adrenocortical carcinoma (ACC), breast invasive carcinoma (BRCA), kidney renal clear cell carcinoma (KIRC), acute myeloid leukemia (LAML), liver hepatocellular carcinoma (LIHC), Mesothelioma (MESO), pheochromocytoma and paraganglioma (PCPG) and sarcoma (SARC) (Figure 3). While in patients with LAML, overexpression of this lncRNA is associated with better clinical outcome, in other types of cancers, its upregulation is associated with lower survival.
Discussion
LINC00511 is a lincRNA being over-expressed in a variety of human tumors and cancer cell lines. Except for a single study in osteosarcoma (Qiao et al., 2020), other studies in this type of cancer and other cancers have reported upregulation of LINC00511 in tumoral tissues compared with their non-tumoral counterparts. Dysregulation of LINC00511 affects cancer pathogenesis through increasing cell proliferation and inhibiting cell apoptosis. It can also increase activity of several cancer-promoting signaling pathways.
A previous meta-analysis has reported association between over-expression of LINC00511 and poor prognosis in different cancers in terms of overall, progression-free or relapse-free survival times (Agbana et al., 2020). Moreover, upregulation of this lncRNA has been associated with larger tumor size, recurrent disorder and metastasis to lymph nodes or distant organs (Agbana et al., 2020).
Mechanistically, LINC00511 can act as a molecular sponge for a variety of miRNAs regulating their targets. miR-185-3p/E2F1, miR-150/MMP13, miR-29c/CDK6, miR-185/STXBP4, miR-573/GSDMC, miR-195-5p/SOX4, miR-124-3p/PDK4, miR-625-5p/NFIX, miR-124-3p/EZH2, miR-625-5p/STAT3, miR-29b/KDM2A, miR-195/EYA1, miR-185-3p/E2F1, miRNA-618/MAEL, miR-195-5p/LRRK1, miR-15a-5p/AEBP1, miR-124-3p/CCND2, miRNA-625-5p/WEE1, miR-29c-3p/NFIA, miR-150-5p/TADA1, miR-524-5p/YB1, miR-625-5p/PKM2, miR-195-5p/GCNT3, miR-182-3p/BIRC5, miR-218-5p/COL1A1, miR-497-5p/MAPK1, miR-153-5p/HIF-1α, miR-324-5p/DRAM1, miR-625/cyclin D1, miR-143-3p/PCMT1, miR-765/LAMC2, miR-625-5p/GSPT1, miR-98-5p/TGFBR1, miR-625/LRRC8E, miR-370-5p/p21, miR-29b-3p/VEGFA and miR-370-5p/ESR1 are examples of miRNA/mRNA axes that are regulated by LINC00511. Molecular axes being regulated by LINC00511 in more than one type of cancer represent better targets for design of anti-cancer therapies since they can be applied in a wider range of malignancies. Therefore, identification of the impact of above-mentioned molecular axes in the progression of different types of cancer is an important step in design of novel therapeutics.
LINC00511 can induce stemness in cancers and facilitate tumor progression and metastasis (Lu et al., 2018). Therefore, LINC00511-modifying modalities can be used as possible strategies for defeating cancer metastasis.
The prognostic role of over-expression of LINC00511 in different cancers has been evaluated thoroughly by various research groups indicating its important effects on survival of affected individuals. Future studies should assess its expression in biofluids to provide a non-invasive route for cancer diagnosis and patients’ follow-up.
Author contributions
MT designed and supervised the study. SG-F wrote the draft and revised it. AS, BH, and SA collected the data and designed the figures and tables. All the authors read the submitted version and approved it.
