- 1School of Public Health, University of South China, Hengyang, China
- 2Department of Radiation Biology, Beijing Key Laboratory for Radiobiology, Beijing Institute of Radiation Medicine, Beijing, China
- 3Department of Cellular and Molecular Biology, Beijing Chest Hospital, Capital Medical University/Beijing Tuberculosis and Thoracic Tumor Research Institute, Beijing, China
DNA-dependent protein kinase catalytic subunit (DNA-PKcs) is a member of the phosphatidylinositol 3-kinase related kinase family, which can phosphorylate more than 700 substrates. As the core enzyme, DNA-PKcs forms the active DNA-PK holoenzyme with the Ku80/Ku70 heterodimer to play crucial roles in cellular DNA damage response (DDR). Once DNA double strand breaks (DSBs) occur in the cells, DNA-PKcs is promptly recruited into damage sites and activated. DNA-PKcs is auto-phosphorylated and phosphorylated by Ataxia-Telangiectasia Mutated at multiple sites, and phosphorylates other targets, participating in a series of DDR and repair processes, which determine the cells’ fates: DSBs NHEJ repair and pathway choice, replication stress response, cell cycle checkpoints, telomeres length maintenance, senescence, autophagy, etc. Due to the special and multi-faceted roles of DNA-PKcs in the cellular responses to DNA damage, it is important to precisely regulate the formation and dynamic of its functional complex and activities for guarding genomic stability. On the other hand, targeting DNA-PKcs has been considered as a promising strategy of exploring novel radiosensitizers and killing agents of cancer cells. Combining DNA-PKcs inhibitors with radiotherapy can effectively enhance the efficacy of radiotherapy, offering more possibilities for cancer therapy.
Introduction
Eukaryotic cells are constantly encountering various endogenous (e.g., DNA replication errors) and exogenous (ionizing radiation, chemical carcinogens, and UV) stresses of genomic DNA damage (Jeggo et al., 2016). DNA damage includes base damage, pyrimidine dimer formation, single or double strand breaks, etc. Among them, DNA double strand break (DSB) is the most serious and dangerous threat that affects the stability of the genome and cell fate (Jackson and Bartek, 2009; Huang and Zhou, 2020). Failure to make timely and precise repairs can lead to accumulation of residual DNA damage, mutations, rearrangements, and/or loss of chromosomes, which can lead to a series of cellular consequences such as cell death, senescence, transformation, mutagenesis, or carcinogenesis (Her and Bunting, 2018). Fortunately, cells have evolved several precise DNA damage response (DDR) and repair machineries to deal with various types of DNA damages. DDRs include DNA damage sensing, initiating DNA damage signaling cascades, remodeling and relaxing chromatin around DSBs, recruiting DNA repair proteins to the damaged site, activating cell cycle checkpoints, and repairing DSB (Jackson and Bartek, 2009; Huang and Zhou, 2020). DNA repair defects are closely related to a series of human diseases and aging (Vijg and Suh, 2013; Longerich et al., 2014; Madabhushi et al., 2014; Shimizu et al., 2014; Chow and Herrup, 2015; Konstantinopoulos et al., 2015; Fang et al., 2016; Uryga et al., 2016; Chatzidoukaki et al., 2020). The DSBs repair pathways of eukaryotic cells mainly include the non-homologous end joining (NHEJ), homologous recombination (HR) and alternative end-joining (Lieber, 2008; Chiruvella et al., 2013; Chang et al., 2017; Shibata and Jeggo, 2020).
DNA-dependent protein kinase (DNA-PK) is an important player in the NHEJ pathway and was also found to function in multiple nodes of DDRs. DNA-activated/or DNA-dependent protein kinase was first reported in 1985 by Walker et al. (1985). They discovered this protein by chance, finding that addition of double-stranded DNA (dsDNA) into the cell extracts increased the phosphorylation of certain proteins. In 1990, Carter et al. (1990) and Lees-Miller et al. (1990) identified the DNA-PK catalytic subunit (DNA-PKcs) with hsp90 or casein as phosphorylation substrate bait. DNA-PKcs is an abundant protein with 50,000–100,000 molecules per cell in humans (Anderson and Lees-Miller, 1992; Blunt et al., 1995; Miller et al., 1995; van der Burg et al., 2009; Woodbine et al., 2013; Chang and Lieber, 2016). PRKDC gene mutations in patients or expression of kinase-dead DNA-PKcs protein in mice causes severe combined immunodeficiency (SCID). DNA-PKcs, Ataxia-Telangiectasia Mutated (ATM), and ATM and RAD3 related (ATR) belong to the family of the phosphatidylinositol 3-kinase related kinase (PIKK), which plays significant roles in DNA damage repair (Blackford and Jackson, 2017; Kantidze et al., 2018). They all harbor similar domain compositions, including a N-terminal HEAT-repeat rich segment followed by the conserved FRAP-ATM-TRRAP (FAT) domain, the kinase domain, the PIKK regulatory domain (PRD) and the FAT C-terminal motif (FATC; Blackford and Jackson, 2017). DNA-PKcs, ATM, and ATR preferentially phosphorylate the S/T-Q motif (serine or threonine residue followed by a glutamine) (Kim et al., 1999). DNA-PKcs and ATM mainly mediate the repair of DNA double-strand breaks through NHEJ and HR, respectively, while ATR responds to the stalled DNA replication forks and DNA single-strand breaks (Falck et al., 2005; Menolfi and Zha, 2020). Whereas, a series of reports indicated that DNA-PKcs is also required for optimal replication stress response (Lin et al., 2014, 2018; Wang et al., 2015; Ying et al., 2016; Kantidze et al., 2018; Joshi et al., 2019; Villafanez et al., 2019).
This review mainly summarized and discussed the updated accomplishments about DNA-PKcs research, including post-translation modifications and activity regulation of DNA-PKcs and its involvements in DDRs. Furthermore, considering the special role of DNA-PKcs in the DDR, we have also reviewed the progress on exploration of DNA-PKcs inhibitors as radiosensitizers for cancer radiotherapy.
Post-translational Modifications and Activation of DNA-PKcs Triggered by DNA Damage Signaling
DNA-dependent protein kinase catalytic subunit belongs to the PIKKs family, and is a type of DNA-activated serine/threonine protein kinase with a molecular weight of approximately 469 kD encoded by the PRKDC or XRCC7 gene (Hartley et al., 1995; Sipley et al., 1995; Lees-Miller, 1996). It forms a holoenzyme DNA-dependent kinase (DNA-PK) with the heterodimer regulatory subunits of Ku70 and Ku80(referred to as Ku together) (Jin and Weaver, 1997; Singleton et al., 1999). Ku70 is encoded by XRCC6 gene and Ku80 is encoded by XRCC5. Ku70 and Ku80 have a strong affinity for DNA ends, and they also provide docking sites for other proteins during DDR (Featherstone and Jackson, 1999).
The DNA-PKcs structure consists of an N-terminal region, a circular cradle unit, and a head unit with the kinase domain between FAT (FRAP, ATM, and TRRAP) and FATC domains (Sibanda et al., 2010, 2017; Sharif et al., 2017; Yin et al., 2017). Both the N-terminal region and the cradle unit contain HEAT (Huntingtin, Elongation factor 3, regulatory subunit A of PP2A, and TOR1) repeats. Both the ABCDE and PQR phosphorylation clusters fall into the cradle unit (Figure 1; Spagnolo et al., 2006; Sibanda et al., 2010; Morris et al., 2011).
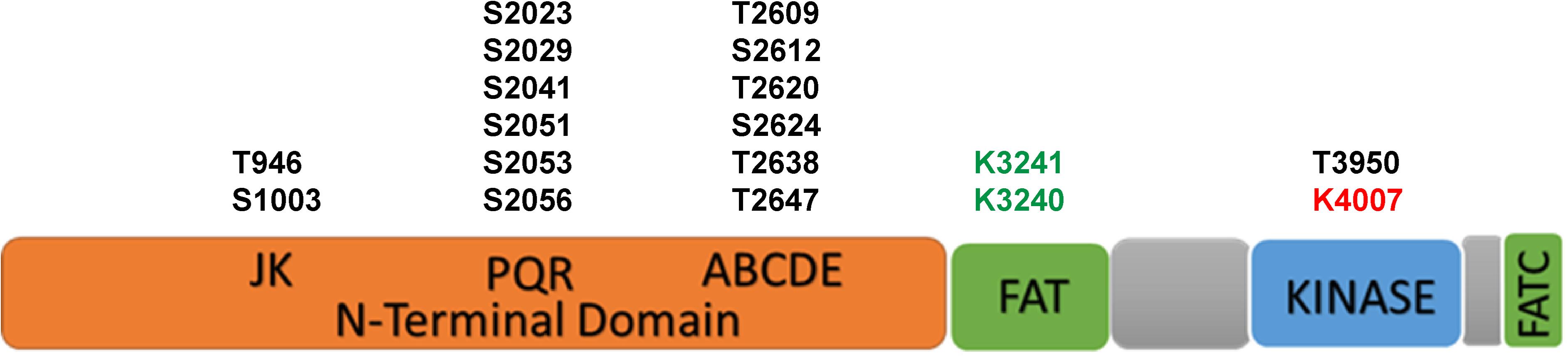
Figure 1. The scheme of functional domains and post-translational modifications of DNA-PKcs. The black font represents phosphorylation sites, green font represents acetylation sites and red font represents the neddylation site.
