- 1Agricultural Resource and Environment Experiment Teaching Center, College of Resource and Environment Science, Nanjing Agricultural University, Nanjing, China
- 2Key Laboratory of Plant Nutrition and Fertilizers, Ministry of Agriculture and Rural Affairs, Institute of Agricultural Resources and Regional Planning, Chinese Academy of Agricultural Sciences, Beijing, China
- 3College of Life and Environmental Sciences, Hangzhou Normal University, Hangzhou, China
Phosphorus (P) is an essential nutrient for plant growth and development. Phosphate transporters (PHTs) are trans-membrane proteins that mediate the uptake and translocation of phosphate (Pi) in green plants. The PHT family including PHT1, PHT2, PHT3 and PHT4 subfamilies are well-studied in land plants; however, PHT genes in green algae are poorly documented and not comprehensively identified. Here, we analyzed the PHTs in a model green alga Chlamydomonas reinhardtii and found 25 putative PHT genes, which can be divided into four subfamilies. The subfamilies of CrPTA, CrPTB, CrPHT3, and CrPHT4 contain four, eleven, one, and nine genes, respectively. The structure, chromosomal distribution, subcellular localization, duplication, phylogenies, and motifs of these genes were systematically analyzed in silico. Expression profile analysis showed that CrPHT genes displayed differential expression patterns under P starvation condition. The expression levels of CrPTA1 and CrPTA3 were down-regulated, while the expression of most CrPTB genes was up-regulated under P starvation, which may be controlled by CrPSR1. The transcript abundance of most CrPHT3 and CrPHT4 genes was not significantly affected by P starvation except CrPHT4-3, CrPHT4-4, and CrPHT4-6. Our results provided basic information for understanding the evolution and features of the PHT family in green algae.
Introduction
Phosphorus (P) is one of the most important macronutrients for algae and land plants. It is a critical component of nucleic acids, phospholipids, and ATP and participates in numerous biochemical pathways, including gene expression, and signal transduction (Lorenzo-Orts et al., 2020). P acquisition is activated by transcriptional induction of genes encoding high- and low-affinity phosphate transporters (PHTs), phosphodiesterases, and phosphatases (Wykoff et al., 1999). In land plants, PHT1 (phosphate transporter 1), PHT2, PHT3, and PHT4 gene families are revealed to encode proteins for uptake and transport of inorganic phosphate (Pi) between the cytoplasm and different subcellular compartments (Wang et al., 2017).
Phosphorus is mainly absorbed as Pi via the proton-coupled H2PO4– symporters PHT1 gene family, which are located in the plasma membrane of cells at the interface with the external environment in land plants (Nussaume et al., 2011; Wang et al., 2017). The PHT2, PHT3, and PHT4 gene families encode proteins that transport Pi between the cytoplasm and different subcellular compartments in land plants (Guo et al., 2008; Zhu et al., 2012; Liu et al., 2019). PHT2 proteins and some PHT4 members, which locate on chloroplast thylakoid membrane or inner envelope, are responsible for Pi homeostasis in chloroplasts (Guo et al., 2008; Liu et al., 2019). The mitochondrial phosphate transporters (PHT3) play crucial roles in respiration by absorbing Pi into the mitochondrial matrix, where Pi is utilized for the conversion of ADP to ATP (Zhu et al., 2012). A subgroup of conserved MYB transcription factors designated as phosphate starvation response (PHR) and PHR1-like (PHL) have been defined as central regulators of Pi starvation signaling in diverse plants (Sun et al., 2016). Through the physical interactions with a cis element, namely, PHR1 binding site (P1BS; GNATATNC), PHR transcription factors are responsible for the transcriptional activation of a considerable proportion of Pi starvation-induced genes, including PHT1 members (Zhou et al., 2008; Bustos et al., 2010; Sun et al., 2016). It has been convinced in paddy rice that OsPHR2 activates OsPHT1;2 expressions by binding to the P1BS motif present in its promoter region (Wang et al., 2014).
In contrast, in diatoms and green algae, P starvation responses are regulated by an MYB family transcription factor phosphorus starvation response 1 (PSR1), which regulates Pi acquisition through the up-regulation of phosphatases and Pi transporters (Wykoff et al., 1999; Kumar Sharma et al., 2020). Green algae resemble the yeast in containing genes encoding both H+/Pi (plant-like, PTA) and the Na+/Pi (animal-like, PTB) transporters. Physiological evidence suggests that Na+ uptake is necessary for Pi influx in Chara corallina under P-deficient conditions (Reid et al., 2000; Mimura et al., 2002). This is related to the sequence conservation and structural similarity of PTB proteins to the metazoan PHT and fungal PHT, which are known to be functional plasma membrane Na+/Pi symporters (Kobayashi et al., 2003; Moseley et al., 2006). PHOSPHATE TRANSPORTER A (PTA) genes were homologous to PHO84 in yeast and encode the high-affinity H+/Pi cotransporter (Moseley et al., 2009). The function of the PHT gene family has been extensively studied in land plants; however, the function of the PHT gene family in green algae is still largely unknown.
In this study, we identified 25 CrPHT genes in a model green alga, Chlamydomonas reinhardtii. We systematically analyzed the gene structures, chromosome localizations, phylogeny, and expression patterns in response to P starvation in both wild-type and psr1 mutant backgrounds. Our work will provide a foundation for the further analysis of PHT genes function in green algae and the different mechanisms of Pi transport between green algae and land plants.
Results
Phylogenetic Relationship of PHTs in Plants
To investigate the phylogenetic relationships among PHTs in green plants, a phylogenetic tree was reconstructed by maximum likelihood method using the PHT protein sequences from green plants, bacteria, and Cyanobacteria (Figure 1). It showed the PHTs in plants form six monophyletic groups. Cluster I (PHT2 subfamily) is sister to the cyanobacteria and bacterial PHT proteins and constitutes a sister group to the Cluster II (PTB subfamily). Cluster III contained the grouping of the PHT4 family. Cluster IV contained the grouping of the PHT3 subfamily. The PHT1 subfamily only includes PHT genes in land plants and constitutes a sister group to the PTA subfamily. PHTs in land plants consisted of PHT1, PHT2, PHT3, and PHT4 subfamilies, while PHTs in algae contained PTA, PTB, PHT3, and PHT4 subfamilies. PHT1 proteins are the H+/Pi symporters that are responsible for Pi uptake in land plants, whereas PTB proteins are predicted to be the Na+/Pi symporters catalyzing Pi uptake in algae. Endosymbiotic theory posits that chloroplasts and mitochondria arose from bacteria. Both organelles have retained their PHTs in membranes. The PHT2, PHT3, and PHT4 gene families encode proteins that transport Pi between the cytoplasm and different subcellular compartments (chloroplasts and mitochondria) in land plants, while how Pi is transported in subcellular compartments in algae was still unclear. To further understand the PHT in algae, we chose a model alga Chlamydomonas to analyze the PHT gene family.