Funding
This study was Financially supported by Shahid Beheshti University of Medical Sciences.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Agbana, Y. L., Abi, M. E., Ni, Y., Xiong, G., Chen, J., Yun, F., et al. (2020). LINC00511 as a prognostic biomarker for human cancers: A systematic review and meta-analysis. BMC Cancer 20 (1), 682. doi:10.1186/s12885-020-07188-3
Azadbakht, N., Doosti, A., and Jami, M. S. (2022). CRISPR/Cas9-mediated LINC00511 knockout strategies, increased apoptosis of breast cancer cells via suppressing antiapoptotic genes. Biol. Proced. Online 24 (1), 8. doi:10.1186/s12575-022-00171-1
Chandrashekar, D. S., Bashel, B., Balasubramanya, S. A. H., Creighton, C. J., Ponce-Rodriguez, I., Chakravarthi, B., et al. (2017). Ualcan: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia 19 (8), 649–658. doi:10.1016/j.neo.2017.05.002
Chen, Y., Bao, C., Zhang, X., Lin, X., and Fu, Y. (2019). Knockdown of LINC00511 promotes radiosensitivity of thyroid carcinoma cells via suppressing JAK2/STAT3 signaling pathway. Cancer Biol. Ther. 20 (9), 1249–1257. doi:10.1080/15384047.2019.1617569
Chen, Z., Wu, H., Zhang, Z., Li, G., and Liu, B. (2019). LINC00511 accelerated the process of gastric cancer by targeting miR-625-5p/NFIX axis. Cancer Cell Int. 19, 351. doi:10.1186/s12935-019-1070-0
Cheng, L., Wang, P., Tian, R., Wang, S., Guo, Q., Luo, M., et al. (2019). LncRNA2Target v2.0: A comprehensive database for target genes of lncRNAs in human and mouse. Nucleic Acids Res. 47 (D1), D140–D144. doi:10.1093/nar/gky1051
Cheng, Y., Wang, S., and Mu, X. (2021). Long non-coding RNA LINC00511 promotes proliferation, invasion, and migration of non-small cell lung cancer cells by targeting miR-625-5p/GSPT1. Transl. Cancer Res. 10 (12), 5159–5173. doi:10.21037/tcr-21-1468
Cui, N., Sun, Q., Liu, H., Li, L., Guo, X., Shi, Y., et al. (2021). Long non-coding RNA LINC00511 regulates the expression of microRNA-625-5p and activates signal transducers and activators of transcription 3 (STAT3) to accelerate the progression of gastric cancer. Bioengineered 12 (1), 2915–2927. doi:10.1080/21655979.2021.1940611
Deng, H., Huang, C., Wang, Y., Jiang, H., Peng, S., and Zhao, X. (2019). LINC00511 promotes the malignant phenotype of clear cell renal cell carcinoma by sponging microRNA-625 and thereby increasing cyclin D1 expression. Aging (Albany NY) 11 (16), 5975–5991. doi:10.18632/aging.102156
Ding, J., Yang, C., and Yang, S. (2018). LINC00511 interacts with miR-765 and modulates tongue squamous cell carcinoma progression by targeting LAMC2. J. Oral Pathol. Med. 47 (5), 468–476. doi:10.1111/jop.12677
Dong, L. M., Zhang, X. L., Mao, M. H., Li, Y. P., Zhang, X. Y., Xue, D. W., et al. (2021). LINC00511/miRNA-143-3p modulates apoptosis and malignant phenotype of bladder carcinoma cells via PCMT1. Front. Cell Dev. Biol. 9, 650999. doi:10.3389/fcell.2021.650999
Du, X., Tu, Y., Liu, S., Zhao, P., Bao, Z., Li, C., et al. (2020). LINC00511 contributes to glioblastoma tumorigenesis and epithelial-mesenchymal transition via LINC00511/miR-524-5p/YB1/ZEB1 positive feedback loop. J. Cell Mol. Med. 24 (2), 1474–1487. doi:10.1111/jcmm.14829
Fang, S., Zhang, L., Guo, J., Niu, Y., Wu, Y., Li, H., et al. (2018). NONCODEV5: A comprehensive annotation database for long non-coding RNAs. Nucleic acids Res. 46 (D1), D308–D314. doi:10.1093/nar/gkx1107
Ghafouri-Fard, S., Abak, A., Shoorei, H., Mohaqiq, M., Majidpoor, J., Sayad, A., et al. (2021). Regulatory role of microRNAs on PTEN signaling. Biomed. Pharmacother. 133, 110986. doi:10.1016/j.biopha.2020.110986