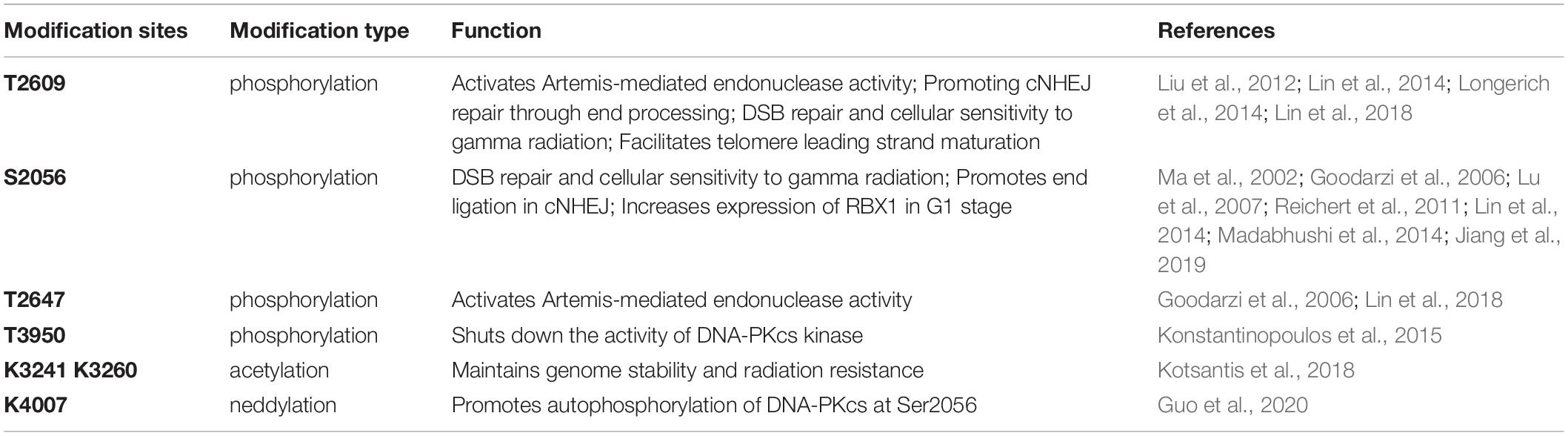
The activity and function of DNA-PKcs in end-ligation or NHEJ aretightly regulated by phosphorylation modification. The N-terminal domain has many phosphorylation sites, DNA-PKcs can be auto-phosphorylated or phosphorylated by ATM and ATR (Chen et al., 2007; Dobbs et al., 2010). The FAT and FATC domains surround the catalytic domain, stabilize the conformational changes of the catalytic center and regulate kinase activity (Rivera-Calzada et al., 2005; Jiang et al., 2006; Spagnolo et al., 2006). As shown in Figure 1, the phosphorylation sites of ABCDE clusters are T2609, S2612, T2620, S2624, T2638, and T2647, and the phosphorylation sites of PQR clusters are S2023, S2029, S2041, S2051, S2053, and S2056 (Douglas et al., 2002). Among them, S2056 and T2609 are two prominent autophosphorylation sites of DNA-PKcs, both are crucial for its activity in DNA repair (Ding et al., 2003; Block et al., 2004; Nagasawa et al., 2011, 2017). The current model suggests that DNA-PKcs S2056 phosphorylation causes conformational changes, thereby promoting DNA-PK disassembly from the DSB site, allowing DNA end ligation (Jiang et al., 2015), while the ABCDE cluster phosphorylation is required for DNA end resection (Shibata et al., 2011). Another autophosphorylation site, T3950, is located in the kinase domain and its phosphorylation was suggested to shut down the activity of DNA-PKcs kinase (Douglas et al., 2007).
In addition to phosphorylation modification, there are also other forms of post-translational modifications on DNA-PKcs (Table 1). Proteomics studies have revealed that DNA-PKcs are widely acetylated (Mori et al., 2016) and ubiquitinated (Ho et al., 2014). Lysine acetylation is an important form of post-translational modifications and is also the most widely studied post-translational modification on histones. Proteomics showed that at least 16 lysine residues were acetylated in the DNA-PKcs. Mori et al. (2016) selected eight of them for further research and have confirmed that K3241 and K3260 were acetylated on DNA-PKcs. In addition, our research group also found that both PARylation (Han et al., 2019) and neddylation (Guo et al., 2020) can regulate DNA-PK activity, a neddylation site was identified at K4007 within the kinase domain. PARP1-dependent DNA-PKcs PARylation can be induced by DNA damage signals and affects DNA-PKcs Ser2056 phosphorylation in cells (Han et al., 2019). DNA-PKcs neddylation occurs in the kinase domain and is catalyzed by HUWE1 ligase (Guo et al., 2020). In 2014, the ring finger protein 144A (RNF144A) was discovered as the first E3 ubiquitin ligase of cytoplasmic DNA-PKcs. Ho et al. (2014) RNF144A is induced in a p53-dependent manner during DNA damage and targets cytoplasmic DNA-PKcs for ubiquitination and degradation.
Considering the close relationship between DNA-PKcs and DSB, it is particularly important to determine the mechanism controlling DNA-PKcs activation. In the cellular response to DSB, Ku heterodimer recognizes and localizes DNA damage sites, and promptly recruits DNA-PKcs. Once DNA-PKcs is recruited, it is activated through phosphorylation in a DNA-dependent manner. The C-terminal 178 amino acid residues of Ku80 is required for Ku80/DNA-PKcs interaction and indispensable for DNA-PKcs activation (Singleton et al., 1999). DNA-PKcs pushes Ku protein inwards onto the DNA, and then phosphorylates the other components nearby, including its own phosphorylation. DNA-PKcs autophosphorylation can be regulated by N-terminal conformational changes of protein (Meek et al., 2012). In addition to the automatic regulation of DNA-PKcs, many other factors are also suggested to be key regulators of DNA-PKcs activity. For example, epidermal growth factor (EGFR) can bind DNA-PKcs and enhance the activity of DNA-PKcs to deal with the damage (Liccardi et al., 2011; Javvadi et al., 2012). The non-kinase regulator protein phosphatase 6 (PP6) and protein phosphatase 1(PP1) is recruited to the DSB site and promotes DNA-PKcs activity through direct interaction and dephosphorylation action (Douglas et al., 2010; Zhu et al., 2017). Thus, the activation of DNA-PKcs is not only the result of binding to the broken DNA ends, but also a complex process affected by many factors. The multiple mechanistic ways of activation suggest that there are many unknown functions waiting to be explored in DNA-PKcs in addition to participating in double-strand break repair.
Regulation of DNA Damage Response by DNA-PKcs
Current knowledge displays that DNA-PKcs can function in multiple pathways of cellular DDRs to maintain the genome stability and cell survival.
DNA-PKcs in Non-homologous End Joining
There are two major repair pathways for DSB: NHEJ and HR (Figure 2). NHEJ repair is an error-prone repair pathway and theoretically executes its function throughout the cell cycle, but it is most important during G1 when no homologous template for recombination is available. HR performs under the guidance of an intact homologous template DNA, and it is known as an error-free repair pathway. HR is most active in the S/G2 phase and nearly absent in G1 phase (Mao et al., 2008; Menon and Povirk, 2016). As a main DSB repair pathway in mammals, NHEJ is initiated by the circular Ku70/Ku80 heterodimer binding to the broken DNA ends. Ku has a high abundance and strong affinity for free DNA. Within a second or less, Ku proteins can associate with any DSB that occurs in the nuclei genomic DNA. The interaction of Ku and broken DNA recruits DNA-PKcs to form DNA-PK complex (Gomes et al., 2017), then a set of NHEJ downstream factors including Artemis, XRCC4, DNA polymerase X family, and DNA ligase IV to join the broken DNA ends. DNA-PKcs is the only active protein kinase described in the NHEJ pathway, it is autophosphorylated in the presence of DNA termini, Mg2+ and ATP. DNA-PKcs forms a tight complex with Artemis to stimulate the nuclease activity of the latter (Kamoi and Mochizuki, 2010; Chang and Lieber, 2016). In this process, DNA-PKcs phosphorylates the C-terminal inhibitory region of Artemis, facilitating the dissociation of the inhibitory region from the N-terminal catalytic domain (Ma et al., 2002; Pannunzio et al., 2018). Ionizing radiation-induced DNA DSB broken ends are usually described as “dirty ends” and incompatibility for ends direct ligation caused by chemical modifications and 5′ or 3′ mismatching overhangs. The activated Artemis processes these “dirty ends” for subsequent ligation. DNA-PKcs kinase activity becomes necessary for direct end-ligation of NHEJ in the presence of DNA-PK protein. The DNA-PKcsKD/KD (kinase-dead mutant) cells show severe end-ligation defects (Jiang et al., 2015).
Usually, NHEJ takes the first action to respond to the DSB (Shibata et al., 2011). If NHEJ cannot be completed, then DSB “cuts,” where one strand of the DNA duplex is degraded to produce a single-stranded DNA overhang suitable for alternative repair pathways, HR occurs (Sallmyr and Tomkinson, 2018). Experiments have shown that NHEJ is much faster than HR and occurs within 30 min (while HR requires 7 h or more), accounting for approximately 75% of repair events (Zhang and Matlashewski, 2019). NHEJ repair does not use sequence homology, regardless of the position or sequence, the DNA break ends are brought together, so this type of repair is prone to errors.
Regulation of DNA-PKcs on the Pathway Choice of DSBs Repair
Homologous recombination repair is initiated by DSB end resection. MRE11 and CtIP together with EXO1 fulfill the DNA end resection process (Syed and Tainer, 2018). It is usually acknowledged that the interaction between 53BP1 and BRCA1 is an important factor to control the end resection (Daley and Sung, 2014; Zimmermann and de Lange, 2014; Mirman and de Lange, 2020). BRCA1 antagonizes certain functions of 53BP1 and promotes HR by reducing 53BP1-mediated NHEJ. After end resection, HR uses the undamaged homologous DNA as a template, and recombinase RAD51 invades the homologous DNA strand, resulting in precise repair (Sun et al., 2020).
How cells select the pathway to execute DSBs repair between HR and NHEJ is a critical issue for efficient and precise repair of DSBs, whereas its mechanism is not fully understood yet (Ceccaldi et al., 2016; Scully et al., 2019). The initiation of DNA end resection is a definite factor that enables the cells to perform HR repair to prevent NHEJ repair (Scully et al., 2019). There are many factors that affect the initiation of end resection, but DNA-PKcs phosphorylation status is a clear factor that impacts cells to choose NHEJ or HR (Neal et al., 2011). A series of reports indicated that DNA-PKcs can regulate the choice of DSBs repair pathways at multiple biochemical nodes in association with the cell cycle. In the G2 phase of cell cycle, autophosphorylation of DNA-PKcs promotes DNA-PKcs dissociation from the DSBs sites, and facilitates the recruitment of end resection enzymes such as EXO1 (Shibata et al., 2011; Sallmyr and Tomkinson, 2018). DNA-PKcs also promotes end-resection from in vitro analyses (Deshpande et al., 2020). The DNA-PKcs mutant that makes autophosphorylation defective at the ABCDE cluster (DNA-PKcs ABCDE 6A) binds DSBs but precludes the completion of NHEJ, significantly reducing DSB end resection at all DSBs (Shibata et al., 2011). Whereas in developing lymphocytes, robust end-resection is detected in both DNA-PKcs kinase-dead or the phosphorylation (ABCDE) mutant (Crowe et al., 2018, 2020). ATM kinase activity can compensate for DNA-PKcs autophosphorylation when DNA-PKcs activity is inhibited and promote resection. The Mre11-Rad50-Nbs1 (MRN) complex further stimulates resection in the presence of Ku and DNA-PKcs by recruiting EXO1 and enhancing DNA-PKcs autophosphorylation, and it also inhibits DNA ligase IV/XRCC4-mediated end rejoining (Zhou and Paull, 2013). In the S phase of cell cycle, BRCA1 interacts with DNA-PKcs and directly blocks DNA-PKcs autophosphorylation, thus priming DNA end resection for HR and HR factors loading on DSBs (Davis et al., 2014; Figure 2). The interaction of TIP60 histone acetyltransferase and DNA-PKcs prompts the autophosphorylation and activation of DNA-PKcs (Jiang et al., 2006). We recently revealed that an increased SUMO2 modification of TIP60 K430 mediated by PISA4 E3 ligase hinders its interaction with DNA-PKcs in S phase cells, leading to impediment of DNA-PKcs S2056 autophosphorylation and preferentially employing the HR pathway for DSBs repair in S phase (Figure 3). TIP60 K430R mutation can recover the interaction of DNA-PKcs and TIP60, resulting in abnormally increased phosphorylation of DNA-PKcs S2056 in S phase and dramatical inhibition of HR efficiency (Gao et al., 2020).
