- Department of Etiology, Shanxi Cancer Hospital, Taiyuan, China
In view of the highly increased prevalence of papillary thyroid carcinoma (PTC) year by year, it is of great importance to explore new molecular targets for anticancer strategies. Emerging evidence indicates that circular RNAs (circRNAs), characterized by a closed-loop structure and high stability, play important roles in tumorigenesis and development of human cancer by regulating multiple complex biological processes, such as cellular proliferation, metastasis, and metabolism. A comprehensive understanding of the roles of circRNAs will facilitate the development of promising future therapeutic strategies for treating cancers, including PTC. In this paper, we review the profile of circRNA in PTC, its regulatory roles, and the pathological mechanism as well as their related clinical significance. In addition, challenges of this specific field are discussed.
Introduction
Thyroid cancer (TC) is the most prevalent endocrine malignancy, accounting for nearly one third of the total head and neck malignancies globally (1, 2). Among all cases, 80%–85% of them are papillary thyroid carcinoma (PTC) (3). Although the overall 5-year survival rate of PTC can reach 97%, the 5-year survival rate of patients with advanced PTC is only 59% (4). PTC can still be life threatening and causes poor prognosis due to its invasiveness and metastasis. Extensive efforts have been conducted on research of the carcinogenesis, progression, and effective therapeutic methods of TC. Despite advances in clinical management, including surgery, radiotherapy, levothyroxine treatment, and target therapy, promising and optimal molecular therapies remain to be further explored. In addition to the DNA mutations, such as the BRAFV600E mutation, which was discovered previously, accumulating evidence indicates that non-coding RNAs (ncRNAs) also participate in the progression and pathogenesis of PTC (5–7). Among them, circular RNAs (circRNAs) have attracted increasing attention. Optimistic exploration of PTC-related circRNA likely will be beneficial to pave the way to improve clinical management.
CircRNAs are a newly identified subclass of ncRNA family, and they are produced cotranscriptionally by the spliceosome at the expense of canonical mRNA isoforms, forming a head-to-tail backsplice characterized by a covalently nonlinear, closed-loop structure that lacks either 5’ to 3’ polarity or a polyadenylated tail (8). Based on the biogenesis of circRNAs in human cells, they are usually classified into three types: exonic circRNAs (ecircRNAs), which are generated from the exons of pre-mRNAs; intronic circRNAs (ciRNAs), which are produced from the intronic region in the pre-mRNAs; and exon-intron circRNAs (EIciRNAs), which consist of both exons and introns from the pre-mRNAs. Due to their closed structures, circRNAs are resistant to RNA degradation and more stable than linear RNA. Emerging evidence shows that dysregulation of circRNAs play important roles in promoting tumorigenesis and tumor progression (9). It is demonstrated that circRNAs serve as competitive endogenous RNAs (ceRNAs) or microRNA sponges, compete with microRNAs (miRNAs), and consequently