Ghafouri-Fard, S., Shoorei, H., and Taheri, M. (2020). Non-coding RNAs are involved in the response to oxidative stress. Biomed. Pharmacother. 127, 110228. doi:10.1016/j.biopha.2020.110228
Guo, W., Yu, Q., Zhang, M., Li, F., Liu, Y., Jiang, W., et al. (2019). Long intergenic non-protein coding RNA 511 promotes the progression of osteosarcoma cells through sponging microRNA 618 to upregulate the expression of maelstrom. Aging (Albany NY) 11 (15), 5351–5367. doi:10.18632/aging.102109
Hu, W. Y., Wei, H. Y., Li, K. M., Wang, R. B., Xu, X. Q., and Feng, R. (2020). LINC00511 as a ceRNA promotes cell malignant behaviors and correlates with prognosis of hepatocellular carcinoma patients by modulating miR-195/EYA1 axis. Biomed. Pharmacother. 121, 109642. doi:10.1016/j.biopha.2019.109642
Hu, Y., Zhang, Y., Ding, M., and Xu, R. (2020). LncRNA LINC00511 acts as an oncogene in colorectal cancer via sponging miR-29c-3p to upregulate NFIA. Onco Targets Ther. 13, 13413–13424. doi:10.2147/OTT.S250377
Huang, H. G., Tang, X. L., Huang, X. S., Zhou, L., Hao, Y. G., and Zheng, Y. F. (2020). Long noncoding RNA LINC00511 promoted cell proliferation and invasion via regulating miR-124-3p/EZH2 pathway in gastric cancer. Eur. Rev. Med. Pharmacol. Sci. 24 (8), 4232–4245. doi:10.26355/eurrev_202004_21003
Hussen, B. M., Kheder, R. K., Abdullah, S. T., Hidayat, H. J., Rahman, H. S., Salihi, A., et al. (2022). Functional interplay between long non-coding RNAs and Breast CSCs. Cancer Cell Int. 22 (1), 233. doi:10.1186/s12935-022-02653-4
Ji, D., Hou, L., Xie, C., Feng, H., Bao, D., Teng, Y., et al. (2022). Deoxyelephantopin suppresses pancreatic cancer progression in vitro and in vivo by targeting linc00511/miR-370-5p/p21 promoter Axis. J. Oncol. 2022, 3855462. doi:10.1155/2022/3855462
Kang, J., Tang, Q., He, J., Li, L., Yang, N., Yu, S., et al. (2022). RNAInter v4.0: RNA interactome repository with redefined confidence scoring system and improved accessibility. Nucleic Acids Res. 50 (D1), D326–D332. doi:10.1093/nar/gkab997
Karagkouni, D., Paraskevopoulou, M. D., Tastsoglou, S., Skoufos, G., Karavangeli, A., Pierros, V., et al. (2020). DIANA-LncBase v3: Indexing experimentally supported miRNA targets on non-coding transcripts. Nucleic Acids Res. 48 (D1), D101–D110. doi:10.1093/nar/gkz1036
Li, C., Li, Z., Yi, H., and Liu, Z. (2022). IncRNA Linc00511 upregulation elevates TGFBR1 and participates in the postoperative distant recurrence of non-small-cell lung cancer by targeting miR-98-5p. Crit. Rev. Eukaryot. Gene Expr. 32 (5), 1–10. doi:10.1615/CritRevEukaryotGeneExpr.2022039498
Li, C., Liu, H., Yang, J., Yang, J., Yang, L., Wang, Y., et al. (2019). Long noncoding RNA LINC00511 induced by SP1 accelerates the glioma progression through targeting miR-124-3p/CCND2 axis. J. Cell Mol. Med. 23 (6), 4386–4394. doi:10.1111/jcmm.14331
Li, J., Li, Y., Meng, F., Fu, L., and Kong, C. (2018). Knockdown of long non-coding RNA linc00511 suppresses proliferation and promotes apoptosis of bladder cancer cells via suppressing Wnt/β-catenin signaling pathway. Biosci. Rep. 38 (4). doi:10.1042/bsr20171701
Li, S., Guo, W., Geng, H., Wang, C., Yang, S., and Xu, X. (2020). LINC00511 exacerbated T-cell acute lymphoblastic leukemia via miR-195-5p/LRRK1 axis. Biosci. Rep. 40 (5). doi:10.1042/BSR20193631
Liu, B., Zhou, F., Liu, H., Wang, Y., Wang, J., Ren, F., et al. (2022). Knockdown of LINC00511 decreased cisplatin resistance in non-small cell lung cancer by elevating miR-625 level to suppress the expression of leucine rich repeat containing eight volume-regulated anion channel subunit E. Hum. Exp. Toxicol. 41, 9603271221089000. doi:10.1177/09603271221089000