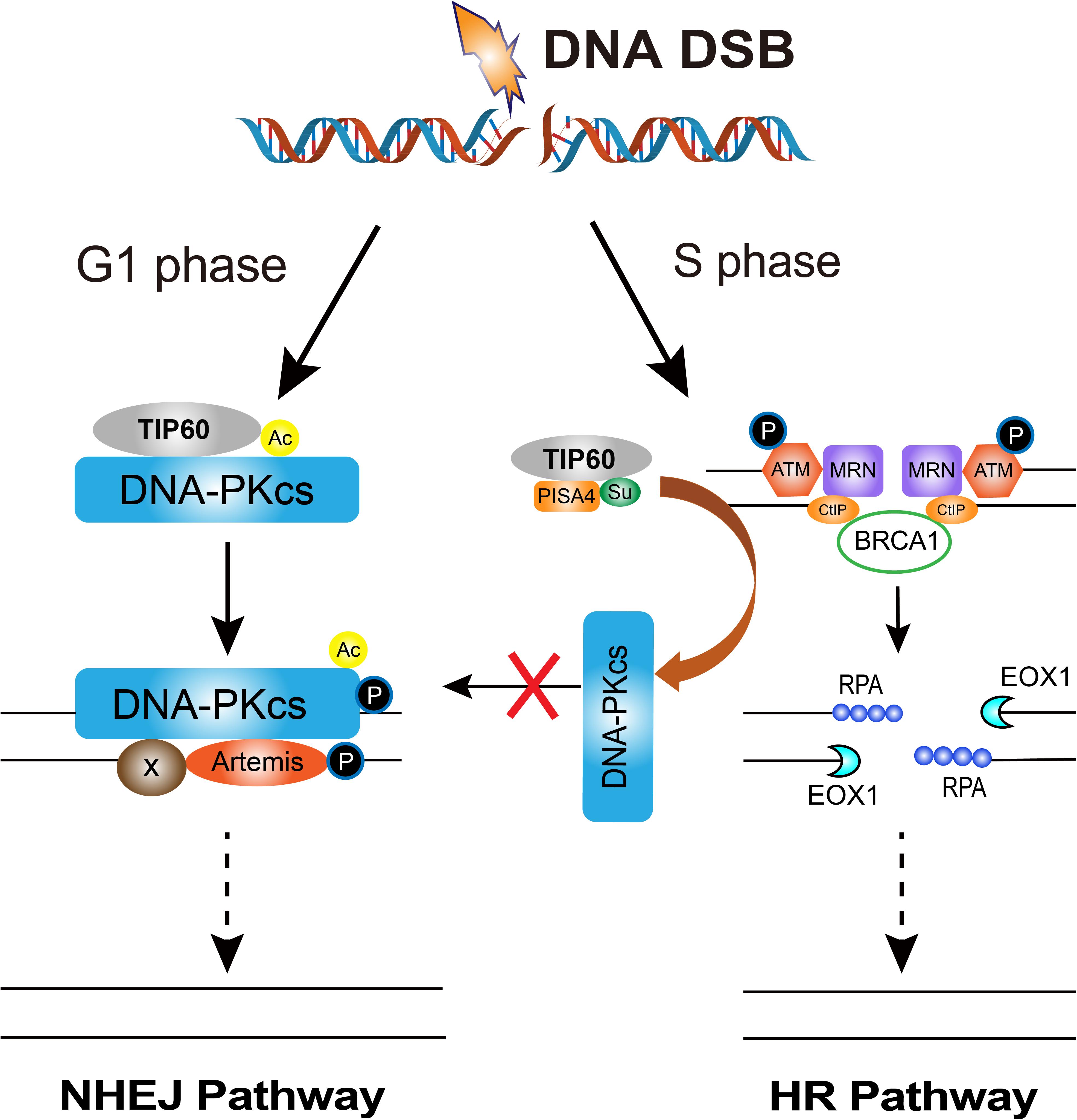
Figure 3. The regulation of DNA-PKcs-interacting TIP60 on the cell cycle-dependent choice of DNA DSBs repair pathway. Upon DNA DSBs signaling, TIP60 interacts with and acetylates DNA-PKcs to activate DNA-PKcs through autophosphorylation at S2056 for executing NHEJ in G1 phase. Whereas, in S phase cells, SUMO2 modification (SU) of TIP60 at K430 is mediated by PISA4 E3 ligase, which hinders the interaction between TIP60 and DNA-PKcs, consequently inactivating DNA-PKcs to give way to HR proteins and facilitates HR repair.
Meanwhile, HR activity in G1 is restricted. Zhou et al. (2017) found that DNA-PKcs phosphorylates ATM directly, inhibiting ATM activity and ATM signaling upon DNA damage, providing a possible mechanism of HR restriction in G1 phase. Moreover, our recent report demonstrated that EXO1 protein is strictly suppressed in G1 phase. Increased expression of RBX1 protein prompts the neddylation and activity of cullin1, a key component of the Skp1-Cullin1-F-box (SCF) ubiquitin 3 ligase, and consequently mediates the ubiquitination degradation of EXO1 in G1 phase. Increased DNA-PKcs activity is responsible for increased RBX1 protein expression, and limiting the formation of DSB-end ssDNA by EXO1 and suppressing the HR repair pathway in G1 cells(Figure 3; Xie et al., 2020).
DNA-PKcs in DNA Replication Stress
In addition to the role in DSB repair, DNA-PKcs is also involved in DNA replication stress response. Cells are particularly susceptible to DNA damage during the process of DNA replication. Almost all forms of DNA damage can disturb the DNA replication and cause replication stress (Bass et al., 2012).
Replication stress is defined as any DNA replication barrier that hinders, prevents or terminates DNA synthesis, and can activate the replication stress response to resolve the damage (Ubhi and Brown, 2019). Replicating stress checkpoint (S-phase checkpoint) involves the gradual activation of damage sensors, mediators and effectors (Allen et al., 2011). Replication protein A (RPA) binds to single-stranded DNA (ssDNA), recruiting numerous sensor proteins including ataxia telangiectasia and Rad3-related (ATR)-interacting protein (ATRIP), the 9-1-1 DNA clamp complex (RAD9-RAD1-HUS1), topoisomerase II binding protein 1 (TOPBP1) and Ewing tumor-associated antigen 1 (ETAA1), etc., triggers ataxia telangiectasia and Rad3-related (ATR) and phosphorylation of RPA32 (a subunit of RPA), leading to Chk1 activation (Zeman and Cimprich, 2014; Kotsantis et al., 2018).
DNA-dependent protein kinase catalytic subunit is necessary for ATR-Chk1 signal transduction. Upon replication stress, the DNA-PKcs is phosphorylated by ATR at the stalled replication forks, leading to transcriptional activation of Claspin expression and Chk1-Claspin complex stability, which is required for the optimal activation of intra S-phase checkpoint (Lin et al., 2014). The apoptosis mediator p53-induced protein with a death domain (PIDD) mediates DNA-PKcs recruitment at the stalled replication forks, and promotes the ATR signaling pathway in the cellular response to replication pressure and the cellular resistance to replication pressure (Lin et al., 2018). Phosphorylation of RPA32 Ser4/Ser8 is a key early step to fully activate ATR in response to replication stress and subsequent replication checkpoint stagnation. DNA-PK is the main kinase that targets Ser4/Ser8 in replication stress. The PIKK phosphorylation of RPA32 plays a key role in replication checkpoint activation, and DNA-PK was considered as an important contributor to this response (Liu et al., 2012). In addition, it was reported that ATR was critical in the early S phase, especially in cells under high replication stress; however, ATR can be circumvented by DNA-PKcs and Chk1 under moderate replication stress. They concluded distinct but concerted roles of ATR, DNA-PK, and Chk1 in countering replication stress (Buisson et al., 2015).
DNA-PKcs in Autophagy
Autophagy is a physiological degradation mechanism of cells through which autophagic vesicles can deliver unfolded proteins and damaged organelles to lysosomes to eliminate them (Leidal et al., 2018). Autophagy plays an important role in different DDR pathways (Hewitt and Korolchuk, 2017). Inhibition of DNA-PKcs by bromovanin, a vanillin derivative, exhibited a potent antiproliferation effect through induction of both apoptosis and autophagy in HepG2 cells (Yan et al., 2007). Daido et al. (2005) reported that relative low-dose IR induced massive autophagic cell death in M059J cells that lack DNA-PKcs. The treatment of M059K cells with DNA-PKcs antisense oligonucleotides caused radiation-induced autophagy and radiosensitized the cells. Similar observations were also obtained by Puustinen et al. (2020) study. Inactivation or depression of DNA-PKcs increases ionizing radiation-induced cell autophagy as well as occurrence of cell apoptosis. It was found that DNA-PKcs interacts with the AMP-dependent protein kinase (AMPK) complex and phosphorylates nucleotide-sensing γ1 subunit (protein kinase AMP-activated non-catalytic subunit gamma) PRKAG1/AMPKγ1 at Ser192 and Thr284, thereby facilitating the AMPK complex activation by STK11 at lysosomal and autophagy.