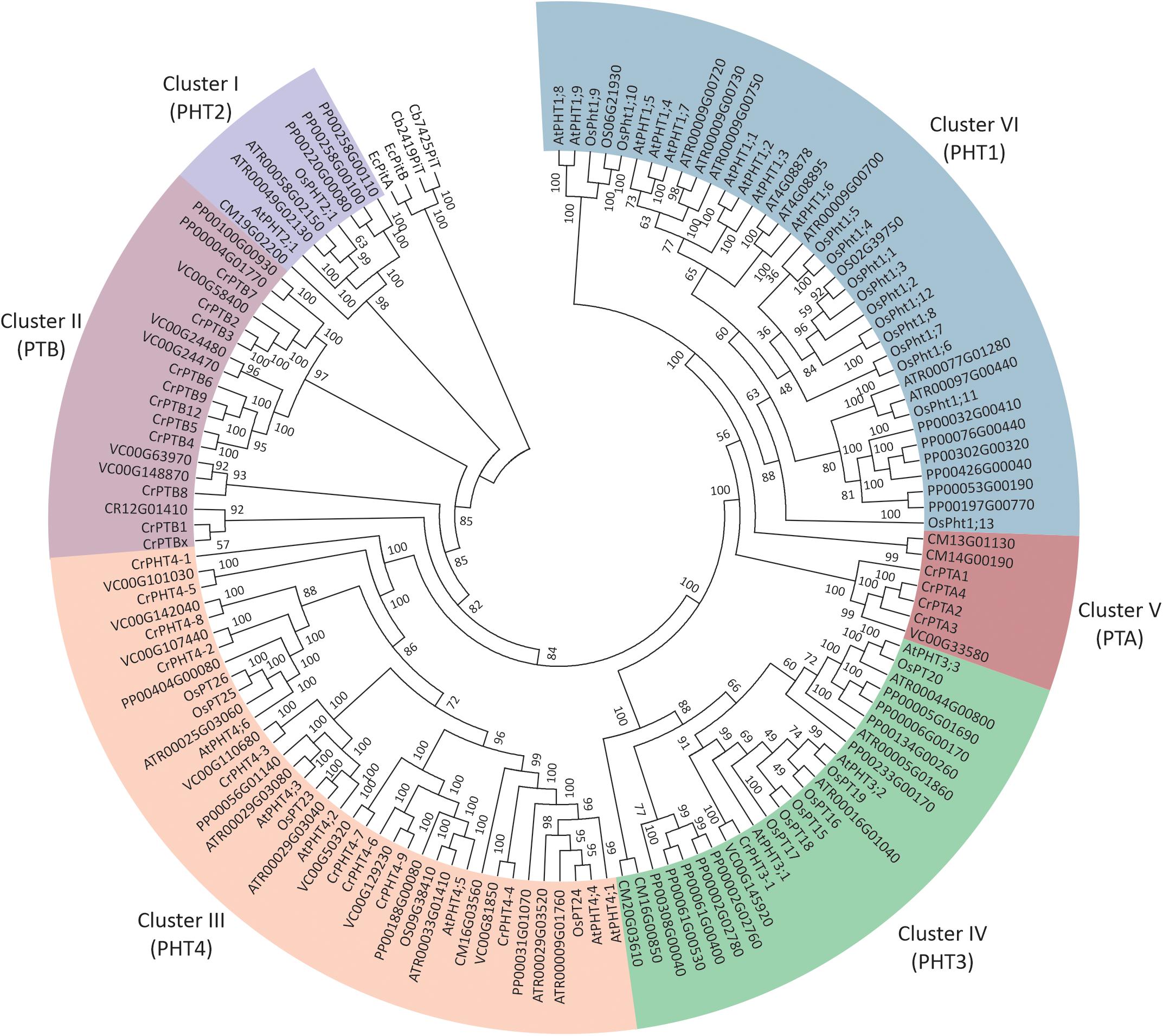
Figure 1. Phylogenetic analyses of phosphate transporter genes in Arabidopsis thaliana (At), Oryza sativa (Os), Amborella trichopoda (Atr), and Volvox carteri (Vc). Physcomitrella patens (Pp) and Chlamydomonas reinhardtii (Cr). Four PHT genes from Escherichia coli and Cyanobacteria were selected as outgroup. Multi-sequence alignments were performed by Clustal Omega, and a phylogenetic tree was reconstructed by IQTREE using the maximum likelihood method. WAG + F + R6 was the best-fitting evolutionary model selected by ModelFinder under Bayesian Information Criterion.
Identification of PHT Gene Family in Chlamydomonas reinhardtii
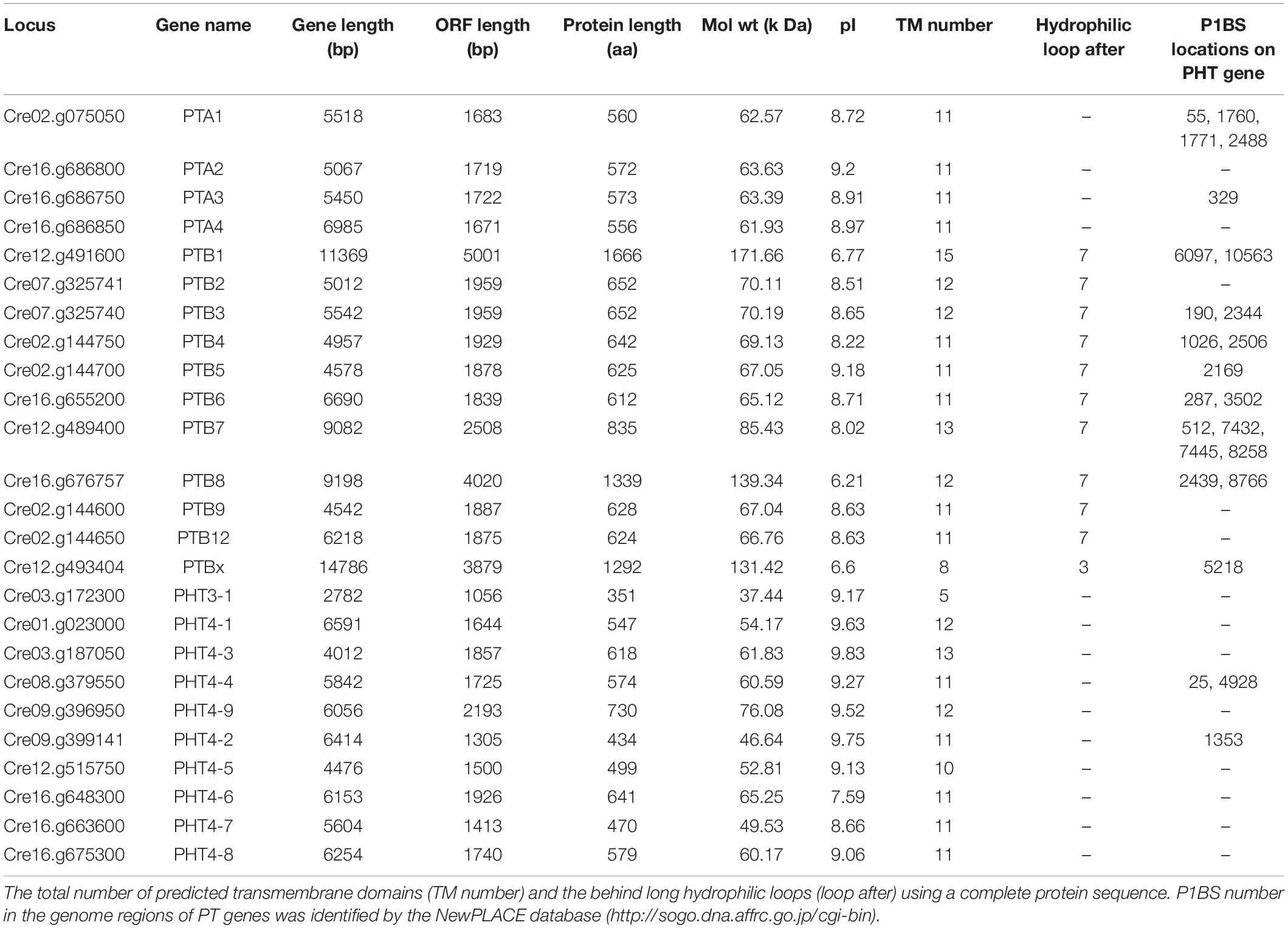
To investigate the PHT genes in Chlamydomonas, we employed PHT protein sequences of Arabidopsis and rice in a global search against Chlamydomonas genomic databases. In all, 25 putative PHT genes were found in Chlamydomonas (Table 1). Details on gene name, locus name, open reading frame (ORF) length, protein length, molecular weight, isoelectric point (pI), predicted transmembrane domain (TM) number, predicted long hydrophilic loop (loop after), and P1BS element are listed in Table 1. The PTA subfamily contains 4 members, PTB contains 11 members, PHT3 contains 1 member, and PHT4 contains 9 members. We conducted blast analysis of the genes against the Pfam database and found that all proteins contain the MFS domain. Values for pI ranged from 6.21 to 9.63. The protein lengths of 25 putative PHTs range from 351 to 1666 aa, and their molecular weights are 37.44 to 171.66 kDa; most of them are around 60 kDa. PHT proteins contain 5 to 15 TM, most of them range from 10 to 12 TM, and most of them contain a large hydrophilic loop between the seventh and eighth TMDs in the PTB family except CrPTBx. The P1BS cis element sequence (GNATATNC) was determined previously as the binding motif for PHR1, and this element is present in the promoter regions of P starvation-induced genes in various higher plants (Zhou et al., 2008; Wang et al., 2014). To identify putative PSR1-binding sites, we searched the GNATATNC sequence in genome sequences of all CrPHT genes (Table 1). P1BS elements were identified in most of the CrPHT genes. P1BS elements were present in the promoter regions of CrPTA1 and CrPTA3. All PTB subfamilies except CrPTB2, CrPTB9, and CrPTB12 have P1BS, while there is no P1BS in the PHT3 family. In the PTB4 family, only CrPHT4-2 and CrPHT4-4 have P1BS elements.