regulate the target gene expression (10). Furthermore, circRNAs are also involved in various physiological and pathophysiological processes, such as modulating alternative splicing (11) and regulating protein–RNA interactions (12). Previous research have profiled the circRNAs expression of PTC and have found significantly differentiated circRNAs in PTC compared with normal thyroid tissue, which may be involved in the pathogenesis of PTC. In the following sections, we highlight the results of recent research efforts, including the profile of markedly dysregulated circRNAs and their related regulatory networks and clinical significance in PTC as well as the current challenges in the field.
Profiled circRNAs and Its Role in PTC
Expression and Biological Function of circRNAs in PTC
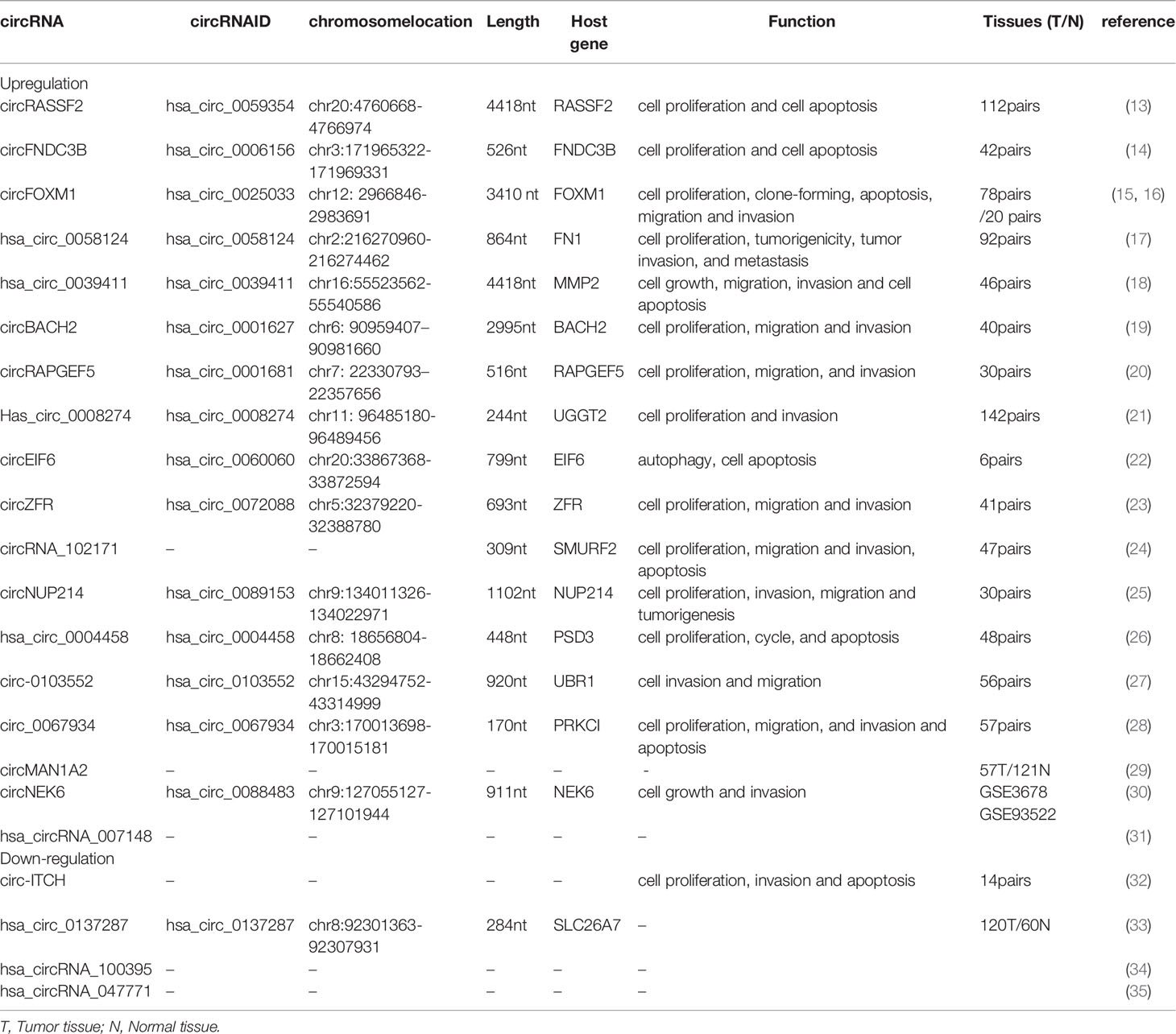
To date, many different circRNAs have been found either upregulated or downregulated in PTC tissues compared with matched adjacent normal tissues (Table 1). In line with tissue expression level, most PTC-related circRNAs are dysregulated in corresponding PTC cell lines versus in normal thyroid cell lines. Based on gain- and loss-function experiments in vivo and in vitro, each identified circRNA displays significantly altered tumor cell biological behavior or cell phenotype in PTC cell lines, such as cell proliferation, cell cycle, apoptosis, migration, and invasion (Table 1), suggesting that the particular circRNA may act as an oncogenic driver or a tumor suppressor. Take cell-cycle regulation as an example; knockdown of circRASSF2, circFNDC3B, and circFOXM1 caused, respectively, significant G1 phase cell-cycle arrest of TPC-1 cells (p < 0.01, p < 0.01, and p < 0.01, respectively). Silenced circRNA_102171 caused G2 phase arrest, and si-circ_0004458 displayed S phase reduction. In contrast, enhanced circRASSF2 expression increased the G2 phase percentage and decreased the G1 phase percentage of K1 cells (p < 0.01). Overexpression of circFNDC3B increased the S-phase percentage and decreased the G0/G1 phase percentage of K1 cells (p < 0.01). circFOXM1 expression increased the S phase percentage and decreased the G0/G1 phase percentage of K1 cells (p < 0.01). On the basis of this series of functional experiments, circRNAs were confirmed to play oncogenic or inhibitory roles in PTC.
Biogenesis, Stability, and Subcellular Location of Profiled circRNAs in PTC
Many reports demonstrate that circRNAs are spliced and derived from the host genes (Table 1), and even some circRNAs may impact the mRNA expression level of their host genes. As shown in Table 1, characteristics of circRNAs are represented, including circRNA ID (http://www.circbase.org), chromosome position, spliced length, and host gene. Among them, most circRNAs are classified as ecircRNAs, such as hsa_circ_0006156 (14), Hsa_circ_0058124 (16), CircBACH2 (19), hsa_circ_0001681 (20), CircRNA_102171 (24), and hsa_circ_0004458 (26). Exceptionally, CircNEK6 is a kind of exonic circRNA encoding the mRNA NEK6. In addition, some circRNAs are not found in circBase because of limited information in current reports, including hsa_circRNA_100395, hsa_circRNA_047771, hsa_circRNA_007148, and circ-ITCH, circMAN1A2.