The Association of DNA-PKcs With Telomeres
Telomeres are DNA–protein complexes located at the ends of chromosomes in eukaryotic cells. The major function of telomeres is to maintain the stability and integrity of chromosomes (Roake and Artandi, 2020). The telomere sequence has unique characteristics, making telomeres susceptible to various DNA damages induced by the external environment and genotoxins (Lazzerini-Denchi and Sfeir, 2016). Any abnormality in telomere function may lead to cell aging/senescence and canceration. Telomeres can sense both intrinsic and extrinsic stresses and its dysfunction drives cell senescence (Victorelli and Passos, 2017). DNA-PKcs plays a role in telomere maintenance and is necessary for telomere capping (Goytisolo et al., 2001; Zhang et al., 2016; Sui et al., 2020). Sui et al. (2015) reported that hTR-mediated DNA-PKcs stimulation and subsequent hnRNP A1 phosphorylation affect the cell cycle-dependent distribution of telomeric repeat-containing RNA (TERRA) on telomeres by promoting the removal of TERRA in telomeres, which is important for the S-phase process, thereby promoting efficient telomere replication and capping (Ting et al., 2009). In cells lacking DNA-PKcs, uncapped telomeres are inappropriately detected and processed as DSB, and therefore not only participate in spontaneous telomere-telomere fusion, but also participate in ionizing radiation induction Telomere-DSB fusion event. The association between accelerated telomere shortening and decreased expression of DDR genes, including DNA-PKcs, Mre11, Xrcc4, etc, was found in the accelerated thymic aging of a rat model of developmental programming (Tarry-Adkins et al., 2019), suggesting DNA-PKcs may affect aging by maintaining telomeres integrity.
DNA-PKcs Functions in Cell Cycle Checkpoints
When DDR occurs, the PIKK kinases ATM, ATR and DNA-PKcs coordinately play the role of maintaining genome integrity (Blackford and Jackson, 2017). These kinases control the repair of the broken DNA ends and transmit the damage signal through the tumor suppressor p53, CHK1/CHK2 and so on to induce cell cycle arrest, apoptosis, or aging. In this process, DNA-PKcs works by affecting G2/M DNA damage checkpoint in ATM deficient cells (Lee et al., 1997; Arlander et al., 2008; Tomimatsu et al., 2009; Shang et al., 2010). In ATM knockdown human mammary epithelial cells, either DNA-PK inhibitor treatment or RNAi knockdown of DNA-PKcs significantly attenuated G2 checkpoint (Arlander et al., 2008). In ATM-deficient AT5BIVA cells, DNA-PKcs inhibition led to a prolonged G2/M arrest (Shang et al., 2010). This phenotype could also be an indirect effect of no repair due to DNA-PKcs inhibitory role in ATM phosphorylation in DDR (Zhou et al., 2017). Inactivation of DNA-PKcs strikingly attenuated the ionizing radiation-induced phosphorylation of Chk1 or Chk2/T68 in ATM-deficient cells.
Ataxia-Telangiectasia Mutated, ATR, and DNA-PKcs can directly phosphorylate p53. ATM and ATR phosphorylate p53 through checkpoint kinases 2 (CHK2) and checkpoint kinases 1 (CHK1), respectively (Roos and Kaina, 2006). p53 stability is mainly regulated by oncoprotein MDM2, a key negative regulator of p53 (Haupt et al., 1997; Kubbutat et al., 1997). Under non-stress conditions, MDM2 and p53 N-terminal domain form a stable complex. MDM2-mediated proteasome degradation keeps p53 at a low level.
Under DNA damage stresses, p53 and MDM2 dissociate and p53 accumulates in the nucleus. p53 functions as a transcription factor to promote the expression of genes involved in DNA repair, cell cycle arrest, apoptosis, and aging to maintain genome integrity. Achanta et al. (2001) found that the DNA-PKcs/Ku/p53 complex may act as a sensor for DNA strand breaks caused by the incorporation of drug molecules, and then transduce signals to trigger apoptosis (Wang et al., 2000). Boehme et al. (2008) found that the activation of DNA-PKcs led to the phosphorylation and activation of Akt/PKB, which subsequently led to the inactivation of GSK-3β. As a result, MDM2 is phosphorylated and p53 accumulation increases. In this regard, DNA-PKcs modulates the p53-dependent apoptosis after DNA damage. At the same time, there are two negative feedback loops between p53 and MDM2 and wild-type p53-induced phosphatase 1 (Wip1) to regulate the level of p53 protein (Lu et al., 2007). Wip1 de-phosphorylates MDM2 and downregulates p53 protein levels by stabilizing MDM2, facilitating its access to p53.
Targeting DNA-PKcs to Reprogram the Cellular Radiosensitivity
In the past century, radiation therapy has become the main treatment for cancer (Baskar et al., 2012). Statistically, about 50–70% of malignant tumor patients in the world are receiving radiotherapy (Huang and Zhou, 2020). However, the existence of radioresistance of cancer cells greatly hampers the efficacy and application of tumor radiotherapy (Kim et al., 2015). Radiation resistance is conducive to normal cells to escape the damage caused by ionizing radiation, but it is not conducive to the radiotherapy of malignant tumors.
The ability of cancer cells to repair DNA damage is a crucial factor that determines their sensitivity to radiation therapy or chemotherapy (Zhu et al., 2009). Inhibiting the repair ability of cancer cells offers a strategy to reduce the radiation resistance of cells and improve the efficacy of radiotherapy.
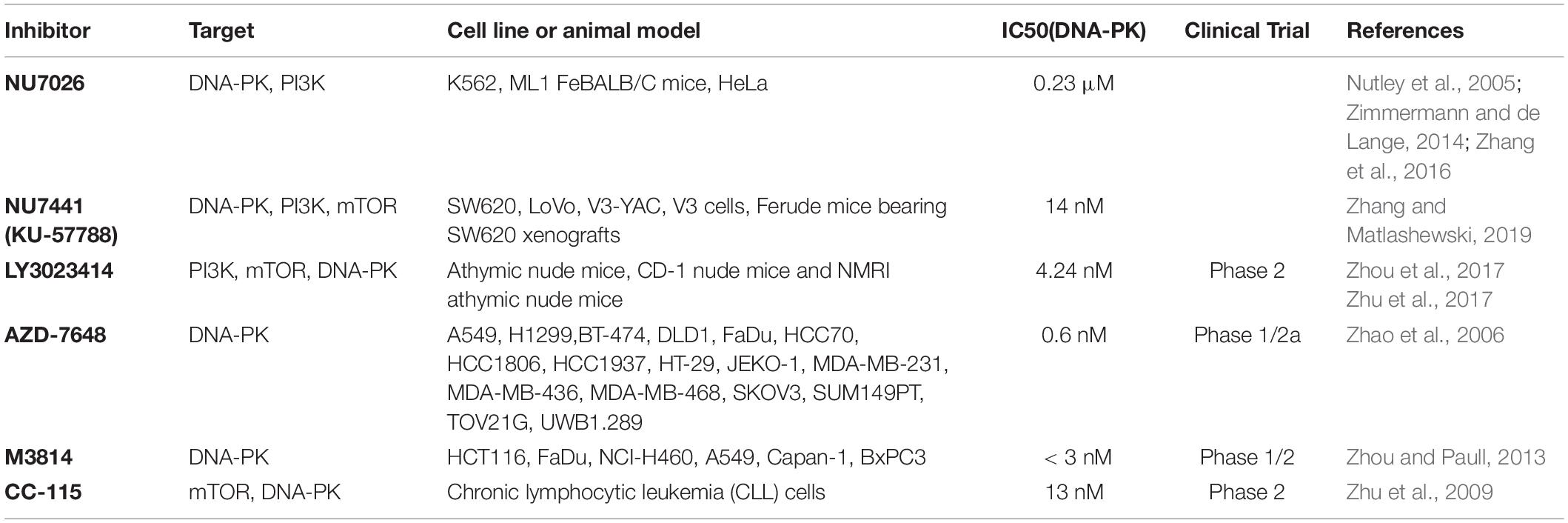
DNA-dependent protein kinase has become an attractive therapeutic target in various cancer treatments, especially when used in combination with genotoxic chemotherapy or ionizing radiation (Huang and Zhou, 2020; Medová et al., 2020). Studies have shown that targeting DNA-PKcs with various inhibitors can effectively enhance radiotherapy, and many small molecule inhibitors of DNA-PKcs have been developed and shown to be effective radiosensitizers in vitro (Table 2). One of the earliest identified inhibitors was wortmannin, which was isolated in 1957. It was defined as an effective non-competitive PI3K irreversible inhibitor in 1993, but it also targets other members of the PIKK family, including DNA-PKcs, ATM, ATR (Ui et al., 1995). The DNA-PK inhibitor NU7026 was reported to potentiate topo II poisons in treatment of leukemia through inhibition of NHEJ and G2/M checkpoint arrest (Willmore et al., 2004). In preclinical evaluation of DNA-PK inhibitor NU7441, it showed sufficient chemosensitization and radiosensitization with either etoposide, doxorubicin or ionizing radiation (Zhao et al., 2006). A highly selective DNA-PK inhibitor, AZD7648 showed efficient sensitization on radiation or doxorubicin in xenograft and patient-derived xenograft (PDX) models (Fok et al., 2019). M3814 inhibits DNA-PK catalytic activity and sensitizes various cancer cell lines to ionizing radiation (IR) and DSB inducers (Zenke et al., 2020). In addition, LY3023414 and CC-115 are both mTOR inhibitors, but they also have inhibitory effects on DNA-PKcs and other PI3KK (Smith et al., 2016; Thijssen et al., 2016; Bendell et al., 2018). It can be envisioned that the inhibition of DNA-PK shows considerable prospects in anti-tumor resistance. However, DNA-PK inhibitors are usually limited by poor pharmacokinetics: these compounds have poor solubility and unstable metabolism in the body, resulting in short serum half-life. Development of new compounds with better performance is a key step in obtaining effective anti-cancer drugs.
Future Perspectives
Over the decades, DNA-PKcs has been revealed to play multi-faceted roles and has been proved to be an essential regulator in the processes of DDR, however, there are still many unresolved questions. How does the interaction of DNA-PKcs and Ku-DNA complex leads to activation of DNA-PK protein kinase activity? There are a large number of sites and forms of post-translation modifications (PTMs) on DNA-PKcs, how these PTMs are inter-related and regulated, and how they influence DNA-PKcs functions. Are there any other DNA-PK substrates yet to be discovered? It is worth to draw the panorama of function-related DNA-PKcs interaction networks. The use of DNA-PK inhibitors for cancer therapy will not only inhibit the activity of DNA-PK in cancer cells, but also inhibit the activity of DNA-PK in normal cells. How to solve this problem? Ongoing exploration of DNA-PKcs’ multi-facet roles in DDR and besides will greatly contribute to our overall understanding of cancer and the discovery of new therapies.