Prediction of Subcellular Targeting of PHT Proteins in Chlamydomonas
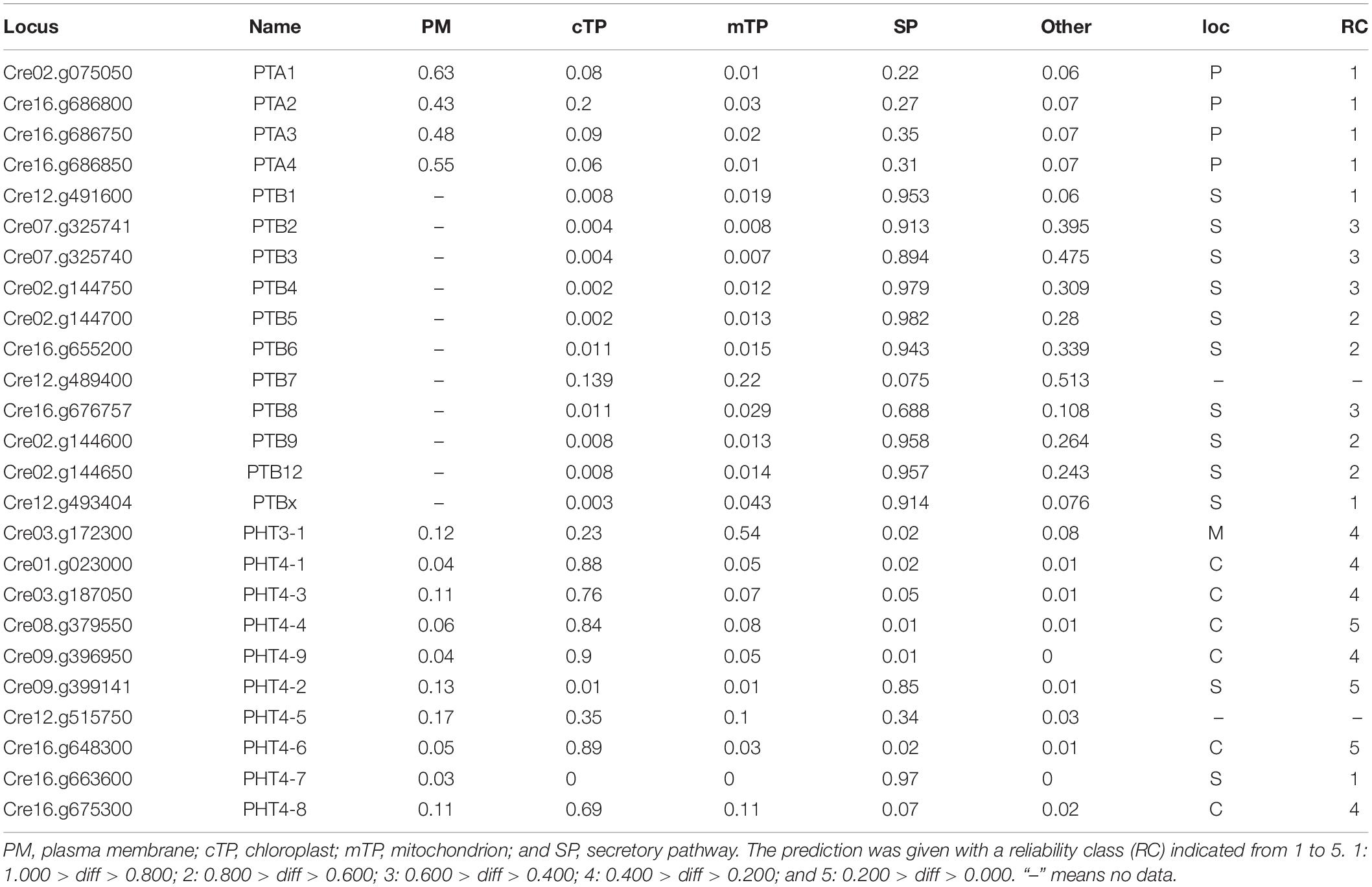
An N-terminal signal peptide targets the protein to the secretory pathway; proteins are inserted into the endoplasmic reticulum (ER), and either remain in the ER or move to the plasma membrane, vacuole, or endomembrane compartments (Chrispeels and Raikhel, 1992). Using four online tools (details in section “Materials and Methods”), which could predict subcellular localization by detecting subcellular targeting motifs, we discovered the signal peptide at the N-terminus of all PHT proteins (Table 2). Among them, the entire PTA family was predicted to locate in the plasma membrane with a very high reliability class. PTB proteins were predicted to target to the secretory pathway. However, based on Bonnot’s report (Bonnot et al., 2017), PTB proteins are plasma membrane proteins, which suggests that they finally move to the plasma membrane through the secretory pathway. PHT3 was predicted to target to the mitochondrion, with a very low reliability class. Most of the PHT4 family proteins were localized in chloroplast with a reliability class between 4 and 5. CrPHT4-2 and CrPHT4-7 were predicted to target to the secretory pathway. The CrPHT4-5 has no signal peptide and did not display predicted localization.
Conserved Motif Analysis of Chlamydomonas PHT Proteins
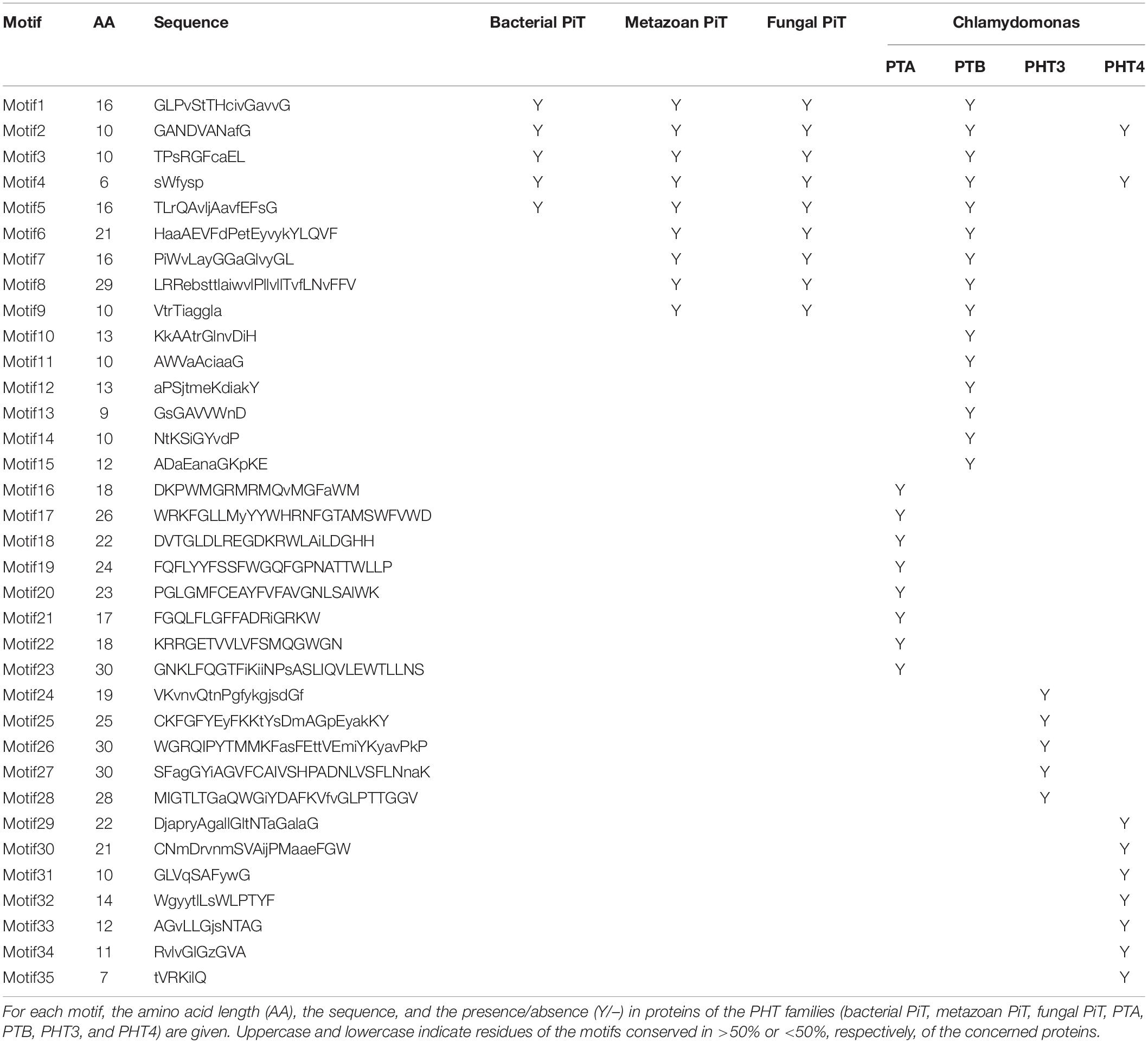
To analyze the conserved motifs and reveal the distinction between green algae and other ancient species, conserved motifs of PHT proteins from Chlamydomonas, bacteria, fungus, and metazoa were identified by MEME (Bailey and Elkan, 1994). The sequences of 4 Chlamydomonas PTA proteins; 11 PTB proteins; 8 PHT4 proteins; 5 PHT3 proteins; fungal PHT proteins ScPho89 and NcPho4; metazoan PHT proteins DmPit, HsPit1, and HsPit2; bacterial PHT proteins TePit, EcPitA, and EcPitB (from Escherichia coli); and 2 cyanobacteria proteins were selected for further analysis (Table 3). We identified a total of 35 conserved motifs among the selected PHT proteins and found that the conserved motifs could distinguish the Chlamydomonas PHT proteins from other members of the PHT proteins. Five motifs (motifs 1–5) were present in bacterial PHT, metazoan PHT, fungal PHT, and PTB family proteins. They contain the PHT signature GANDVAN domains (motif 2). Four motifs (motif 6–9) were common to the metazoan, fungal PHT, and PTB proteins. Twelve motifs (motifs 1–12) were present in PTB proteins. Eight motifs (motifs 16–23) were specific to the PTA proteins, and five motifs (motifs 24–28) were specific to the PHT3 proteins. Motifs 2, 4, and 29–35 were present in all PHT4 proteins. All motifs identified in the bacterial, metazoan, and fungal PHT proteins were present in the PTB proteins (Table 3). Meanwhile, motifs specific to the PTA and PHT3 family were found. Together, the conserved and specific motifs found in the different subfamilies of CrPHT proteins suggest their conservation in evolution and their divergence of functions.