Generally, stability of the circRNA is critical for exerting its function. Analysis of stability for circRNA and its host gene in PTC cells, treated with transcription inhibitor actinomycin D, reveals that the half-life of circRASSF2 exceeds 24 h, whereas that of RASSF2 mRNA is only about 3 h in TPC-1 cells (13). Similarly, the half-life of circFNDC3B and circFOXM1 transcript exceeds 24 h, much more stable than the corresponding host genes FNDC3B and FOXM1 (14, 15), respectively. Furthermore, circRNA is resistant to RNase R digestion. This proves that circRNAs are extremely more stable than their mRNA level. Given their stability, circRNAs are appropriate for future clinical applications for PTC.
In addition, subcellular location may be related to the distinct molecular roles of various kinds of circRNAs in cells. EcircRNAs are predominantly localized in the cytoplasm (35), and ciRNAs and ElciRNAs are preferred in the nucleus (36). Subcellular location by cell fraction assay and FISH analysis indicates that circFNDC3B (14), circBACH2 (19), circRAPGEF5 (20), and circNUP214 (25) are predominantly localized in the cytoplasm of PTC cells, and hsa_circ_0058124 primarily appears in the nucleus and also exists in cytoplasm (17). In brief, it is essential to get the properties of circRNAs to facilitate the following pathological mechanism.
Pathological Mechanism of circRNAs in PTC
CircRNAs are widely involved in human physiological and pathological processes and can be used in various manners (37), including (1) serving as microRNA (miRNA) or protein sponges; (2) interacting with proteins, such as recruiting specific proteins, enhancing protein function, and functioning as protein scaffolding; and (3) translating into peptides. Highly abundant circRNAs have been found to contain many competing miRNA binding sites. Therefore, they can be used as RNA “sponges” to cooperatively adsorb miRNAs, thereby regulating the expression of downstream target genes that are inhibited by miRNAs through competing with endogenous RNAs (38). In cancer research, the use of circRNAs as miRNA sponges to regulate downstream target genes is widely reported.
CircRNA Serves as ceRNA Involved in PTC Progression
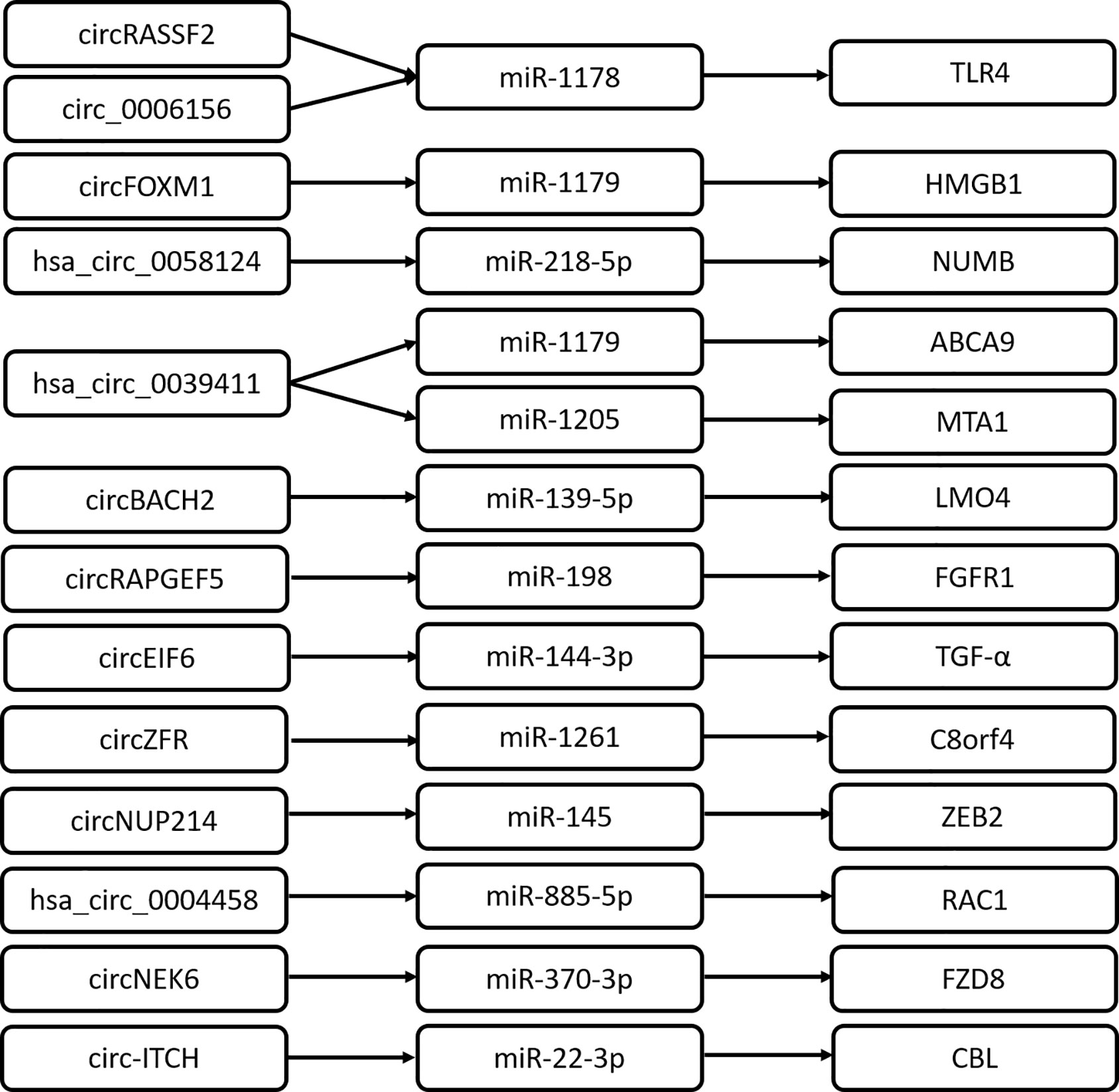
CircRNAs are important transcriptional regulators of gene expression, relieving the association between miRNA and target genes involved in the pathogenesis of various diseases. It is reported that circRNAs could act as miRNA sponges and regulate the expression of downstream target genes. Previous studies show that an increasing number of circRNA/miRNA/mRNA axes are identified to promote PTC progression (Figure 1). This well depicts the interactional network between circRNA and RNA for a better understanding of the transcriptional regulation mechanism by circRNA. Of note, some RNA regulatory network mediated by circRNA remains to be further improved. For example, circ_0025033/miR-1231 and miR-1304 (16), circ-0103552/miR-127 (27), hsa_circ100395/miR-141-3p/miR-200a-3p (34).