Author Contributions
P-KZ conceived and designed this study. XY drafted the initial manuscript. CB and DX reviewed and commented the manuscript. TM and P-KZ critically revised and finalized the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from the National Natural Science Foundation of China (11705283 and 31870847).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Achanta, G., Pelicano, H., Feng, L., Plunkett, W., and Huang, P. (2001). Interaction of p53 and DNA-PK in response to nucleoside analogues: potential role as a sensor complex for DNA damage. Cancer Res. 61, 8723–8729.
Allen, C., Ashley, A. K., Hromas, R., and Nickoloff, J. A. (2011). More forks on the road to replication stress recovery. J. Mol. Cell Biol. 3, 4–12. doi: 10.1093/jmcb/mjq049
Anderson, C. W., and Lees-Miller, S. P. (1992). The nuclear serine/threonine protein kinase DNA-PK. Crit. Rev. Eukaryot. Gene Expr. 2, 283–314.
Arlander, S. J., Greene, B. T., Innes, C. L., and Paules, R. S. (2008). DNA protein kinase-dependent G2 checkpoint revealed following knockdown of ataxia-telangiectasia mutated in human mammary epithelial cells. Cancer Res. 68, 89–97. doi: 10.1158/0008-5472.Can-07-0675
Baskar, R., Lee, K. A., Yeo, R., and Yeoh, K. W. (2012). Cancer and radiation therapy: current advances and future directions. Int. J. Med. Sci. 9, 193–199. doi: 10.7150/ijms.3635
Bass, K. L., Murray, J. M., and O’Connell, M. J. (2012). Brc1-dependent recovery from replication stress. J. Cell Sci. 125(Pt 11), 2753–2764. doi: 10.1242/jcs.103119
Bendell, J. C., Varghese, A. M., Hyman, D. M., Bauer, T. M., Pant, S., Callies, S., et al. (2018). A first-in-human phase 1 study of LY3023414, an oral PI3K/mTOR dual inhibitor, in patients with advanced cancer. Clin. Cancer Res. 24, 3253–3262. doi: 10.1158/1078-0432.Ccr-17-3421
Blackford, A. N., and Jackson, S. P. (2017). ATM, ATR, and DNA-PK: the trinity at the heart of the DNA damage response. Mol. Cell 66, 801–817. doi: 10.1016/j.molcel.2017.05.015
Block, W. D., Yu, Y., Merkle, D., Gifford, J. L., Ding, Q., Meek, K., et al. (2004). Autophosphorylation-dependent remodeling of the DNA-dependent protein kinase catalytic subunit regulates ligation of DNA ends. Nucleic Acids Res. 32, 4351–4357. doi: 10.1093/nar/gkh761
Blunt, T., Finnie, N. J., Taccioli, G. E., Smith, G. C., Demengeot, J., Gottlieb, T. M., et al. (1995). Defective DNA-dependent protein kinase activity is linked to V(D)J recombination and DNA repair defects associated with the murine scid mutation. Cell 80, 813–823. doi: 10.1016/0092-8674(95)90360-7
Boehme, K. A., Kulikov, R., and Blattner, C. (2008). p53 stabilization in response to DNA damage requires Akt/PKB and DNA-PK. Proc. Natl. Acad. Sci. U.S.A. 105, 7785–7790. doi: 10.1073/pnas.0703423105
Buisson, R., Boisvert, J. L., Benes, C. H., and Zou, L. (2015). Distinct but concerted roles of ATR, DNA-PK, and Chk1 in countering replication STRESS during S phase. Mol. Cell 59, 1011–1024. doi: 10.1016/j.molcel.2015.07.029
Carter, T., Vancurová, I., Sun, I., Lou, W., and DeLeon, S. (1990). A DNA-activated protein kinase from HeLa cell nuclei. Mol. Cell. Biol. 10, 6460–6471. doi: 10.1128/mcb.10.12.6460
Ceccaldi, R., Rondinelli, B., and D’Andrea, A. D. (2016). Repair pathway choices and consequences at the double-strand break. Trends Cell Biol. 26, 52–64. doi: 10.1016/j.tcb.2015.07.009
Chang, H. H. Y., Pannunzio, N. R., Adachi, N., and Lieber, M. R. (2017). Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat. Rev. Mol. Cell Biol. 18, 495–506. doi: 10.1038/nrm.2017.48
Chang, H. H., and Lieber, M. R. (2016). Structure-specific nuclease activities of Artemis and the Artemis: DNA-PKcs complex. Nucleic Acids Res. 44, 4991–4997. doi: 10.1093/nar/gkw456
Chatzidoukaki, O., Goulielmaki, E., Schumacher, B., and Garinis, G. A. (2020). DNA damage response and metabolic reprogramming in health and disease. Trends Genet. 36, 777–791. doi: 10.1016/j.tig.2020.06.018
Chen, B. P., Uematsu, N., Kobayashi, J., Lerenthal, Y., Krempler, A., Yajima, H., et al. (2007). Ataxia telangiectasia mutated (ATM) is essential for DNA-PKcs phosphorylations at the Thr-2609 cluster upon DNA double strand break. J. Biol. Chem. 282, 6582–6587. doi: 10.1074/jbc.M611605200
Chiruvella, K. K., Liang, Z., and Wilson, T. E. (2013). Repair of double-strand breaks by end joining. Cold Spring Harb. Perspect. Biol. 5:a012757. doi: 10.1101/cshperspect.a012757
Chow, H. M., and Herrup, K. (2015). Genomic integrity and the ageing brain. Nat. Rev. Neurosci. 16, 672–684. doi: 10.1038/nrn4020
Crowe, J. L., Shao, Z., Wang, X. S., Wei, P. C., Jiang, W., Lee, B. J., et al. (2018). Kinase-dependent structural role of DNA-PKcs during immunoglobulin class switch recombination. Proc. Natl. Acad. Sci. U.S.A. 115, 8615–8620. doi: 10.1073/pnas.1808490115
Crowe, J. L., Wang, X. S., Shao, Z., Lee, B. J., Estes, V. M., and Zha, S. (2020). DNA-PKcs phosphorylation at the T2609 cluster alters the repair pathway choice during immunoglobulin class switch recombination. Proc. Natl. Acad. Sci. U.S.A. 117, 22953–22961. doi: 10.1073/pnas.2007455117
Daido, S., Yamamoto, A., Fujiwara, K., Sawaya, R., Kondo, S., and Kondo, Y. (2005). Inhibition of the DNA-dependent protein kinase catalytic subunit radiosensitizes malignant glioma cells by inducing autophagy. Cancer Res. 65, 4368–4375. doi: 10.1158/0008-5472.Can-04-4202
Daley, J. M., and Sung, P. (2014). 53BP1, BRCA1, and the choice between recombination and end joining at DNA double-strand breaks. Mol. Cell. Biol. 34, 1380–1388. doi: 10.1128/mcb.01639-13
Davis, A. J., Chi, L., So, S., Lee, K. J., Mori, E., Fattah, K., et al. (2014). BRCA1 modulates the autophosphorylation status of DNA-PKcs in S phase of the cell cycle. Nucleic Acids Res. 42, 11487–11501. doi: 10.1093/nar/gku824
Deshpande, R. A., Myler, L. R., Soniat, M. M., Makharashvili, N., Lee, L., Lees-Miller, S. P., et al. (2020). DNA-dependent protein kinase promotes DNA end processing by MRN and CtIP. Sci. Adv. 6:eaay0922. doi: 10.1126/sciadv.aay0922
Ding, Q., Reddy, Y. V., Wang, W., Woods, T., Douglas, P., Ramsden, D. A., et al. (2003). Autophosphorylation of the catalytic subunit of the DNA-dependent protein kinase is required for efficient end processing during DNA double-strand break repair. Mol. Cell. Biol. 23, 5836–5848. doi: 10.1128/mcb.23.16.5836-5848.2003
Dobbs, T. A., Tainer, J. A., and Lees-Miller, S. P. (2010). A structural model for regulation of NHEJ by DNA-PKcs autophosphorylation. DNA Repair (Amst.) 9, 1307–1314. doi: 10.1016/j.dnarep.2010.09.019
Douglas, P., Cui, X., Block, W. D., Yu, Y., Gupta, S., Ding, Q., et al. (2007). The DNA-dependent protein kinase catalytic subunit is phosphorylated in vivo on threonine 3950, a highly conserved amino acid in the protein kinase domain. Mol. Cell. Biol. 27, 1581–1591. doi: 10.1128/mcb.01962-06
Douglas, P., Sapkota, G. P., Morrice, N., Yu, Y., Goodarzi, A. A., Merkle, D., et al. (2002). Identification of in vitro and in vivo phosphorylation sites in the catalytic subunit of the DNA-dependent protein kinase. Biochem. J. 368(Pt 1), 243–251. doi: 10.1042/bj20020973
Douglas, P., Zhong, J., Ye, R., Moorhead, G. B., Xu, X., and Lees-Miller, S. P. (2010). Protein phosphatase 6 interacts with the DNA-dependent protein kinase catalytic subunit and dephosphorylates gamma-H2AX. Mol. Cell. Biol. 30, 1368–1381. doi: 10.1128/mcb.00741-09
Falck, J., Coates, J., and Jackson, S. P. (2005). Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature 434, 605–611. doi: 10.1038/nature03442
Fang, E. F., Scheibye-Knudsen, M., Chua, K. F., Mattson, M. P., Croteau, D. L., and Bohr, V. A. (2016). Nuclear DNA damage signalling to mitochondria in ageing. Nat. Rev. Mol. Cell Biol. 17, 308–321. doi: 10.1038/nrm.2016.14
Featherstone, C., and Jackson, S. P. (1999). Ku, a DNA repair protein with multiple cellular functions? Mutat. Res. 434, 3–15. doi: 10.1016/s0921-8777(99)00006-3
Fok, J. H. L., Ramos-Montoya, A., Vazquez-Chantada, M., Wijnhoven, P. W. G., Follia, V., James, N., et al. (2019). AZD7648 is a potent and selective DNA-PK inhibitor that enhances radiation, chemotherapy and olaparib activity. Nat. Commun. 10:5065. doi: 10.1038/s41467-019-12836-9