Chromosomal Localization and Gene Duplication of CrPHT Genes
The chromosomal distributions of CrPHT genes were determined. As shown in Figure 2,25 PHT genes are distributed on 8 Chlamydomonas chromosomes. Out of the 25 predicted PHT genes, 18 are distributed across the 3 Chlamydomonas chromosomes (chromosome 2, 12 and 16) (Figure 2). Eight PHT genes scattered in clusters in chromosome 16. Chromosomes 1 and 8 each contain one gene. Chromosome 3, 7, and 9 each contain two genes. We further performed chromosome mapping to determine the gene duplication of PHT genes on the Chlamydomonas chromosomes. As shown in Supplementary Table 1, gene duplication of CrPHT genes contained four types, including tandem duplication (TD), proximal duplication (PD), transposed duplication (TRD), and dispersed duplication (DSD). Almost all the PHT genes were identified as duplicated genes (Supplementary Table 1). As shown in Figure 2, three groups of PHT genes can be identified as TD genes including group I (CrPTA2, CrPTA3, and CrPTA4) located on chromosome 16, group II (CrPTB4, CrPTB5, CrPTB9, and CrPTB12) located on chromosome 2, and group III (CrPTB2 and CrPTB3) located on chromosome 7, implying that the high density of PHT genes on this chromosome was mainly due to the TD events.
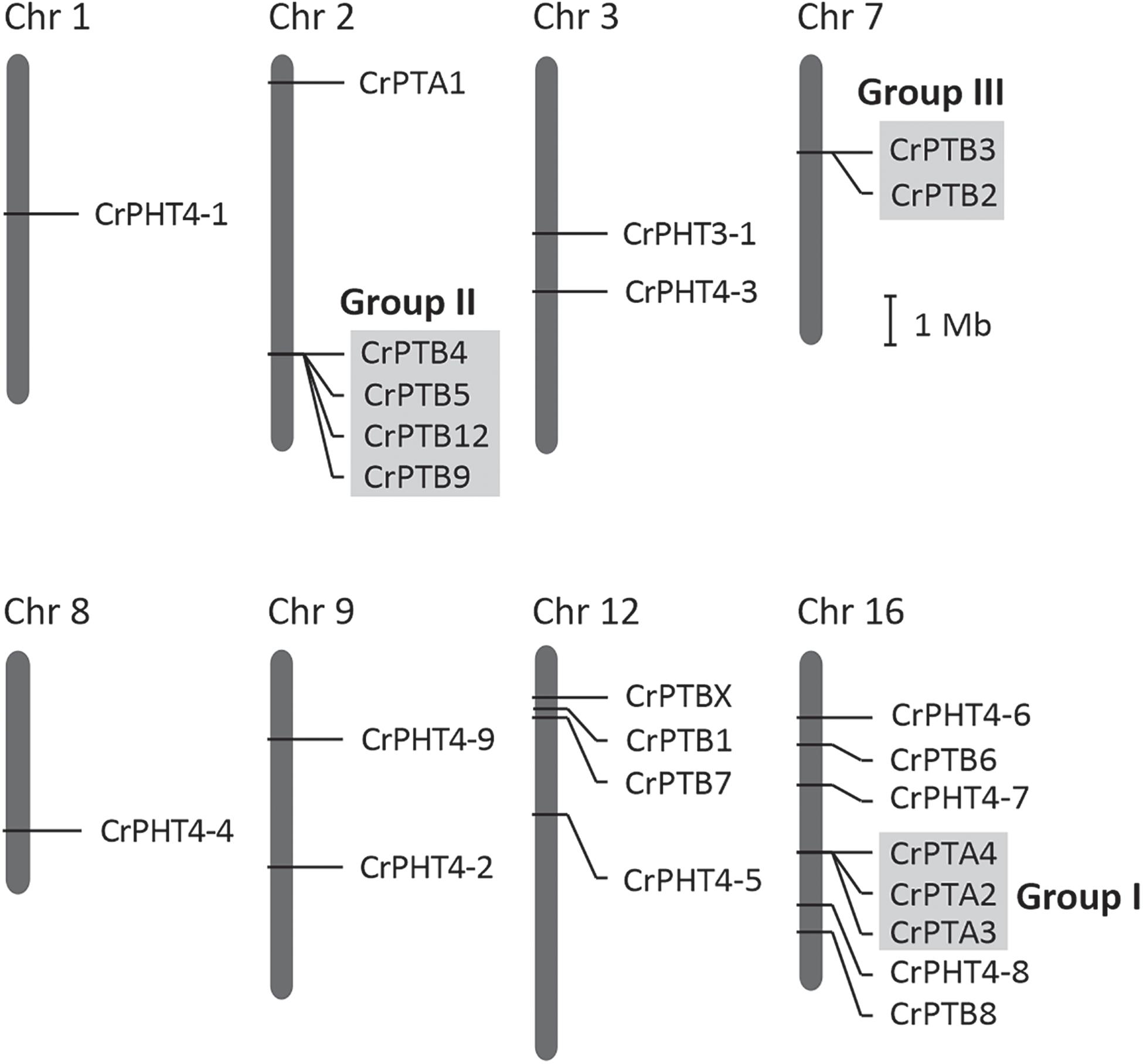
Figure 2. Chromosomal location and gene duplication of PHT proteins in Chlamydomonas. Chromosome numbers are shown at the top of each bar. Recent tandem duplicated genes were marked with a gray background. The scale presented on the left indicates chromosome sizes in megabases (Mb).