Signaling Pathway Modulated by PTC-Related circRNAs
CircRNAs also exert their regulatory roles to modulate signaling pathways in cancer, for instance, the wnt/β-catenin signaling pathway (39–40), AMPK/mTOR signaling pathway (41), PI3K/AKT signaling pathways (42–44), and NOTCH pathway (45, 46). As a classical pathway, the wnt signaling pathway is involved in many phases of vertebrate embryonic development and contributes to tumorigenesis. Its aberrant activation could facilitate the progression of various human cancers. CircRNA_102171 directly interacts with CTNNBIP1 and impairs the formation of CTNNBIP1/β-catenin complex (24). Consequently, circRNA_102171 promotes the interaction of β-catenin with TCF proteins; significantly enhances the expression of corresponding target genes, such as CCND1, CCND2, MYC, and SOX4; and activates the Wnt/β-catenin pathway in a CTNNBIP1-dependent manner (24). Frizzled class receptor 8 (FZD8) is reported to be one of the cell surface receptors of the Wnt signaling pathway, which belongs to the Frizzled family of serpentine proteins. Chen F et al. find that circNECK6 binds target miR-370-3p to inhibit FZD8 degradation and the upregulated FZD8 activates the wnt signaling pathway (30). Wang M et al. reveals a novel mechanism regulating the wnt pathway by circRNA (32). Circ-ITCH sponges miR-22-3p to elevate CBL (an E3 ligase of nuclear β-catenin) expression, which leads to the inactivation of the Wnt/β-catenin pathway and consequently attenuates PTC progression. Moreover, Yao Y et al. reports that hsa_circ_0058124 plays an oncogenic driver in PTC by downregulating the NOTCH3 signaling pathway. hsa_circ_0058124 may exert its biological effects in PTC through hsa_circ_0058124/miR-218-5p/NUMB, subsequently with repression of the NOTCH3/GATAD2A axis because NUMB is a strong suppressor of the NOTCH pathway (17). Collectively, circRNAs modulate various pathways to activate the PTC progression program.
circRNAs Act as Tumor Biomarkers in PTC
The Relationship Between circRNAs and Clinicopathological Parameters in PTC
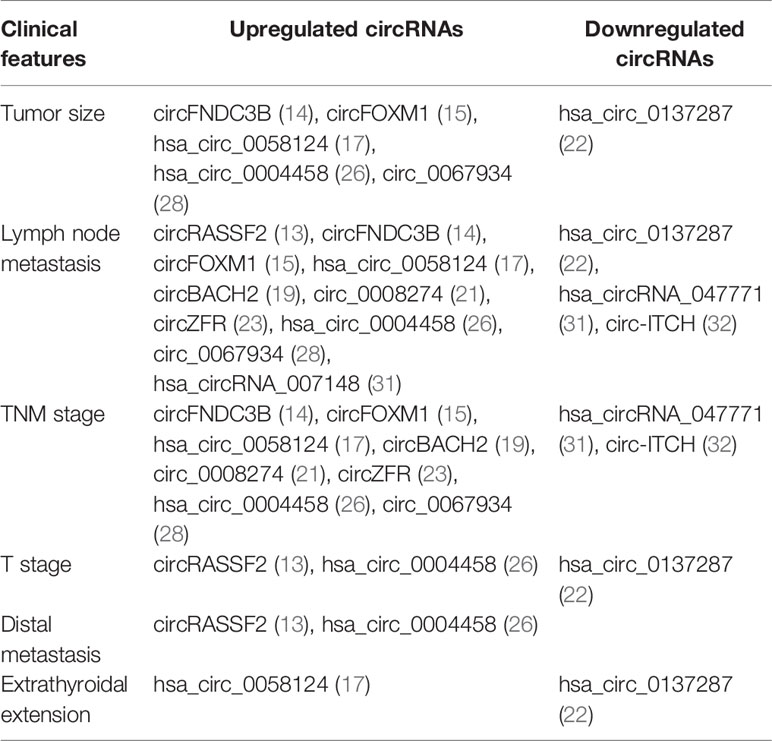
Clinical analysis reveals that dysregulated circRNAs correlate with aggressive clinicopathological characteristics of PTC, including tumor size, TNM stage, lymph node metastasis, T stage, distal metastasis, and extrathyroidal extension (Table 2). Among them, highly expressed circRASSF2 (13), circFNDC3B (14), circFOXM1 (15), hsa_circ_0058124 (17), circBACH2 (19), circ_0008274 (21), circZFR (23), hsa_circ_0004458 (26), circ_0067934 (28), and hsa_circRNA_007148 (31) positively correlate with a few aggressive features, whereas lower levels of hsa_circ_0137287 (22), hsa_circRNA_047771 (31), and circ-ITCH (32) negatively correlate with some clinical features. Of note, tumor size is classified by different groups in different research. For example, downregulation of hsa_circ_0137287 correlates with tumor size >2 cm. Upregulated circ_0067934 and circ_0006156 correlate with tumor size >1 cm, hsa_circ_0004458 with tumor size ≥3 cm, hsa_circ_0058124 with tumor size >2 cm, and circFOXM1 with tumor size >3 cm. Generally, PTC is often combined with other types of thyroid disease, such as Hashimoto’s thyroiditis (HT), nodular goiter (NG), and so on. It is reported that the level of circFOXM1 is significantly associated with NG (P = 0.009) (15). In addition, a great deal of previous research indicates that the BRAFV600E mutation is identified as an essential genetic factor in PTC progression. The BRAFV600E mutation, which can cause activation of MAPK pathway signaling, is significantly associated with more aggressive characteristics of PTCs and facilitates risk stratification and the management of patients with thyroid nodules. A decreased hsa_circRNA_047771 expression level is associated with the BRAFV600E mutation (P < 0.05) (31). Collectively, the association between circRNAs and aggressive clinicopathological characteristics supports that circRNAs can serve as prognostic factors for PTC patients.