Gao, S. S., Guan, H., Yan, S., Hu, S., Song, M., Guo, Z. P., et al. (2020). TIP60 K430 SUMOylation attenuates its interaction with DNA-PKcs in S-phase cells: facilitating homologous recombination and emerging target for cancer therapy. Sci. Adv. 6:eaba7822. doi: 10.1126/sciadv.aba7822
Gomes, L. R., Menck, C. F. M., and Leandro, G. S. (2017). Autophagy roles in the modulation of DNA repair pathways. Int. J. Mol. Sci. 18:2351. doi: 10.3390/ijms18112351
Goodarzi, A. A., Yu, Y., Riballo, E., Douglas, P., Walker, S. A., Ye, R., et al. (2006). DNA-PK autophosphorylation facilitates Artemis endonuclease activity. Embo J. 25, 3880–3889. doi: 10.1038/sj.emboj.7601255
Goytisolo, F. A., Samper, E., Edmonson, S., Taccioli, G. E., and Blasco, M. A. (2001). The absence of the DNA-dependent protein kinase catalytic subunit in mice results in anaphase bridges and in increased telomeric fusions with normal telomere length and G-strand overhang. Mol. Cell. Biol. 21, 3642–3651. doi: 10.1128/mcb.21.11.3642-3651.2001
Guo, Z., Wang, S., Xie, Y., Han, Y., Hu, S., Guan, H., et al. (2020). HUWE1-dependent DNA-PKcs neddylation modulates its autophosphorylation in DNA damage response. Cell Death Dis. 11:400. doi: 10.1038/s41419-020-2611-0
Han, Y., Jin, F., Xie, Y., Liu, Y., Hu, S., Liu, X. D., et al. (2019). DNA-PKcs PARylation regulates DNA-PK kinase activity in the DNA damage response. Mol. Med. Rep. 20, 3609–3616. doi: 10.3892/mmr.2019.10640
Hartley, K. O., Gell, D., Smith, G. C., Zhang, H., Divecha, N., Connelly, M. A., et al. (1995). DNA-dependent protein kinase catalytic subunit: a relative of phosphatidylinositol 3-kinase and the ataxia telangiectasia gene product. Cell 82, 849–856. doi: 10.1016/0092-8674(95)90482-4
Haupt, Y., Maya, R., Kazaz, A., and Oren, M. (1997). Mdm2 promotes the rapid degradation of p53. Nature 387, 296–299. doi: 10.1038/387296a0
Her, J., and Bunting, S. F. (2018). How cells ensure correct repair of DNA double-strand breaks. J. Biol. Chem. 293, 10502–10511. doi: 10.1074/jbc.TM118.000371
Hewitt, G., and Korolchuk, V. I. (2017). Repair, reuse, recycle: the expanding role of autophagy in genome maintenance. Trends Cell Biol. 27, 340–351. doi: 10.1016/j.tcb.2016.11.011
Ho, S. R., Mahanic, C. S., Lee, Y. J., and Lin, W. C. (2014). RNF144A, an E3 ubiquitin ligase for DNA-PKcs, promotes apoptosis during DNA damage. Proc. Natl. Acad. Sci. U.S.A. 111, E2646–E2655. doi: 10.1073/pnas.1323107111
Huang, R. X., and Zhou, P. K. (2020). DNA damage response signaling pathways and targets for radiotherapy sensitization in cancer. Signal Transduct. Target. Ther. 5:60. doi: 10.1038/s41392-020-0150-x
Jackson, S. P., and Bartek, J. (2009). The DNA-damage response in human biology and disease. Nature 461, 1071–1078. doi: 10.1038/nature08467
Javvadi, P., Makino, H., Das, A. K., Lin, Y. F., Chen, D. J., Chen, B. P., et al. (2012). Threonine 2609 phosphorylation of the DNA-dependent protein kinase is a critical prerequisite for epidermal growth factor receptor-mediated radiation resistance. Mol. Cancer Res. 10, 1359–1368. doi: 10.1158/1541-7786.Mcr-12-0482-t
Jeggo, P. A., Pearl, L. H., and Carr, A. M. (2016). DNA repair, genome stability and cancer: a historical perspective. Nat. Rev. Cancer 16, 35–42. doi: 10.1038/nrc.2015.4
Jiang, W., Crowe, J. L., Liu, X., Nakajima, S., Wang, Y., Li, C., et al. (2015). Differential phosphorylation of DNA-PKcs regulates the interplay between end-processing and end-ligation during nonhomologous end-joining. Mol. Cell 58, 172–185. doi: 10.1016/j.molcel.2015.02.024
Jiang, W., Estes, V. M., Wang, X. S., Shao, Z., Lee, B. J., Lin, X., et al. (2019). Phosphorylation at S2053 in murine (S2056 in human) DNA-PKcs is dispensable for lymphocyte development and class switch recombination. J. Immunol. 203, 178–187. doi: 10.4049/jimmunol.1801657
Jiang, X., Sun, Y., Chen, S., Roy, K., and Price, B. D. (2006). The FATC domains of PIKK proteins are functionally equivalent and participate in the Tip60-dependent activation of DNA-PKcs and ATM. J. Biol. Chem. 281, 15741–15746. doi: 10.1074/jbc.M513172200
Jin, S., and Weaver, D. T. (1997). Double-strand break repair by Ku70 requires heterodimerization with Ku80 and DNA binding functions. Embo J. 16, 6874–6885. doi: 10.1093/emboj/16.22.6874
Joshi, R. R., Ali, S. I., and Ashley, A. K. (2019). DNA ligase IV prevents replication fork stalling and promotes cellular proliferation in triple negative breast cancer. J. Nucleic Acids 2019:9170341. doi: 10.1155/2019/9170341
Kamoi, K., and Mochizuki, M. (2010). Phaco forward-chop technique for managing posterior nuclear plate of hard cataract. J. Cataract Refract. Surg. 36, 9–12. doi: 10.1016/j.jcrs.2009.07.047
Kantidze, O. L., Velichko, A. K., Luzhin, A. V., Petrova, N. V., and Razin, S. V. (2018). Synthetically lethal interactions of ATM, ATR, and DNA-PKcs. Trends Cancer 4, 755–768. doi: 10.1016/j.trecan.2018.09.007
Kim, B. M., Hong, Y., Lee, S., Liu, P., Lim, J. H., Lee, Y. H., et al. (2015). Therapeutic implications for overcoming radiation resistance in cancer therapy. Int. J. Mol. Sci. 16, 26880–26913. doi: 10.3390/ijms161125991
Kim, S. T., Lim, D. S., Canman, C. E., and Kastan, M. B. (1999). Substrate specificities and identification of putative substrates of ATM kinase family members. J. Biol. Chem. 274, 37538–37543. doi: 10.1074/jbc.274.53.37538
Konstantinopoulos, P. A., Ceccaldi, R., Shapiro, G. I., and D’Andrea, A. D. (2015). Homologous recombination deficiency: exploiting the fundamental vulnerability of ovarian cancer. Cancer Discov. 5, 1137–1154. doi: 10.1158/2159-8290.Cd-15-0714
Kotsantis, P., Petermann, E., and Boulton, S. J. (2018). Mechanisms of oncogene-induced replication stress: jigsaw falling into place. Cancer Discov. 8, 537–555. doi: 10.1158/2159-8290.Cd-17-1461
Kubbutat, M. H., Jones, S. N., and Vousden, K. H. (1997). Regulation of p53 stability by Mdm2. Nature 387, 299–303. doi: 10.1038/387299a0
Lazzerini-Denchi, E., and Sfeir, A. (2016). Stop pulling my strings – what telomeres taught us about the DNA damage response. Nat. Rev. Mol. Cell Biol. 17, 364–378. doi: 10.1038/nrm.2016.43
Lee, S. E., Mitchell, R. A., Cheng, A., and Hendrickson, E. A. (1997). Evidence for DNA-PK-dependent and –independent DNA double-strand break repair pathways in mammalian cells as a function of the cell cycle. Mol. Cell. Biol. 17, 1425–1433. doi: 10.1128/mcb.17.3.1425
Lees-Miller, S. P. (1996). The DNA-dependent protein kinase, DNA-PK: 10 years and no ends in sight. Biochem. Cell Biol. 74, 503–512. doi: 10.1139/o96-054
Lees-Miller, S. P., Chen, Y. R., and Anderson, C. W. (1990). Human cells contain a DNA-activated protein kinase that phosphorylates simian virus 40 T antigen, mouse p53, and the human Ku autoantigen. Mol. Cell. Biol. 10, 6472–6481. doi: 10.1128/mcb.10.12.6472
Leidal, A. M., Levine, B., and Debnath, J. (2018). Autophagy and the cell biology of age-related disease. Nat. Cell Biol. 20, 1338–1348. doi: 10.1038/s41556-018-0235-8
Liccardi, G., Hartley, J. A., and Hochhauser, D. (2011). EGFR nuclear translocation modulates DNA repair following cisplatin and ionizing radiation treatment. Cancer Res. 71, 1103–1114. doi: 10.1158/0008-5472.Can-10-2384
Lieber, M. R. (2008). The mechanism of human nonhomologous DNA end joining. J. Biol. Chem. 283, 1–5. doi: 10.1074/jbc.R700039200
Lin, Y. F., Shih, H. Y., Shang, Z. F., Kuo, C. T., Guo, J., Du, C., et al. (2018). PIDD mediates the association of DNA-PKcs and ATR at stalled replication forks to facilitate the ATR signaling pathway. Nucleic Acids Res. 46, 1847–1859. doi: 10.1093/nar/gkx1298
Lin, Y. F., Shih, H. Y., Shang, Z., Matsunaga, S., and Chen, B. P. (2014). DNA-PKcs is required to maintain stability of Chk1 and Claspin for optimal replication stress response. Nucleic Acids Res. 42, 4463–4473. doi: 10.1093/nar/gku116
Liu, S., Opiyo, S. O., Manthey, K., Glanzer, J. G., Ashley, A. K., Amerin, C., et al. (2012). Distinct roles for DNA-PK, ATM and ATR in RPA phosphorylation and checkpoint activation in response to replication stress. Nucleic Acids Res. 40, 10780–10794. doi: 10.1093/nar/gks849
Longerich, S., Li, J., Xiong, Y., Sung, P., and Kupfer, G. M. (2014). Stress and DNA repair biology of the Fanconi anemia pathway. Blood 124, 2812–2819. doi: 10.1182/blood-2014-04-526293