Expression Profiles of CrPHT Genes During Pi Starvation
To uncover the expression profiles of CrPHT genes under deprivation starvation and whether they are controlled by CrPSR1, we reanalyzed the RNA-seq data from the database1 (Bajhaiya et al., 2016). Phylogenetic trees constructed by the NJ method showed that these CrPHT proteins could be divided into four clades (Figure 3). Transcription levels of CrPHT genes varied under Pi starvation (Figure 3 and Supplementary Table 2). CrPTA1 and CrPTA3 transcripts declined following exposure of nutrient-replete cells to P starvation, and this decline is less significant in the psr1 mutant. This characteristic makes CrPTA1 and CrPTA3 candidates for the low-affinity Pi uptake system that operates in cells that are P-replete. The abundance of CrPTA2 and CrPTA4 transcripts is not significantly affected by P starvation. Upon Pi starvation, expressions of CrPTB2, CrPTB3, CrPTB4, CrPTB5, CrPTB7, CrPTB8, and CrPTB12 were induced, and this induction does not occur in the psr1 mutant, indicating that these genes were regulated by PSR1. The abundance of CrPTB1, CrPTB9, CrPTB12, and CrPTBx transcripts were not significantly affected by P starvation. The expression levels of genes in the CrPHT3 and CrPHT4 subfamilies were not significantly affected by P starvation, except CrPHT4-3, CrPHT4-4, and CrPHT4-6.
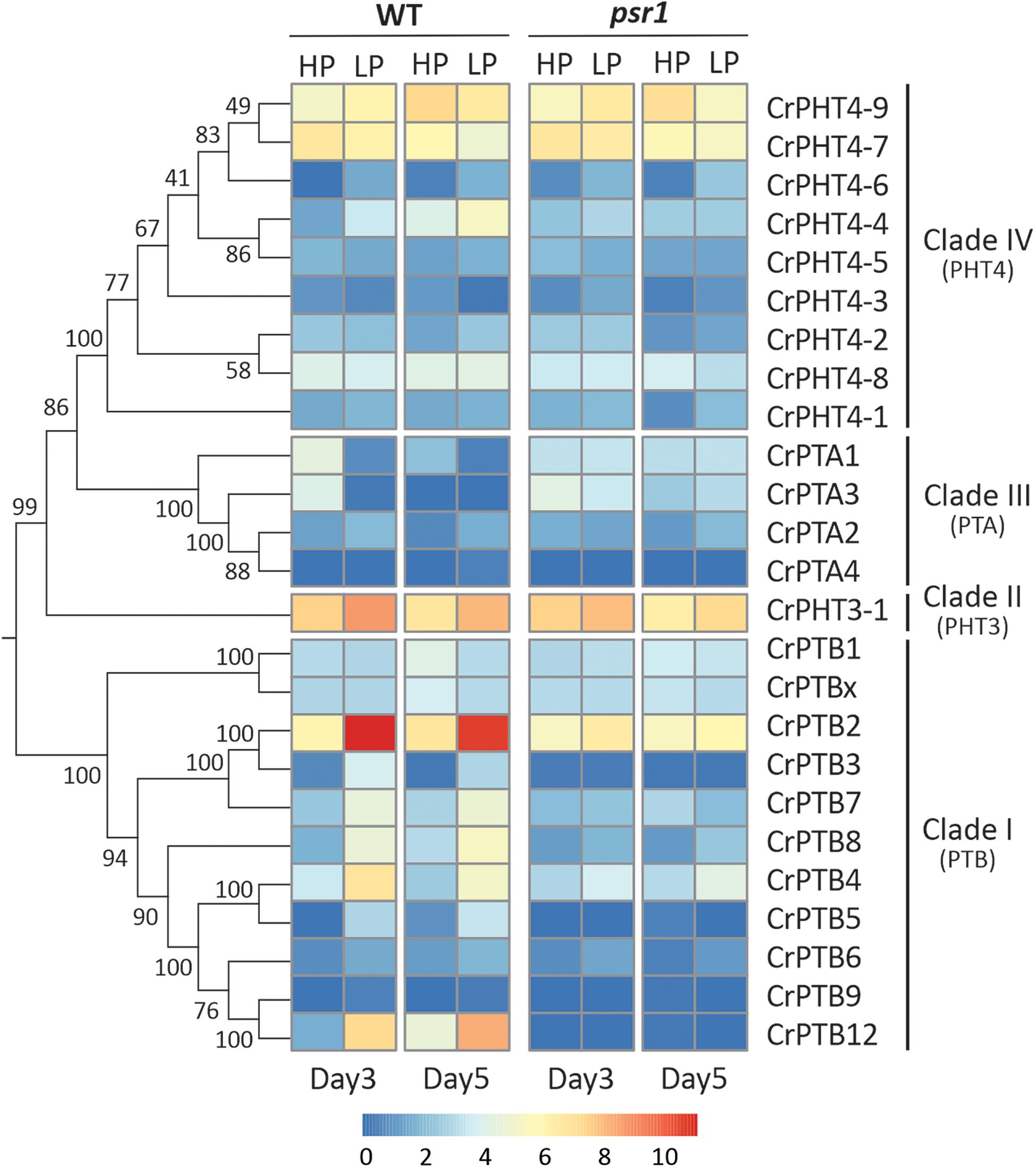
Figure 3. Expression profiles of Chlamydomonas PHT genes under phosphate-replete and starvation conditions. The phylogenetic tree of PHT proteins, showing in the left panel, was constructed by the neighbor-joining method with 1000 bootstrap replications using MEGA 7.0 software. Bootstrap values were shown on the branches. Heat map showing the expression of phosphate transporter genes under high-P (HP) and low-P (LP) conditions in day 3 and day 5 which display change in expression relative to WT HP conditions or psr1 HP. Each value corresponds to log2 of normalized counts. Day 3 and day 5 mean equivalence to the onset of P starvation and 48 h after the onset of P starvation, respectively.
Noticeably, the expression profiles of genes in each TD group (Figure 2) are variable. In Group I, CrPTA2 and CrPTA4 are poorly expressed, while CrPTA3 is highly expressed in nutrient-replete cells and its expression responded to phosphate starvation. This indicates that CrPTA2 and CrPTA4 are likely degenerated or redundant in function after duplication, compared to CrPTA3. In another duplicated group (Group III), the expression level of CrPTB2 is much higher than CrPTB3 under all the conditions, suggesting that the regulation of CrPTB2 and CrPTB3 are different. In Group III, CrPTB4, CrPTB5, and CrPTB12 were sharply up-regulated under phosphate starvation, while CrPTB9 is barely expressed under phosphate starvation in both WT and psr1 mutant. Different from other genes in Group III, CrPTB4 still showed some expression in the psr1 mutant. Together, functions of duplicated genes in each group are divergent and the gene duplication and subsequent divergence of CrPHT genes make up the complex network for P transport to adapt to a variable Pi environment.
qRT-PCR Analyses of CrPHT Genes Under P Stress Conditions
To explore the expression profiles of CrPHT genes under deprivation starvation, qRT-PCR analysis was performed (Figure 4). We firstly investigated the growth of WT and psr1 mutant under P-replete or depleted conditions. In P-replete conditions, the growth of the psr1 mutant is similar to that of WT. However, the psr1 mutant was unable to acclimate properly to P starvation and exhibited much less growth than wild-type strains on the TA medium (Shimogawara et al., 1999). The psr1 mutant showed growth defect compared to the WT in P-depleted conditions (Figure 4A). We further analyzed the steady-state transcript levels of genes encoding the phosphatase CrPHOX; the PHTs CrPTB2, CrPTB4, CrPTB5 CrPTB12, and CrPTA1; and the transcription factor CrPSR1 in the WT and psr1 mutant (Figure 4). Consistent with previous report (Shimogawara et al., 1999), the expression of CrPSR1 was significantly induced by P starvation (Figure 4B). CrPHOX was induced by P starvation in WT and not in the psr1 mutant (Figure 4C). Under Pi starvation conditions, the expressions of CrPTB2, CrPTB4, CrPTB5, and CrPTB12 were induced in the WT while they showed no effect in the psr1 mutant. The CrPTA1 transcript reduced following exposure of nutrient-replete cells to P starvation, and this decline does not occur in the psr1 mutant. These results suggest that different CrPHT genes have different expression patterns and CrPSR1 displays an important role in regulating Pi starvation-dependent signaling in Chlamydomonas.