Diagnostic Value of PTC-Related circRNAs
Pathological diagnosis is a gold standard method for the preoperative evaluation of thyroid nodules; however, cytology remains indeterminate for up to 30% of nodules that cannot be definitively diagnosed (47). Except for the BRAFV600E mutation, a novel molecular biomarker is required in favor of clinical diagnosis and risk stratification, especially for efficient management of cN0 papillary thyroid microcarcinoma (PTMC). Extensive exploration in recent years reveals that ncRNAs, such as miRNAs, lncRNAs, and circRNAs, could function as a promising diagnostic biomarker for PTC patients (48, 49). A receiver operating characteristic (ROC) curve was used to evaluate the diagnostic value of circRNAs in PTC tissues compared with paratumor tissues, and it was found that the area under the ROC curve (AUC) of circFNDC3B was 0.891 (95% CI = 0.820–0.961, P < 0.0001) (14) and of circBACH2 was 0.8631 (95% CI = 0.7774–0.9489, P < 0.0001) (19). More importantly, circRNAs also serve as postsurgical diagnostic biomarkers. Lan X et al. find that hsa_circ_0137287 has a potential diagnostic value in predicting malignancy (AUC = 0.8973, 95% CI = 0.8452–0.9494, P < 0.0001), extrathyroidal extension (AUC = 0.6885, 95% CI = 0.5908–0.7862, P = 0.0009), and lymph node metastasis (AUC = 0.6691, 95% CI = 0.5641–0.7742, P = 0.0034), respectively (33). Additionally, hsa_circRNA_047771 (AUC = 0.876, 95% CI = 0.78–0.94, sensitivity = 87.5%, specificity = 80.0%) and hsa_circRNA_007148 (AUC = 0.846, 95% CI = 0.75–0.96, sensitivity = 82.5%, specificity = 77.5%) may be candidate diagnostic biomarkers for PTC (31). In view of, so far, limited exploration, further studies are required to discover more optimal biomarkers for diagnosis of PTC.
Predicting Roles of circRNAs for Prognosis in PTC
Previous follow-up studies indicate that most PTC patients have a good prognosis: 85% of PTC cases are highly curable for innocent biological behavior. However, it is necessary to carefully observe the recurrence and metastasis, especially for advanced PTC patients. As with other coding genes (BRAFV600E, RAS, etc.) and noncoding genes (miRNA, lncRNA, etc.), circRNAs may be potential predictors for prognosis of PTC. Kaplan–Meier survival curve analysis reveals that PTC patients with low expression of circFNDC3B display obviously longer overall survival (OS) times than those with high expression of circFNDC3B (P < 0.05) (14). Similar to circFNDC3B, downregulated circBACH2 had relatively longer OS (P < 0.05) (19), a higher expression of circZFR in PTC patients is correlated with worse prognosis (23), and patients with high expression of circ_0067934 show lower survival rates (28). Moreover, Cox proportional hazards regression model analysis also indicates that circ_0067934 is an independent risk factor for prognosis (RR = 4.385, 95% CI = 1.087–17.544, P = 0.038) (28), like the circ-ITCH as well (32). More importantly, it is necessary to monitor relapse and progression by reliable biomarkers in long-term follow-up studies. In addition, the relationship between circRNAs, such as circFND3B, circBACH2, and circZFR, and prognosis-predicting roles reveals that it is insufficient to confirm its predicting role for prognosis due to limited survival analysis. Maybe it will be more convincing if performing further analysis by Cox proportional hazards regression models. Even the researcher could observe the relationship between circRNA and recurrence and metastasis in PTC for fine management of PTC, to fully elucidate the prognostic value of circRNAs for PTC.