Lu, X., Ma, O., Nguyen, T. A., Jones, S. N., Oren, M., and Donehower, L. A. (2007). The Wip1 Phosphatase acts as a gatekeeper in the p53-Mdm2 autoregulatory loop. Cancer Cell 12, 342–354. doi: 10.1016/j.ccr.2007.08.033
Ma, Y., Pannicke, U., Schwarz, K., and Lieber, M. R. (2002). Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell 108, 781–794. doi: 10.1016/s0092-8674(02)00671-2
Madabhushi, R., Pan, L., and Tsai, L. H. (2014). DNA damage and its links to neurodegeneration. Neuron 83, 266–282. doi: 10.1016/j.neuron.2014.06.034
Mao, Z., Bozzella, M., Seluanov, A., and Gorbunova, V. (2008). DNA repair by nonhomologous end joining and homologous recombination during cell cycle in human cells. Cell Cycle 7, 2902–2906. doi: 10.4161/cc.7.18.6679
Medová, M., Medo, M., Hovhannisyan, L., Maldonado, C. M., Aebersold, D. M., and Zimmer, Y. (2020). DNA-PK in human malignant disorders: mechanisms and implications for pharmacological interventions. Pharmacol. Ther. 215:107617. doi: 10.1016/j.pharmthera.2020.107617
Meek, K., Lees-Miller, S. P., and Modesti, M. (2012). N-terminal constraint activates the catalytic subunit of the DNA-dependent protein kinase in the absence of DNA or Ku. Nucleic Acids Res. 40, 2964–2973. doi: 10.1093/nar/gkr1211
Menolfi, D., and Zha, S. (2020). ATM, ATR and DNA-PKcs kinases-the lessons from the mouse models: inhibition ≠ deletion. Cell Biosci. 10:8. doi: 10.1186/s13578-020-0376-x
Menon, V., and Povirk, L. F. (2016). End-processing nucleases and phosphodiesterases: an elite supporting cast for the non-homologous end joining pathway of DNA double-strand break repair. DNA Repair (Amst.) 43, 57–68. doi: 10.1016/j.dnarep.2016.05.011
Miller, R. D., Hogg, J., Ozaki, J. H., Gell, D., Jackson, S. P., and Riblet, R. (1995). Gene for the catalytic subunit of mouse DNA-dependent protein kinase maps to the scid locus. Proc. Natl. Acad. Sci. U.S.A. 92, 10792–10795. doi: 10.1073/pnas.92.23.10792
Mirman, Z., and de Lange, T. (2020). 53BP1: a DSB escort. Genes Dev. 34, 7–23. doi: 10.1101/gad.333237.119
Mori, E., Davis, A. J., Hasegawa, M., and Chen, D. J. (2016). Lysines 3241 and 3260 of DNA-PKcs are important for genomic stability and radioresistance. Biochem. Biophys. Res. Commun. 477, 235–240. doi: 10.1016/j.bbrc.2016.06.048
Morris, E. P., Rivera-Calzada, A., da Fonseca, P. C., Llorca, O., Pearl, L. H., and Spagnolo, L. (2011). Evidence for a remodelling of DNA-PK upon autophosphorylation from electron microscopy studies. Nucleic Acids Res. 39, 5757–5767. doi: 10.1093/nar/gkr146
Nagasawa, H., Lin, Y. F., Kato, T. A., Brogan, J. R., Shih, H. Y., Kurimasa, A., et al. (2017). Coordination of the Ser2056 and Thr2609 clusters of DNA-PKcs in regulating gamma rays and extremely low fluencies of alpha-particle irradiation to G(0)/G(1) phase cells. Radiat. Res. 187, 259–267. doi: 10.1667/rr14679.1
Nagasawa, H., Little, J. B., Lin, Y. F., So, S., Kurimasa, A., Peng, Y., et al. (2011). Differential role of DNA-PKcs phosphorylations and kinase activity in radiosensitivity and chromosomal instability. Radiat. Res. 175, 83–89. doi: 10.1667/rr2092.1
Neal, J. A., Dang, V., Douglas, P., Wold, M. S., Lees-Miller, S. P., and Meek, K. (2011). Inhibition of homologous recombination by DNA-dependent protein kinase requires kinase activity, is titratable, and is modulated by autophosphorylation. Mol. Cell. Biol. 31, 1719–1733. doi: 10.1128/mcb.01298-10
Nutley, B. P., Smith, N. F., Hayes, A., Kelland, L. R., Brunton, L., Golding, B. T., et al. (2005). Preclinical pharmacokinetics and metabolism of a novel prototype DNA-PK inhibitor NU7026. Br. J. Cancer 93, 1011–1018. doi: 10.1038/sj.bjc.6602823
Pannunzio, N. R., Watanabe, G., and Lieber, M. R. (2018). Nonhomologous DNA end-joining for repair of DNA double-strand breaks. J. Biol. Chem. 293, 10512–10523. doi: 10.1074/jbc.TM117.000374
Puustinen, P., Keldsbo, A., Corcelle-Termeau, E., Ngoei, K., Sønder, S. L., Farkas, T., et al. (2020). DNA-dependent protein kinase regulates lysosomal AMP-dependent protein kinase activation and autophagy. Autophagy 16, 1871–1888. doi: 10.1080/15548627.2019.1710430
Reichert, S., Rödel, C., Mirsch, J., Harter, P. N., Tomicic, M. T., Mittelbronn, M., et al. (2011). Survivin inhibition and DNA double-strand break repair: a molecular mechanism to overcome radioresistance in glioblastoma. Radiother. Oncol. 101, 51–58. doi: 10.1016/j.radonc.2011.06.037
Rivera-Calzada, A., Maman, J. D., Spagnolo, L., Pearl, L. H., and Llorca, O. (2005). Three-dimensional structure and regulation of the DNA-dependent protein kinase catalytic subunit (DNA-PKcs). Structure 13, 243–255. doi: 10.1016/j.str.2004.12.006
Roake, C. M., and Artandi, S. E. (2020). Regulation of human telomerase in homeostasis and disease. Nat. Rev. Mol. Cell Biol. 21, 384–397. doi: 10.1038/s41580-020-0234-z
Roos, W. P., and Kaina, B. (2006). DNA damage-induced cell death by apoptosis. Trends Mol. Med. 12, 440–450. doi: 10.1016/j.molmed.2006.07.007
Sallmyr, A., and Tomkinson, A. E. (2018). Repair of DNA double-strand breaks by mammalian alternative end-joining pathways. J. Biol. Chem. 293, 10536–10546. doi: 10.1074/jbc.TM117.000375
Scully, R., Panday, A., Elango, R., and Willis, N. A. (2019). DNA double-strand break repair-pathway choice in somatic mammalian cells. Nat. Rev. Mol. Cell Biol. 20, 698–714. doi: 10.1038/s41580-019-0152-0
Shang, Z. F., Huang, B., Xu, Q. Z., Zhang, S. M., Fan, R., Liu, X. D., et al. (2010). Inactivation of DNA-dependent protein kinase leads to spindle disruption and mitotic catastrophe with attenuated checkpoint protein 2 Phosphorylation in response to DNA damage. Cancer Res. 70, 3657–3666. doi: 10.1158/0008-5472.Can-09-3362
Sharif, H., Li, Y., Dong, Y., Dong, L., Wang, W. L., Mao, Y., et al. (2017). Cryo-EM structure of the DNA-PK holoenzyme. Proc. Natl. Acad. Sci. U.S.A. 114, 7367–7372. doi: 10.1073/pnas.1707386114
Shibata, A., and Jeggo, P. A. (2020). Canonical DNA non-homologous end-joining; capacity versus fidelity. Br. J. Radiol. 93:20190966. doi: 10.1259/bjr.20190966
Shibata, A., Conrad, S., Birraux, J., Geuting, V., Barton, O., Ismail, A., et al. (2011). Factors determining DNA double-strand break repair pathway choice in G2 phase. Embo J. 30, 1079–1092. doi: 10.1038/emboj.2011.27
Shimizu, I., Yoshida, Y., Suda, M., and Minamino, T. (2014). DNA damage response and metabolic disease. Cell Metab. 20, 967–977. doi: 10.1016/j.cmet.2014.10.008
Sibanda, B. L., Chirgadze, D. Y., and Blundell, T. L. (2010). Crystal structure of DNA-PKcs reveals a large open-ring cradle comprised of HEAT repeats. Nature 463, 118–121. doi: 10.1038/nature08648
Sibanda, B. L., Chirgadze, D. Y., Ascher, D. B., and Blundell, T. L. (2017). DNA-PKcs structure suggests an allosteric mechanism modulating DNA double-strand break repair. Science 355, 520–524. doi: 10.1126/science.aak9654
Singleton, B. K., Torres-Arzayus, M. I., Rottinghaus, S. T., Taccioli, G. E., and Jeggo, P. A. (1999). The C terminus of Ku80 activates the DNA-dependent protein kinase catalytic subunit. Mol. Cell. Biol. 19, 3267–3277. doi: 10.1128/mcb.19.5.3267
Sipley, J. D., Menninger, J. C., Hartley, K. O., Ward, D. C., Jackson, S. P., and Anderson, C. W. (1995). Gene for the catalytic subunit of the human DNA-activated protein kinase maps to the site of the XRCC7 gene on chromosome 8. Proc. Natl. Acad. Sci. U.S.A. 92, 7515–7519. doi: 10.1073/pnas.92.16.7515
Smith, M. C., Mader, M. M., Cook, J. A., Iversen, P., Ajamie, R., Perkins, E., et al. (2016). Characterization of LY3023414, a novel PI3K/mTOR dual inhibitor eliciting transient target modulation to impede tumor growth. Mol. Cancer Ther. 15, 2344–2356. doi: 10.1158/1535-7163.Mct-15-0996
Spagnolo, L., Rivera-Calzada, A., Pearl, L. H., and Llorca, O. (2006). Three-dimensional structure of the human DNA-PKcs/Ku70/Ku80 complex assembled on DNA and its implications for DNA DSB repair. Mol. Cell 22, 511–519. doi: 10.1016/j.molcel.2006.04.013
Sui, J., Lin, Y. F., Xu, K., Lee, K. J., Wang, D., and Chen, B. P. (2015). DNA-PKcs phosphorylates hnRNP-A1 to facilitate the RPA-to-POT1 switch and telomere capping after replication. Nucleic Acids Res. 43, 5971–5983. doi: 10.1093/nar/gkv539