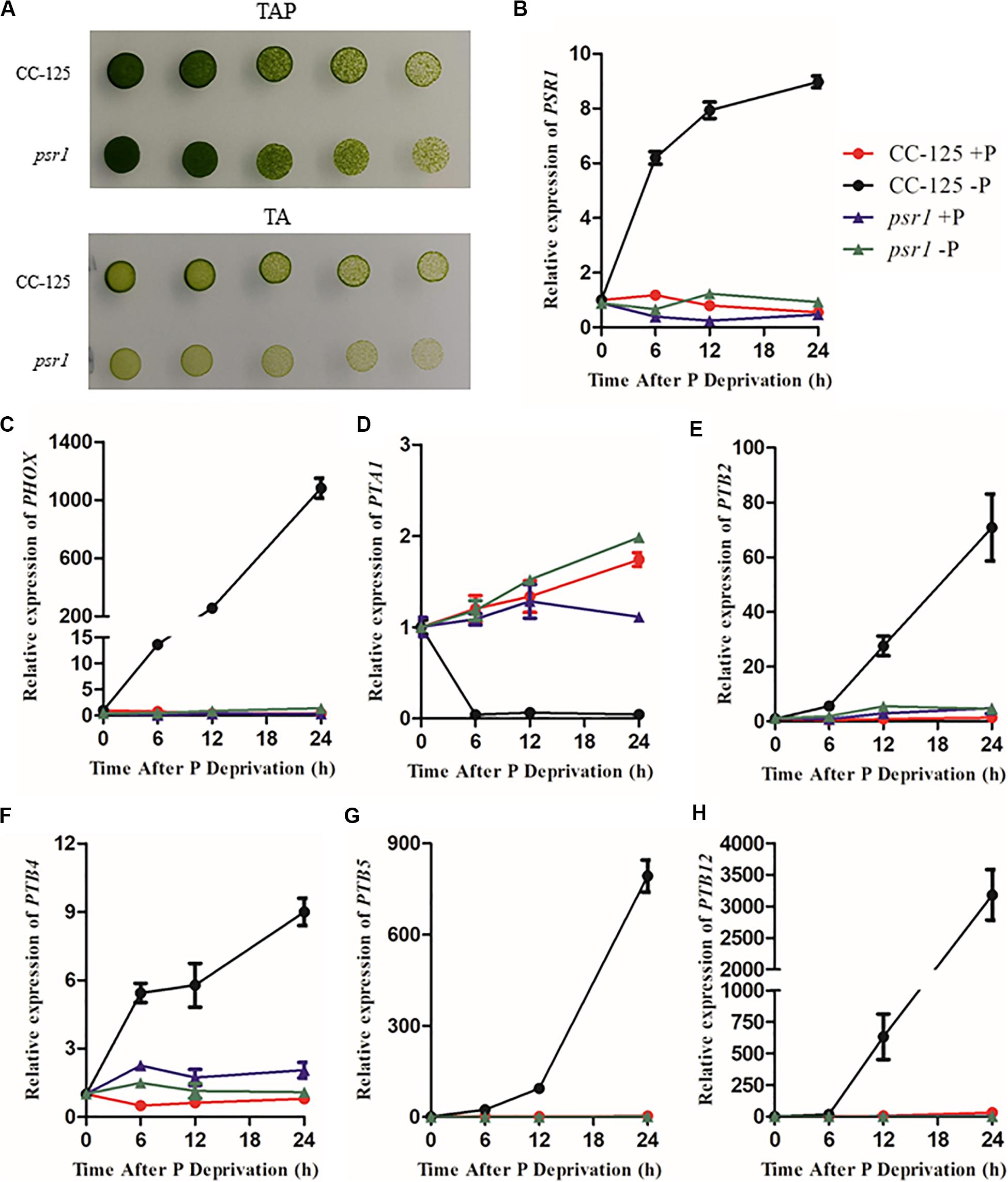
Figure 4. The relative expression of Chlamydomonas PHT genes under phosphate replete and starvation conditions. (A) Growth of CC-125 and psr1 in the TAP or TA plate. (B) to (H), The CrPSR1 (B), CrPHOX (C), CrPTA1 (D), CrPTB2 (E), CrPTB4 (F), CrPTB5 (G), and CrPTB12 (H) mRNA levels were quantified by qRT-PCR over a period of 0, 6, 12, and 24 h of phosphate deprivation. CBLP was used as the internal control. For B–H, data are the means ± s.d. All experiments were repeated three times, and similar results were obtained.
Discussion
Phosphate is one of the most important nutrients that significantly affect plant growth and metabolism (Raghothama, 1999). Although the evolution and phylogenetic relationship of PHTs in different plant species have been reported, none of these studies addressed the possible origin and early evolution of plant PHTs. In this study, we identified members of the PHT family in Chlamydomonas and characterized their evolutionary and expression profiles under P starvation conditions. Interestingly, based on phylogenetic analyses of PHT family members among different plant species, we found that two Pi uptake systems exist in chlorophytes, including H+/Pi symport and Na+/Pi symport (Figure 1), while higher plants only contain a H+/Pi symport (Nussaume et al., 2011). PTB proteins are hypothesized to be Na+/Pi symporters, which facilitate Pi uptake in chlorophytes, whereas PHT1 proteins are H+/Pi symporters that facilitate Pi uptake in angiosperms and all streptophyte lineages (Ayadi et al., 2015; Bonnot et al., 2017). We suggest that the PTA subfamily in algae may also facilitate Pi uptake through H+/Pi symport.
In this study, 25 CrPHT genes were identified in the Chlamydomonas genome, comprising all four major types of plant PHT genes. Chromosome location analysis revealed that almost all the PHT genes were derived from multiple gene duplication events in Chlamydomonas. We found 4 CrPTA genes in Chlamydomonas, in which CrPTA1 and CrPTA3 are through gene duplication. All PTA proteins were predicted to be located in the plasma membrane, similar to the PHT1 in land plants. PHT1 proteins are the well-documented plant Pi transporters. These proteins have a conserved structure containing 12 transmembrane (TM) domains with a large hydrophilic loop between TM6 and TM7 and both hydrophilic N and C termini located in the cytoplasm (Raghothama, 1999; Nussaume et al., 2011). All plant PHT1 proteins have the conserved PHT1 signature GGDYPLSATIxSE and in fungi it is GGDYPLSxxIxSE (Karandashov and Bucher, 2005). In angiosperms, proteins in the PHT1 family catalyze the uptake of Pi from the environment through H+/Pi symport (Ayadi et al., 2015). We propose that the origin of PHT1 genes in land plants is worth more studies, to uncover the mechanism of PHT type bias. All PTB genes were located in chromosomes 2, 7, 12, and 16 and formed 4 clusters. We found a signal peptide in PTB proteins, and all PTB were predicted to the secretory pathway. For example, MpPTB proteins traffic to the plasma membrane through the secretory pathway and function as the plasma membrane PHT for Pi uptake (Bonnot et al., 2017). It indicated that CrPTB may also locate on the plasma membrane and control the Pi uptake in Chlamydomonas.
The topology of the phylogenetic tree showed that the PHT2 subfamily is sister to the bacterial PHT proteins, together with a sister group to the PTB subfamily. We inferred that the PTB protein may move and locate to the chloroplast during evolution because we did not find PHT2 in chlorophyte algae, while in land plants, such as rice, PHT2;1 localized at the chloroplast envelope and functioned as a low-affinity Pi transporter (Liu et al., 2019). PHT3 proteins have two TM α-helices separated by a hydrophilic extramembrane loop, which are conserved in all analyzed mitochondrial transporter proteins, being essential for mitochondrial targeting (Zhu et al., 2012). There are three members in Arabidopsis, as well as in a basic angiosperm, Amborella trichopoda, while six transporters in rice and nine in Physcomitrella patens were clustered with AtPHT3 proteins in the phylogenetic tree (Wang et al., 2017). We only found one PHT3 gene in Chlamydomonas. Therefore, PHT3 family rapidly expanded in land plants, especially in mosses and monocots, through gene duplication during the evolution of plant lineages. In contrast to the rapid expansion of PHT3 in land plants, the members of the PHT4 family are ancient and relatively consistent. The PHT4 family was first characterized and designated by Guo et al. (2008) in Arabidopsis (Guo et al., 2008). There are six members in the PHT4 family in Arabidopsis and seven putative PHT4 members in rice (Wang et al., 2017). PHT4 proteins share a similarity with SLC17/type I Pi transporters. We found nine PHT4 genes in Chlamydomonas which contained 11–13 TM and mainly located in the chloroplast.
Transcription of CrPHT genes varies in response to changes of phosphate levels. We found that, upon Pi starvation, expression of CrPTA1 and CrPTA3 is reduced in the cells. This characteristic makes CrPTA1 and CrPTA3 candidates for the low-affinity Pi uptake system that operates in cells under P-replete conditions. Most Arabidopsis PHT1 family genes, which encode H+/Pi symporters, are up-regulated in phosphate-deprived conditions (Mudge et al., 2002). We also found that P starvation led to enhanced expression levels of CrPTB2, CrPTB3, CrPTB4, CrPTB5, CrPTB6, CrPTB7, and CrPTB8, indicating that they may be high-affinity PHTs operating at low Pi conditions. We also found that the reduction of CrPTA and induction of CrPTB transcription does not occur in psr1 mutant (Figure 4), consistent with that the P1BS element is present in these genes (Table 1). Our results suggest that they may be regulated by CrPSR1.