Challenges and Prospects
To date, a handful of ncRNAs have been identified, and many have shown oncogenic or tumor-suppressive roles in human cancer, especially lncRNAs and circRNAs. However, it is just like the tip of an iceberg. Despite advances in the relationship between circRNAs and PTC, current research still has a few limitations. For example, the sample size and histological types of TC are limited. Except for circEIF6 (22), most TC-related circRNA research does not include other TCs such as anaplastic thyroid carcinoma (ATC) and medullary thyroid carcinoma (MTC) due to their low incidence. However, it is necessary to explore further by prolonging the observation period and performing multicenter clinical studies.
Furthermore, the molecular mechanism of circRNAs in the PTC pathological process needs to be further clarified to establish RNA regulatory networks. Currently, most studies focus on the “molecular sponge” function or ceRNA role of certain circRNAs. According to ceRNA theory, artificial circRNAs engineered with diverse methods can act as potential and promising therapeutic molecular tools. Nevertheless, circRNAs represent diversity in functions. Therefore, other functions of circRNAs in TC should be explored for a more comprehensive landscape and better understanding of the mechanism in the future, such as alternative splicing, regulation of gene transcription, and crosstalk with RBPs. More importantly, it needs a series of sufficient and logically scientific proofs outside of the molecular mechanism research for a reliable but not farfetched explanation.
Additionally, in view of the clinical applications of circRNAs, further studies should pay more attention to evaluating the diagnostic and prognostic value of circRNAs and the associations with clinical drug resistance. Notably, few reports examine PTC-related circRNAs involved in this field. Liu F et al. demonstrates that circEIF6 associates with chemo-resistance (cisplatin-resistance) by influencing cell autophagy (22). More importantly, circRNAs could be secreted into blood, saliva (50), and even exosomes (51), which play important roles in the tumor microenvironment, suggesting that the circRNA level in body liquid and FNAB samples could facilitate clinical management, such as serum circMAN1A2 (29), serum exosomal circRASSF2 (13), and circ_0006156 (14).
Taken together, it is expected to identify more promising RNA signatures and unveil the underlying mechanism of circRNAs for better understanding of the etiology and pathological progress in TC, which sheds light on the potential applications of circRNAs for translational medicines.
Author Contributions
XX drafted the manuscript. JJ supervised and revised the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Kitahara CM, Sosa JA. The changing incidence of thyroid cancer. Nat Rev Endocrinol (2016) 12(11):646–53. doi: 10.1038/nrendo.2016.110
2. Cabanillas ME, McFadden DG, Durante C. Thyroid cancer. Lancet (2016) 388(10061):2783–95. doi: 10.1016/S0140-6736(16)30172-6
3. Fagin JA, Wells SA Jr. Biologic and clinical perspectives on thyroid Cancer. N Engl J Med (2016) 375(11):1054–67. doi: 10.1056/NEJMra1501993
4. Kunavisarut T. Diagnostic biomarkers of differentiated thyroid cancer. Endocrine (2013) 44(3):616–22. doi: 10.1007/s12020-013-9974-2
5. Tallini G, de Biase D, Durante C, Acquaviva G, Bisceglia M, Bruno R, et al. BRAF V600E and risk stratification of thyroid microcarcinoma: a multicenter pathological and clinical study. Mod Pathol (2015) 28(10):1343–59. doi: 10.1038/modpathol.2015.92
6. Fuziwara CS, Kimura ET. MicroRNAs in thyroid development, function and tumorigenesis. Mol Cell Endocrinol (2016) 456:44–50. doi: 10.1016/j.mce.2016.12.017
7. Liyanarachchi S, Li W, Yan P, Bundschuh R, Brock P, Senter L, et al. Genome-Wide Expression Screening Discloses Long Noncoding RNAs Involved in Thyroid Carcinogenesis. J Clin Endocrinol Metab (2016) 101(11):4005–13. doi: 10.1210/jc.2016-1991
8. Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol (2014) 32(5):453–61. doi: 10.1038/nbt.2890
9. He J, Xie Q, Xu H, Li J, Li Y. Circular RNAs and cancer. Cancer Lett (2017) 396:138–44. doi: 10.1016/j.canlet.2017.03.027
10. Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet (2016) 17(5):272–83. doi: 10.1038/nrg.2016.20
11. Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell (2014) 56(1):55–66. doi: 10.1016/j.molcel.2014.08.019
12. Ebbesen KK, Kjems J, Hansen TB. Circular RNAs: identification, biogenesis and function. Biochim Biophys Acta (2016) 1859(1):163–8. doi: 10.1016/j.bbagrm.2015.07.007
13. Wu G, Zhou W, Lin X, Sun Y, Li J, Xu H, et al. circRASSF2 Acts as ceRNA and Promotes Papillary Thyroid Carcinoma Progression through miR-1178/TLR4 Signaling Pathway. Mol Ther Nucleic Acids (2020) 19:1153–63. doi: 10.1016/j.omtn.2019.11.037
14. Wu G, Zhou W, Pan X, Sun Z, Sun Y, Xu H, et al. Circular RNA Profiling Reveals Exosomal circ_0006156 as a Novel Biomarker in Papillary Thyroid Cancer. Mol Ther Nucleic Acids (2020) 19:1134–44. doi: 10.1016/j.omtn.2019.12.025
15. Ye M, Hou H, Shen M, Dong S, Zhang T. Circular RNA circFOXM1 Plays a Role in Papillary Thyroid Carcinoma by Sponging miR-1179 and Regulating HMGB1 Expression. Mol Ther Nucleic Acids (2020) 19:741–50. doi: 10.1016/j.omtn.2019.12.014
16. Pan Y, Xu T, Liu Y, Li W, Zhang W. Upregulated circular RNA circ_0025033 promotes papillary thyroid cancer cell proliferation and invasion via sponging miR-1231 and miR-1304. Biochem Biophys Res Commun (2019) 510(2):334–8. doi: 10.1016/j.bbrc.2019.01.108
17. Yao Y, Chen X, Yang H, Chen W, Qian Y, Yan Z, et al. Hsa_circ_0058124 promotes papillary thyroid cancer tumorigenesis and invasiveness through the NOTCH3/GATAD2A axis. J Exp Clin Cancer Res (2019) 38(1):318. doi: 10.1186/s13046-019-1321-x
18. Yang Y, Ding L, Li Y, Xuan C. Hsa_circ_0039411 promotes tumorigenesis and progression of papillary thyroid cancer by miR-1179/ABCA9 and miR-1205/MTA1 signaling pathways. J Cell Physiol (2020) 235(2):1321–9. doi: 10.1002/jcp.29048
19. Cai X, Zhao Z, Dong J, Lv Q, Yun B, Liu J, et al. Circular RNA circBACH2 plays a role in papillary thyroid carcinoma by sponging miR-139-5p and regulating LMO4 expression. Cell Death Dis (2019) 10(3):184. doi: 10.1038/s41419-019-1439-y
20. Liu W, Zhao J, Jin M, Zhou M. circRAPGEF5 Contributes to Papillary Thyroid Proliferation and Metastatis by Regulation miR-198/FGFR1. Mol Ther Nucleic Acids (2019) 14:609–16. doi: 10.1016/j.omtn.2019.01.003
21. Zhou GK, Zhang GY, Yuan ZN, Pei R, Liu DM. Has_circ_0008274 promotes cell proliferation and invasion involving AMPK/mTOR signaling pathway in papillary thyroid carcinoma. Eur Rev Med Pharmacol Sci (2018) 22(24):8772–80. doi: 10.26355/eurrev_201812_16644
22. Liu F, Zhang J, Qin L, Yang Z, Xiong J, Zhang Y, et al. Circular RNA EIF6 (Hsa_circ_0060060) sponges miR-144-3p to promote the cisplatin-resistance of human thyroid carcinoma cells by autophagy regulation. Aging (Albany NY) (2018) 10(12):3806–20. doi: 10.18632/aging.101674
23. Wei H, Pan L, Tao D, Li R. Circular RNA circZFR contributes to papillary thyroid cancer cell proliferation and invasion by sponging miR-1261 and facilitating C8orf4 expression. Biochem Biophys Res Commun (2018) 503(1):56–61. doi: 10.1016/j.bbrc.2018.05.174
24. Bi W, Huang J, Nie C, Liu B, He G, Han J, et al. CircRNA circRNA_102171 promotes papillary thyroid cancer progression through modulating CTNNBIP1-dependent activation of β-catenin pathway. J Exp Clin Cancer Res (2018) 37(1):275. doi: 10.1186/s13046-018-0936-7
25. Li X, Tian Y, Hu Y, Yang Z, Zhang L, Luo J. CircNUP214 sponges miR-145 to promote the expression of ZEB2 in thyroid cancer cells. Biochem Biophys Res Commun (2018) 507(1-4):168–72. doi: 10.1016/j.bbrc.2018.10.200
26. Jin X, Wang Z, Pang W, Zhou J, Liang Y, Yang J, et al. Upregulated hsa_circ_0004458 Contributes to Progression of Papillary Thyroid Carcinoma by Inhibition of miR-885-5p and Activation of RAC1. Med Sci Monit (2018) 24:5488–500. doi: 10.12659/MSM.911095