Liu, C., Xu, Y., Liu, X., Fu, Y., Zhu, K., Niu, Z., et al. (2021). Upregulation of LINC00511 expression by DNA hypomethylation promotes the progression of breast cancer. Gland. Surg. 10 (4), 1418–1430. doi:10.21037/gs-21-84
Liu, L., Zhu, Y., Liu, A. M., Feng, Y., and Chen, Y. (2019). Long noncoding RNA LINC00511 involves in breast cancer recurrence and radioresistance by regulating STXBP4 expression via miR-185. Eur. Rev. Med. Pharmacol. Sci. 23 (17), 7457–7468. doi:10.26355/eurrev_201909_18855
Liu, Z., Tao, B., Li, L., Liu, P., Xia, K., and Zhong, C. (2021). LINC00511 knockdown suppresses glioma cell malignant progression through miR-15a-5p/AEBP1 axis. Brain Res. Bull. 173, 82–96. doi:10.1016/j.brainresbull.2021.05.010
Lu, G., Li, Y., Ma, Y., Lu, J., Chen, Y., Jiang, Q., et al. (2018). Long noncoding RNA LINC00511 contributes to breast cancer tumourigenesis and stemness by inducing the miR-185-3p/E2F1/Nanog axis. J. Exp. Clin. Cancer Res. 37 (1), 289. doi:10.1186/s13046-018-0945-6
Lu, M., Gao, Q., Wang, Y., Ren, J., and Zhang, T. (2022). LINC00511 promotes cervical cancer progression by regulating the miR-497-5p/MAPK1 axis. Apoptosis 27, 800–811. doi:10.1007/s10495-022-01768-3
Lu, Y., Yu, Y., Liu, F., Han, Y., Xue, H., Sun, X., et al. (2021). LINC00511-dependent inhibition of IL-24 contributes to the oncogenic role of HNF4α in colorectal cancer. Am. J. Physiol. Gastrointest. Liver Physiol. 320 (3), G338–G350. doi:10.1152/ajpgi.00243.2020
Mahmoud, M. M., Sanad, E. F., Elshimy, R. A. A., and Hamdy, N. M. (2021). Competitive endogenous role of the linc00511/miR-185-3p Axis and miR-301a-3p from liquid biopsy as molecular markers for breast cancer diagnosis. Front. Oncol. 11, 749753. doi:10.3389/fonc.2021.749753
Mao, B. D., Xu, P., Xu, P., Zhong, Y., Ding, W. W., and Meng, Q. Z. (2019). LINC00511 knockdown prevents cervical cancer cell proliferation and reduces resistance to paclitaxel. J. Biosci. 44 (2), 44. doi:10.1007/s12038-019-9851-0
Melé, M., Mattioli, K., Mallard, W., Shechner, D. M., Gerhardinger, C., and Rinn, J. L. (2017). Chromatin environment, transcriptional regulation, and splicing distinguish lincRNAs and mRNAs. Genome Res. 27 (1), 27–37. doi:10.1101/gr.214205.116
Peng, X., Li, X., Yang, S., Huang, M., Wei, S., Ma, Y., et al. (2021). LINC00511 drives invasive behavior in hepatocellular carcinoma by regulating exosome secretion and invadopodia formation. J. Exp. Clin. Cancer Res. 40 (1), 183. doi:10.1186/s13046-021-01990-y
Qian, X., Jiang, C., Zhu, Z., Han, G., Xu, N., Ye, J., et al. (2022). Long non-coding RNA LINC00511 facilitates colon cancer development through regulating microRNA-625-5p to target WEE1. Cell Death Discov. 8 (1), 233. doi:10.1038/s41420-021-00790-9
Qiao, S., Qi, K., Liu, C., Xu, C., Ma, J., Xu, X., et al. (2020). Long intergenic non-coding RNA 511 correlates with improved prognosis, and hinders osteosarcoma progression both in vitro and in vivo. J. Clin. Lab. Anal. 34 (5), e23164. doi:10.1002/jcla.23164
Rouillard, A. D., Gundersen, G. W., Fernandez, N. F., Wang, Z., Monteiro, C. D., McDermott, M. G., et al. (2016). The harmonizome: A collection of processed datasets gathered to serve and mine knowledge about genes and proteins. Database (Oxford) 2016, baw100. doi:10.1093/database/baw100