Materials and Methods
Sequence Collection and Phylogenetic Analyses
To illuminate the evolutionary relationship of PHT gene families in plant lineages, the representative PHT genes from Arabidopsis thaliana (At), Oryza sativa (Os), Amborella trichopoda (Atr), and Volvox carteri (Vc). Physcomitrella patens (Pp), Chlamydomonas reinhardtii (Cr), and Cyanidioschyzon merolae (Cm) were selected for constructing a phylogenetic tree with PHT genes from Escherichia coli (Ec) and Cyanobacteria as outgroup. Multi-sequence alignments were performed by Clustal Omega (Sievers et al., 2011), and a phylogenetic tree was reconstructed by IQTREE using the maximum likelihood method (Nguyen et al., 2015). WAG + F + R6 was the best-fitting evolutionary model selected by ModelFinder under Bayesian Information Criterion (Kalyaanamoorthy et al., 2017). The gene name referred to Liu’s report (Liu et al., 2011).
Sequence Analysis and Structural Characterization
Information of candidate PHT genes in Chlamydomonas was obtained via searching the Phytozome database2 (Goodstein et al., 2012), including chromosome locations, genomic sequences, coding sequences (CDS), and amino acid sequences. Conserved motifs were identified by MEME (Bailey and Elkan, 1994). Molecular weight, theoretical isoelectric point (theoretical pI), and instability index (II; with the value > 40 classified as unstable) of CrPHT proteins were analyzed by the ProtParam tool3. Hydropathy profiles were predicted using TMPRED4. P1BS was identified by the NewPLACE database5 (Higo et al., 1999).
Subcellular Localization and Conserved Motif Analysis of CrPHT
Four Online tools were employed to predict subcellular localization, including Wolf TargetP6 (Emanuelsson et al., 2007), Psort7, MultiLoc8, and YLoc9. The prediction of the signal peptide and TM was performed with the SMART program10. The putative protein sequences were subjected to MEME program11 to investigate conserved motifs with the following parameters: site distribution—any number of repetitions, number of motifs—35, the motif width between 5 and 30 (Bailey and Elkan, 1994).
Chromosome Localization and Gene Duplication
Chromosome mapping of the candidate CrPHT genes was viewed using the software MapDraw V2.1 (Yu et al., 2016). Gene duplication was identified based on plantDGD12 (Qiao et al., 2019).
Expression Profile of CrPHT Genes Under P Stress Conditions
To get an insight into the expression profiles of the CrPHT gene family under P deprivation, we reanalyzed the RNA-seq sequence data in the ArrayExpress database13 under the accession number E-MTAB-2556 (Bajhaiya et al., 2016). In detail, wild-type (CC125) and psr1 mutant cells were cultured until day 3 and day 5 in high-P and low-P TAP media. For the low-P wild-type cells, day 3 and day 5 were equivalent to onset of P starvation and 48 h post-P starvation, respectively. Normalized log2 values of expression levels are shown in Supplementary Table 2.
Algae Growth Conditions, RNA Extraction, cDNA Synthesis, and Expression Analysis
The C. reinhardtii WT strain CC-125 and CC-4350 (Crpsr1-1) mutants were obtained from the Chlamydomonas Resource Center (Li et al., 2016). Cells were cultured in standard Tris–acetate-phosphate TAP medium at pH 7.0 under continuous illumination (50 mmol photons m–2 s–1) on a rotating platform (150 rpm) at 24°C. For Pi deprivation, cells in the mid-logarithmic phase (5–8 × 106 cells mL–1) were pelleted by centrifugation (2,000 g, 5 min), washed twice with TA in which 1.5 mM potassium chloride which was substituted for 1 mM potassium phosphate (Quisel et al., 1996), and then resuspended in TA medium.
Total RNA was extracted from frozen cell pellets using the RNeasy Mini Kit (Qiagen) and reverse transcribed to complementary DNA after DNase I treatment following the manufacturer’s instructions (NEB). Quantitative real-time PCR was performed using a SYBR Premix kit (Roche) on a QuantStudio 6 Flex machine (Life Technologies). The CBLP gene was used as an internal control (Chang et al., 2005). The primer pairs used for RT-qPCR are given in the Supplementary Table 3.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
YZ and HYZ conceived and supervised the project. LW, LX, YZ, and HYZ designed the research. LW and LX performed the experiments. LW, HY, LX, GC, and HZ analyzed the data. LW, HZ, and HYZ wrote the manuscript. All authors discussed the results and commented on the manuscript.
Funding
This work was supported by the National Key R&D Program of China (2017YFD0200200/0200204/0200206) and National Natural Science Foundation (31801924).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Dr. Lei Xu for helpful discussion.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2020.590947/full#supplementary-material
Supplementary Table 1 | List of gene duplication pairs identified in CrPHT genes.
Supplementary Table 2 | Expression level of CrPHT genes under phosphate starvation in the WT and psr1 mutant.
Supplementary Table 3 | List of all primers used in this study.
Footnotes
- ^ http://www.ebi.ac.uk/arrayexpress
- ^ https://phytozome.jgi.doe.gov/pz/portal.html
- ^ http://web.expasy.org/protparam/
- ^ http://www.ch.embnet.org/software/TMPRED_form.html
- ^ http://sogo.dna.affrc.go.jp/cgi-bin/
- ^ http://www.cbs.dtu.dk/services/TargetP/
- ^ http://www.genscript.com/psort/wolf_psort.html
- ^ http://abi.inf.unituebingen.de/Services/MultiLoc2
- ^ https://abi-services.informatik.uni-tuebingen.de/yloc/webloc.cgi
- ^ http://smart.embl-heidelberg.de/smart/batch.pl
- ^ http://meme-suite.org/tools/meme
- ^ http://pdgd.njau.edu.cn:8080
- ^ http://www.ebi.ac.uk/arrayexpress
References
Ayadi, A., David, P., Arrighi, J. F., Chiarenza, S., Thibaud, M. C., Nussaume, L., et al. (2015). Reducing the genetic redundancy of Arabidopsis PHOSPHATE TRANSPORTER1 transporters to study phosphate uptake and signaling. Plant Physiol. 167, 1511–1526. doi: 10.1104/pp.114.252338
Bailey, T. L., and Elkan, C. (1994). Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2, 28–36.
Bajhaiya, A. K., Dean, A. P., Zeef, L. A., Webster, R. E., and Pittman, J. K. (2016). PSR1 is a global transcriptional regulator of phosphorus deficiency responses and carbon storage metabolism in Chlamydomonas reinhardtii. Plant Physiol. 170, 1216–1234. doi: 10.1104/pp.15.01907
Bonnot, C., Proust, H., Pinson, B., Colbalchini, F. P. L., Lesly-Veillard, A., Breuninger, H., et al. (2017). Functional PTB phosphate transporters are present in streptophyte algae and early diverging land plants. New Phytol. 214, 1158–1171. doi: 10.1111/nph.14431
Bustos, R., Castrillo, G., Linhares, F., Puga, M. I., Rubio, V., Perez-Perez, J., et al. (2010). A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis. PLoS Genet. 6:e001102. doi: 10.1371/journal.pgen.1001102
Chang, C. W., Moseley, J. L., Wykoff, D., and Grossman, A. R. (2005). The LPB1 gene is important for acclimation of Chlamydomonas reinhardtii to phosphorus and sulfur deprivation. Plant Physiol. 138, 319–329. doi: 10.1104/pp.105.059550
Chrispeels, M. J., and Raikhel, N. V. (1992). Short peptide domains target proteins to plant vacuoles. Cell 68, 613–616. doi: 10.1016/0092-8674(92)90134-x
Emanuelsson, O., Brunak, S., Von Heijne, G., and Nielsen, H. (2007). Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2, 953–971. doi: 10.1038/nprot.2007.131
Goodstein, D. M., Shu, S., Howson, R., Neupane, R., Hayes, R. D., Fazo, J., et al. (2012). Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40, D1178–D1186.