Sui, J., Zhang, S., and Chen, B. P. C. (2020). DNA-dependent protein kinase in telomere maintenance and protection. Cell. Mol. Biol. Lett. 25:2. doi: 10.1186/s11658-020-0199-0
Sun, Y., McCorvie, T. J., Yates, L. A., and Zhang, X. (2020). Structural basis of homologous recombination. Cell. Mol. Life Sci. 77, 3–18. doi: 10.1007/s00018-019-03365-1
Syed, A., and Tainer, J. A. (2018). The MRE11-RAD50-NBS1 complex conducts the orchestration of damage signaling and outcomes to stress in DNA replication and repair. Annu. Rev. Biochem. 87, 263–294. doi: 10.1146/annurev-biochem-062917-012415
Tarry-Adkins, J. L., Aiken, C. E., Ashmore, T. J., Fernandez-Twinn, D. S., Chen, J. H., and Ozanne, S. E. (2019). A suboptimal maternal diet combined with accelerated postnatal growth results in an altered aging profile in the thymus of male rats. Faseb J. 33, 239–253. doi: 10.1096/fj.201701350RR
Thijssen, R., Ter Burg, J., Garrick, B., van Bochove, G. G., Brown, J. R., Fernandes, S. M., et al. (2016). Dual TORK/DNA-PK inhibition blocks critical signaling pathways in chronic lymphocytic leukemia. Blood 128, 574–583. doi: 10.1182/blood-2016-02-700328
Ting, N. S., Pohorelic, B., Yu, Y., Lees-Miller, S. P., and Beattie, T. L. (2009). The human telomerase RNA component, hTR, activates the DNA-dependent protein kinase to phosphorylate heterogeneous nuclear ribonucleoprotein A1. Nucleic Acids Res. 37, 6105–6115. doi: 10.1093/nar/gkp636
Tomimatsu, N., Mukherjee, B., and Burma, S. (2009). Distinct roles of ATR and DNA-PKcs in triggering DNA damage responses in ATM-deficient cells. EMBO Rep. 10, 629–635. doi: 10.1038/embor.2009.60
Ubhi, T., and Brown, G. W. (2019). Exploiting DNA replication stress for cancer treatment. Cancer Res. 79, 1730–1739. doi: 10.1158/0008-5472.Can-18-3631
Ui, M., Okada, T., Hazeki, K., and Hazeki, O. (1995). Wortmannin as a unique probe for an intracellular signalling protein, phosphoinositide 3-kinase. Trends Biochem. Sci. 20, 303–307. doi: 10.1016/s0968-0004(00)89056-8
Uryga, A., Gray, K., and Bennett, M. (2016). DNA damage and repair in vascular disease. Annu. Rev. Physiol. 78, 45–66. doi: 10.1146/annurev-physiol-021115-105127
van der Burg, M., Ijspeert, H., Verkaik, N. S., Turul, T., Wiegant, W. W., Morotomi-Yano, K., et al. (2009). A DNA-PKcs mutation in a radiosensitive T-B- SCID patient inhibits Artemis activation and nonhomologous end-joining. J. Clin. Invest. 119, 91–98. doi: 10.1172/jci37141
Victorelli, S., and Passos, J. F. (2017). Telomeres and cell senescence – size matters not. EBioMedicine 21, 14–20. doi: 10.1016/j.ebiom.2017.03.027
Vijg, J., and Suh, Y. (2013). Genome instability and aging. Annu. Rev. Physiol. 75, 645–668. doi: 10.1146/annurev-physiol-030212-183715
Villafanez, F., Garcia, I. A., Carbajosa, S., Pansa, M. F., Mansilla, S., Llorens, M. C., et al. (2019). AKT inhibition impairs PCNA ubiquitylation and triggers synthetic lethality in homologous recombination-deficient cells submitted to replication stress. Oncogene 38, 4310–4324. doi: 10.1038/s41388-019-0724-7
Walker, A. I., Hunt, T., Jackson, R. J., and Anderson, C. W. (1985). Double-stranded DNA induces the phosphorylation of several proteins including the 90 000 mol. wt. heat-shock protein in animal cell extracts. Embo J. 4, 139–145. doi: 10.1002/j.1460-2075.1985.tb02328.x
Wang, C. Y., Huang, E. Y., Huang, S. C., and Chung, B. C. (2015). DNA-PK/Chk2 induces centrosome amplification during prolonged replication stress. Oncogene 34, 1263–1269. doi: 10.1038/onc.2014.74
Wang, S., Guo, M., Ouyang, H., Li, X., Cordon-Cardo, C., Kurimasa, A., et al. (2000). The catalytic subunit of DNA-dependent protein kinase selectively regulates p53-dependent apoptosis but not cell-cycle arrest. Proc. Natl. Acad. Sci. U.S.A. 97, 1584–1588. doi: 10.1073/pnas.97.4.1584
Willmore, E., de Caux, S., Sunter, N. J., Tilby, M. J., Jackson, G. H., Austin, C. A., et al. (2004). A novel DNA-dependent protein kinase inhibitor, NU7026, potentiates the cytotoxicity of topoisomerase II poisons used in the treatment of leukemia. Blood 103, 4659–4665. doi: 10.1182/blood-2003-07-2527
Woodbine, L., Neal, J. A., Sasi, N. K., Shimada, M., Deem, K., Coleman, H., et al. (2013). PRKDC mutations in a SCID patient with profound neurological abnormalities. J. Clin. Invest. 123, 2969–2980. doi: 10.1172/jci67349
Xie, Y., Liu, Y. K., Guo, Z. P., Guan, H., Liu, X. D., Xie, D. F., et al. (2020). RBX1 prompts degradation of EXO1 to limit the homologous recombination pathway of DNA double-strand break repair in G1 phase. Cell Death Differ. 27, 1383–1397. doi: 10.1038/s41418-019-0424-4
Yan, Y. Q., Zhang, B., Wang, L., Xie, Y. H., Peng, T., Bai, B., et al. (2007). Induction of apoptosis and autophagic cell death by the vanillin derivative 6-bromine-5-hydroxy-4-methoxybenzaldehyde is accompanied by the cleavage of DNA-PKcs and rapid destruction of c-Myc oncoprotein in HepG2 cells. Cancer Lett. 252, 280–289. doi: 10.1016/j.canlet.2007.01.007
Yin, X., Liu, M., Tian, Y., Wang, J., and Xu, Y. (2017). Cryo-EM structure of human DNA-PK holoenzyme. Cell Res. 27, 1341–1350. doi: 10.1038/cr.2017.110
Ying, S., Chen, Z., Medhurst, A. L., Neal, J. A., Bao, Z., Mortusewicz, O., et al. (2016). DNA-PKcs and PARP1 bind to unresected stalled DNA replication forks where they recruit XRCC1 to mediate repair. Cancer Res. 76, 1078–1088. doi: 10.1158/0008-5472.Can-15-0608
Zeman, M. K., and Cimprich, K. A. (2014). Causes and consequences of replication stress. Nat. Cell Biol. 16, 2–9. doi: 10.1038/ncb2897
Zenke, F. T., Zimmermann, A., Sirrenberg, C., Dahmen, H., Kirkin, V., Pehl, U., et al. (2020). Pharmacologic inhibitor of DNA-PK, M3814, potentiates radiotherapy and regresses human tumors in mouse models. Mol. Cancer Ther. 19, 1091–1101. doi: 10.1158/1535-7163.Mct-19-0734
Zhang, S., Matsunaga, S., Lin, Y. F., Sishc, B., Shang, Z., Sui, J., et al. (2016). Spontaneous tumor development in bone marrow-rescued DNA-PKcs(3A/3A) mice due to dysfunction of telomere leading strand deprotection. Oncogene 35, 3909–3918. doi: 10.1038/onc.2015.459
Zhang, W. W., and Matlashewski, G. (2019). Single-strand annealing plays a major role in double-strand DNA break repair FOLLOWING CRISPR-Cas9 cleavage in Leishmania. mSphere 4:e00408–19. doi: 10.1128/mSphere.00408-19
Zhao, Y., Thomas, H. D., Batey, M. A., Cowell, I. G., Richardson, C. J., Griffin, R. J., et al. (2006). Preclinical evaluation of a potent novel DNA-dependent protein kinase inhibitor NU7441. Cancer Res. 66, 5354–5362. doi: 10.1158/0008-5472.Can-05-4275
Zhou, Y., and Paull, T. T. (2013). DNA-dependent protein kinase regulates DNA end resection in concert with Mre11-Rad50-Nbs1 (MRN) and ataxia telangiectasia-mutated (ATM). J. Biol. Chem. 288, 37112–37125. doi: 10.1074/jbc.M113.514398
Zhou, Y., Lee, J. H., Jiang, W., Crowe, J. L., Zha, S., and Paull, T. T. (2017). Regulation of the DNA damage response by DNA-PKcs inhibitory phosphorylation of ATM. Mol. Cell 65, 91–104. doi: 10.1016/j.molcel.2016.11.004
Zhu, S., Fisher, L. A., Bessho, T., and Peng, A. (2017). Protein phosphatase 1 and phosphatase 1 nuclear targeting subunit-dependent regulation of DNA-dependent protein kinase and non-homologous end joining. Nucleic Acids Res. 45, 10583–10594. doi: 10.1093/nar/gkx686
Zhu, Y., Hu, J., Hu, Y., and Liu, W. (2009). Targeting DNA repair pathways: a novel approach to reduce cancer therapeutic resistance. Cancer Treat. Rev. 35, 590–596. doi: 10.1016/j.ctrv.2009.06.005
Keywords: DNA-PKcs, DNA damage response, DNA repair, genomic instability, radiosensitization
Citation: Yue X, Bai C, Xie D, Ma T and Zhou PK (2020) DNA-PKcs: A Multi-Faceted Player in DNA Damage Response. Front. Genet. 11:607428. doi: 10.3389/fgene.2020.607428
Received: 17 September 2020; Accepted: 01 December 2020;
Published: 23 December 2020.
Edited by:
Chunlong Chen, Institut Curie, FranceCopyright © 2020 Yue, Bai, Xie, Ma and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Teng Ma, mateng82913@163.com; Ping-Kun Zhou, zhoupk@bmi.ac.cn; birm4th@163.com
 Xiaoqiao Yue
Xiaoqiao Yue Chenjun Bai2
Chenjun Bai2 Teng Ma
Teng Ma Ping-Kun Zhou
Ping-Kun Zhou