Shi, G., Cheng, Y., Zhang, Y., Guo, R., Li, S., and Hong, X. (2021). Long non-coding RNA LINC00511/miR-150/MMP13 axis promotes breast cancer proliferation, migration and invasion. Biochim. Biophys. Acta Mol. Basis Dis. 1867 (3), 165957. doi:10.1016/j.bbadis.2020.165957
Shi, Y., Liu, M., Huang, Y., Zhang, J., and Yin, L. (2020). Promotion of cell autophagy and apoptosis in cervical cancer by inhibition of long noncoding RNA LINC00511 via transcription factor RXRA-regulated PLD1. J. Cell Physiol. 235 (10), 6592–6604. doi:10.1002/jcp.29529
Statello, L., Guo, C-J., Chen, L-L., and Huarte, M. (2021). Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 22 (2), 96–118. doi:10.1038/s41580-020-00315-9
Sun, C. B., Wang, H. Y., Han, X. Q., Liu, Y. N., Wang, M. C., Zhang, H. X., et al. (2020). LINC00511 promotes gastric cancer cell growth by acting as a ceRNA. World J. Gastrointest. Oncol. 12 (4), 394–404. doi:10.4251/wjgo.v12.i4.394
Sun, C. C., Li, S. J., Li, G., Hua, R. X., Zhou, X. H., and Li, D. J. (2016). Long intergenic noncoding RNA 00511 acts as an oncogene in non-small-cell lung cancer by binding to EZH2 and suppressing p57. Mol. Ther. Nucleic Acids 5 (11), e385. doi:10.1038/mtna.2016.94
Sun, K., Chen, R. X., Li, J. Z., and Luo, Z. X. (2022). LINC00511/hsa-miR-573 axis-mediated high expression of Gasdermin C associates with dismal prognosis and tumor immune infiltration of breast cancer. Sci. Rep. 12 (1), 14788. doi:10.1038/s41598-022-19247-9
Sun, S., Xia, C., and Xu, Y. (2020). HIF-1α induced lncRNA LINC00511 accelerates the colorectal cancer proliferation through positive feedback loop. Biomed. Pharmacother. 125, 110014. doi:10.1016/j.biopha.2020.110014
Wang, D., Liu, K., and Chen, E. (2020). LINC00511 promotes proliferation and invasion by sponging miR-515-5p in gastric cancer. Cell Mol. Biol. Lett. 25, 4. doi:10.1186/s11658-020-0201-x
Wang, K., Zhu, G., Bao, S., and Chen, S. (2019). Long non-coding RNA LINC00511 mediates the effects of ESR1 on proliferation and invasion of ovarian cancer through miR-424-5p and miR-370-5p. Cancer Manag. Res. 11, 10807–10819. doi:10.2147/CMAR.S232140
Wang, Q., Mao, X., Luo, F., and Wang, J. (2021). LINC00511 promotes gastric cancer progression by regulating SOX4 and epigenetically repressing PTEN to activate PI3K/AKT pathway. J. Cell Mol. Med. 25 (19), 9112–9127. doi:10.1111/jcmm.16656
Wang, R. P., Jiang, J., Jiang, T., Wang, Y., and Chen, L. X. (2019). Increased long noncoding RNA LINC00511 is correlated with poor prognosis and contributes to cell proliferation and metastasis by modulating miR-424 in hepatocellular carcinoma. Eur. Rev. Med. Pharmacol. Sci. 23 (8), 3291–3301. doi:10.26355/eurrev_201904_17691
Wang, W. J., Wang, D., Zhao, M., Sun, X. J., Li, Y., Lin, H., et al. (2020). Serum lncRNAs (CCAT2, LINC01133, LINC00511) with squamous cell carcinoma antigen panel as novel non-invasive biomarkers for detection of cervical squamous carcinoma. Cancer Manag. Res. 12, 9495–9502. doi:10.2147/CMAR.S259586
Wang, Y., Mei, X., Song, W., Wang, C., and Qiu, X. (2022). LncRNA LINC00511 promotes COL1A1-mediated proliferation and metastasis by sponging miR-126-5p/miR-218-5p in lung adenocarcinoma. BMC Pulm. Med. 22 (1), 272. doi:10.1186/s12890-022-02070-3
Wu, Y., Li, L., Wang, Q., Zhang, L., He, C., Wang, X., et al. (2020). LINC00511 promotes lung squamous cell carcinoma proliferation and migration via inhibiting miR-150-5p and activating TADA1. Transl. Lung Cancer Res. 9 (4), 1138–1148. doi:10.21037/tlcr-19-701
Xiang, J., Guan, Y., Bhandari, A., Xia, E., Wen, J., and Wang, O. (2020). LINC00511 influences cellular proliferation through cyclin-dependent kinases in papillary thyroid carcinoma. J. Cancer 11 (2), 450–459. doi:10.7150/jca.35364