Guo, B., Jin, Y., Wussler, C., Blancaflor, E. B., Motes, C. M., and Versaw, W. K. (2008). Functional analysis of the Arabidopsis PHT4 family of intracellular phosphate transporters. New Phytol. 177, 889–898. doi: 10.1111/j.1469-8137.2007.02331.x
Higo, K., Ugawa, Y., Iwamoto, M., and Korenaga, T. (1999). Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 27, 297–300. doi: 10.1093/nar/27.1.297
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., Von Haeseler, A., and Jermiin, L. S. (2017). ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589. doi: 10.1038/nmeth.4285
Karandashov, V., and Bucher, M. (2005). Symbiotic phosphate transport in arbuscular mycorrhizas. Trends Plant Sci. 10, 22–29. doi: 10.1016/j.tplants.2004.12.003
Kobayashi, I., Fujiwara, S., Shimogawara, K., Kaise, T., Usuda, H., and Tsuzuki, M. (2003). Insertional mutagenesis in a homologue of a Pi transporter gene confers arsenate resistance on Chlamydomonas. Plant Cell Physiol. 44, 597–606. doi: 10.1093/pcp/pcg081
Kumar Sharma, A., Muhlroth, A., Jouhet, J., Marechal, E., Alipanah, L., Kissen, R., et al. (2020). The Myb-like transcription factor phosphorus starvation response (PtPSR) controls conditional P acquisition and remodelling in marine microalgae. New Phytol. 225, 2380–2395. doi: 10.1111/nph.16248
Li, X., Zhang, R., Patena, W., Gang, S. S., Blum, S. R., Ivanova, N., et al. (2016). An indexed, mapped mutant library enables reverse genetics studies of biological processes in Chlamydomonas reinhardtii. Plant Cell 28, 367–387. doi: 10.1105/tpc.15.00465
Liu, F., Chang, X.-J., Ye, Y., Xie, W.-B., Wu, P., Lian, X.-M., et al. (2011). Comprehensive sequence and whole-life-cycle expression profile analysis of the phosphate transporter gene family in rice. Mol. Plant. 4, 1105–1122. doi: 10.1093/mp/ssr058
Liu, X. L., Wang, L., Wang, X. W., Yan, Y., Yang, X. L., Xie, M. Y., et al. (2019). Mutation of the chloroplast-localized phosphate transporter OsPHT2;1 reduces flavonoids accumulation and UV tolerance in rice. Plant J. 102, 53–67. doi: 10.1111/tpj.14611
Lorenzo-Orts, L., Couto, D., and Hothorn, M. (2020). Identity and functions of inorganic and inositol polyphosphates in plants. New Phytol. 225, 637–652. doi: 10.1111/nph.16129
Mimura, T., Reid, R. J., Ohsumi, Y., and Smith, F. A. (2002). Induction of the Na+/Pi cotransport system in the plasma membrane of Chara corallina requires external Na+ and low levels of Pi. Plant Cell Environ. 25, 1475–1481. doi: 10.1046/j.1365-3040.2002.00921.x
Moseley, J. L., Chang, C. W., and Grossman, A. R. (2006). Genome-based approaches to understanding phosphorus deprivation responses and PSR1 control in Chlamydomonas reinhardtii. Eukaryot. Cell 5, 26–44. doi: 10.1128/ec.5.1.26-44.2006
Moseley, J. L., Gonzalez-Ballester, D., Pootakham, W., Bailey, S., and Grossman, A. R. (2009). Genetic interactions between regulators of Chlamydomonas phosphorus and sulfur deprivation responses. Genetics 181, 889–905. doi: 10.1534/genetics.108.099382
Mudge, S. R., Rae, A. L., Diatloff, E., and Smith, F. W. (2002). Expression analysis suggests novel roles for members of the Pht1 family of phosphate transporters in Arabidopsis. Plant J. 31, 341–353. doi: 10.1046/j.1365-313x.2002.01356.x
Nguyen, L. T., Schmidt, H. A., Von Haeseler, A., and Minh, B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Nussaume, L., Kanno, S., Javot, H., Marin, E., Pochon, N., Ayadi, A., et al. (2011). Phosphate import in plants: focus on the PHT1 transporters. Front. Plant Sci. 2:83. doi: 10.3389/fpls.2011.00083
Qiao, X., Li, Q., Yin, H., Qi, K., Li, L., Wang, R., et al. (2019). Gene duplication and evolution in recurring polyploidization-diploidization cycles in plants. Genome Biol. 20:38.
Quisel, J. D., Wykoff, D. D., and Grossman, A. R. (1996). Biochemical characterization of the extracellular phosphatases produced by phosphorus-deprived Chlamydomonas reinhardtii. Plant Physiol. 111, 839–848. doi: 10.1104/pp.111.3.839
Raghothama, K. G. (1999). Phosphate acquisition. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 665–693.
Reid, R. J., Mimura, T., Ohsumi, Y., Walker, N. A., and Smith, F. A. (2000). Phosphate uptake in chara: membrane transport via Na/Pi cotransport. Plant Cell Environ. 23, 223–228. doi: 10.1046/j.1365-3040.2000.00524.x
Shimogawara, K., Wykoff, D. D., Usuda, H., and Grossman, A. R. (1999). Chlamydomonas reinhardtii mutants abnormal in their responses to phosphorus deprivation. Plant Physiol. 120, 685–694. doi: 10.1104/pp.120.3.685
Sievers, F., Wilm, A., Dineen, D., Gibson, T. J., Karplus, K., Li, W. Z., et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7:539. doi: 10.1038/msb.2011.75
Sun, L., Song, L., Zhang, Y., Zheng, Z., and Liu, D. (2016). Arabidopsis PHL2 and PHR1 act redundantly as the key components of the central regulatory system controlling transcriptional responses to phosphate starvation. Plant Physiol. 170, 499–514. doi: 10.1104/pp.15.01336
Wang, D., Lv, S., Jiang, P., and Li, Y. (2017). Roles, regulation, and agricultural application of plant phosphate transporters. Front. Plant Sci. 8:817. doi: 10.3389/fpls.2011.00817
Wang, Z. Y., Ruan, W. Y., Shi, J., Zhang, L., Xiang, D., Yang, C., et al. (2014). Rice SPX1 and SPX2 inhibit phosphate starvation responses through interacting with PHR2 in a phosphate-dependent manner. Proc. Natl. Acad. Sci. U.S.A. 111, 14953–14958. doi: 10.1073/pnas.1404680111
Wykoff, D. D., Grossman, A. R., Weeks, D. P., Usuda, H., and Shimogawara, K. (1999). Psr1, a nuclear localized protein that regulates phosphorus metabolism in Chlamydomonas. Proc. Natl. Acad. Sci. U.S.A. 96, 15336–15341. doi: 10.1073/pnas.96.26.15336
Yu, J., Cheng, Y., Feng, K., Ruan, M., Ye, Q., Wang, R., et al. (2016). Genome-wide identification and expression profiling of tomato Hsp20 gene family in response to biotic and abiotic stresses. Front. Plant Sci. 7:1215. doi: 10.3389/fpls.2011.01215
Zhou, J., Jiao, F., Wu, Z., Li, Y., Wang, X., He, X., et al. (2008). OsPHR2 is involved in phosphate-starvation signaling and excessive phosphate accumulation in shoots of plants. Plant Physiol. 146, 1673–1686. doi: 10.1104/pp.107.111443
Keywords: phosphate transporters, phylogenetic analysis, expression pattern, Chlamydomonas, green algae
Citation: Wang L, Xiao L, Yang H, Chen G, Zeng H, Zhao H and Zhu Y (2020) Genome-Wide Identification, Expression Profiling, and Evolution of Phosphate Transporter Gene Family in Green Algae. Front. Genet. 11:590947. doi: 10.3389/fgene.2020.590947
Received: 03 August 2020; Accepted: 07 September 2020;
Published: 08 October 2020.
Edited by:
Hongjian Wan, ZheJiang Academy of Agricultural Sciences, ChinaReviewed by:
Jian Yang, Sichuan University, ChinaZhong-Chang Wu, Zhejiang University, China
Fuquan Liu, Queen’s University Belfast, United Kingdom
Copyright © 2020 Wang, Xiao, Yang, Chen, Zeng, Zhao and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yiyong Zhu, Yiyong1973@njau.edu.cn; Hongyu Zhao, zhy200869@163.com
†These authors have contributed equally to this work
 Long Wang
Long Wang Liang Xiao1†
Liang Xiao1† Houqing Zeng
Houqing Zeng Yiyong Zhu
Yiyong Zhu