Xu, J., Chen, G., Zhang, Y., Huang, Z., Cheng, X., Gu, H., et al. (2020). LINC00511 promotes osteosarcoma tumorigenesis and invasiveness through the miR-185-3p/E2F1 Axis. Biomed. Res. Int. 2020, 1974506. doi:10.1155/2020/1974506
Xue, J., and Zhang, F. (2020). LncRNA LINC00511 plays an oncogenic role in lung adenocarcinoma by regulating PKM2 expression via sponging miR-625-5p. Thorac. Cancer 11 (9), 2570–2579. doi:10.1111/1759-7714.13576
Yan, L., Wu, X., Liu, Y., and Xian, W. (2018). LncRNA Linc00511 promotes osteosarcoma cell proliferation and migration through sponging miR-765. J. Cell Biochem. 120, 7248–7256. doi:10.1002/jcb.27999
Yang, F., Lyu, S., Dong, S., Liu, Y., Zhang, X., and Wang, O. (2016). Expression profile analysis of long noncoding RNA in HER-2-enriched subtype breast cancer by next-generation sequencing and bioinformatics. Onco Targets Ther. 9, 761–772. doi:10.2147/OTT.S97664
Yu, C. L., Xu, X. L., and Yuan, F. (2019). LINC00511 is associated with the malignant status and promotes cell proliferation and motility in cervical cancer. Biosci. Rep. 39 (9). doi:10.1042/BSR20190903
Zhang, J., Sui, S., Wu, H., Zhang, J., Zhang, X., Xu, S., et al. (2019). The transcriptional landscape of lncRNAs reveals the oncogenic function of LINC00511 in ER-negative breast cancer. Cell Death Dis. 10 (8), 599–602. doi:10.1038/s41419-019-1835-3
Zhang, X., Wang, Y., Zhao, A., Kong, F., Jiang, L., and Wang, J. (2020). Long non-coding RNA LINC00511 accelerates proliferation and invasion in cervical cancer through targeting miR-324-5p/DRAM1 Axis. Onco Targets Ther. 13, 10245–10256. doi:10.2147/OTT.S255067
Zhang, Y., Xiao, P., and Hu, X. (2022). LINC00511 enhances LUAD malignancy by upregulating GCNT3 via miR-195-5p. BMC Cancer 22 (1), 389. doi:10.1186/s12885-022-09459-7
Zhao, X., Liu, Y., Li, Z., Zheng, S., Wang, Z., Li, W., et al. (2018). Linc00511 acts as a competing endogenous RNA to regulate VEGFA expression through sponging hsa-miR-29b-3p in pancreatic ductal adenocarcinoma. J. Cell Mol. Med. 22 (1), 655–667. doi:10.1111/jcmm.13351
Zhao, Y., Chen, X., Jiang, J., Wan, X., Wang, Y., and Xu, P. (2020). Epigallocatechin gallate reverses gastric cancer by regulating the long noncoding RNA LINC00511/miR-29b/KDM2A axis. Biochim. Biophys. Acta Mol. Basis Dis. 1866 (10), 165856. doi:10.1016/j.bbadis.2020.165856
Zhu, F. Y., Zhang, S. R., Wang, L. H., Wu, W. D., and Zhao, H. (2019). LINC00511 promotes the progression of non-small cell lung cancer through downregulating LATS2 and KLF2 by binding to EZH2 and LSD1. Eur. Rev. Med. Pharmacol. Sci. 23 (19), 8377–8390. doi:10.26355/eurrev_201910_19149
Keywords: LINC00511, cancer, biomarker, expression, diagnostic
Citation: Ghafouri-Fard S, Safarzadeh A, Hussen BM, Taheri M and Ayatollahi SA (2023) A review on the role of LINC00511 in cancer. Front. Genet. 14:1116445. doi: 10.3389/fgene.2023.1116445
Received: 05 December 2022; Accepted: 05 April 2023;
Published: 14 April 2023.
Edited by:
Yujing Li, Emory University, United StatesCopyright © 2023 Ghafouri-Fard, Safarzadeh, Hussen, Taheri and Ayatollahi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mohammad Taheri, mohammad.taheri@uni-jena.de; Seyed Abdulmajid Ayatollahi, majid_ayatollahi@yahoo.com
 Soudeh Ghafouri-Fard
Soudeh Ghafouri-Fard Arash Safarzadeh
Arash Safarzadeh Bashdar Mahmud Hussen
Bashdar Mahmud Hussen Mohammad Taheri
Mohammad Taheri Seyed Abdulmajid Ayatollahi6*
Seyed Abdulmajid Ayatollahi6*