27. Zheng FB, Chen D, Ding YY, Wang SR, Shi DD, Zhu ZP. Circular RNA circ_0103552 promotes the invasion and migration of thyroid carcinoma cells by sponging miR-127. Eur Rev Med Pharmacol Sci (2020) 24(5):2572–8. doi: 10.26355/eurrev_202003_20526
28. Wang H, Yan X, Zhang H, Zhan X. CircRNA circ_0067934 Overexpression Correlates with Poor Prognosis and Promotes Thyroid Carcinoma Progression. Med Sci Monit (2019) 25:1342–9. doi: 10.12659/MSM.913463
29. Fan CM, Wang JP, Tang YY, Zhao J, He SY, Xiong F, et al. circMAN1A2 could serve as a novel serum biomarker for malignant tumors. Cancer Sci (2019) 110(7):2180–8. doi: 10.1111/cas.14034
30. Chen F, Feng Z, Zhu J, Liu P, Yang C, Huang R, et al. Emerging roles of circRNA_NEK6 targeting miR-370-3p in the proliferation and invasion of thyroid cancer via Wnt signaling pathway. Cancer Biol Ther (2018) 19(12):1139–52. doi: 10.1080/15384047.2018.1480888
31. Ren H, Liu Z, Liu S, Zhou X, Wang H, Xu J, et al. Profile and clinical implication of circular RNAs in human papillary thyroid carcinoma. PeerJ (2018) 6:e5363. doi: 10.7717/peerj.5363
32. Wang M, Chen B, Ru Z, Cong L. CircRNA circ-ITCH suppresses papillary thyroid cancer progression through miR-22-3p/CBL/β-catenin pathway. Biochem Biophys Res Commun (2018) 504(1):283–8. doi: 10.1016/j.bbrc.2018.08.175
33. Lan X, Cao J, Xu J, Chen C, Zheng C, Wang J, et al. Decreased expression of hsa_circ_0137287 predicts aggressive clinicopathologic characteristics in papillary thyroid carcinoma. J Clin Lab Anal (2018) 32(8):e22573. doi: 10.1002/jcla.22573
34. Peng N, Shi L, Zhang Q, Hu Y, Wang N, Ye H. Microarray profiling of circular RNAs in human papillary thyroid carcinoma. PloS One (2017) 12(3):e0170287. doi: 10.1371/journal.pone.0170287
35. Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA (2013) 19(2):141–57. doi: 10.1261/rna.035667.112
36. Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, et al. Circular intronic long noncoding RNAs. Mol Cell (2013) 51(6):792–806. doi: 10.1016/j.molcel.2013.08.017
37. Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet (2019) 20(11):675–91. doi: 10.1038/s41576-019-0158-7
38. Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient microRNA sponges. Nature (2013) 495:384–8. doi: 10.1038/nature11993
39. Liang WC, Wong CW, Liang PP, Shi M, Cao Y, Rao ST, et al. Translation of the circular RNA circβ-catenin promotes liver cancer cell growth through activation of the Wnt pathway. Genome Biol (2019) 20(1):84. doi: 10.1186/s13059-019-1685-4
40. Yao Y, Hua Q, Zhou Y, Shen H. CircRNA has_circ_0001946 promotes cell growth in lung adenocarcinoma by regulating miR-135a-5p/SIRT1 axis and activating Wnt/β-catenin signaling pathway. BioMed Pharmacother (2019) 111:1367–75. doi: 10.1016/j.biopha.2018.12.120
41. Shang J, Chen WM, Liu S, Wang ZH, Wei TN, Chen ZZ, et al. CircPAN3 contributes to drug resistance in acute myeloid leukemia through regulation of autophagy. Leuk Res (2019) 85:106198. doi: 10.1016/j.leukres.2019.106198
42. He Y, Mingyan E, Wang C, Liu G, Shi M, Liu S. CircVRK1 regulates tumor progression and radioresistance in esophageal squamous cell carcinoma by regulating miR-624-3p/PTEN/PI3K/AKT signaling pathway. Int J Biol Macromol (2019) 125:116–23. doi: 10.1016/j.ijbiomac.2018.11.273
43. Lin Q, Ling YB, Chen JW, Zhou CR, Chen J, Li X, et al. Circular RNA circCDK13 suppresses cell proliferation, migration and invasion by modulating the JAK/STAT and PI3K/AKT pathways in liver cancer. Int J Oncol (2018) 53(1):246–56. doi: 10.3892/ijo.2018.4371
44. Pan H, Li T, Jiang Y, Pan C, Ding Y, Huang Z, et al. Overexpression of Circular RNA ciRS-7 Abrogates the Tumor Suppressive Effect of miR-7 on Gastric Cancer via PTEN/PI3K/AKT Signaling Pathway. J Cell Biochem (2018) 119(1):440–6. doi: 10.1002/jcb.26201
45. Xu H, Zhang Y, Qi L, Ding L, Jiang H, Yu H. NFIX Circular RNA Promotes Glioma Progression by Regulating miR-34a-5p via Notch Signaling Pathway. Front Mol Neurosci (2018) 11:225. doi: 10.3389/fnmol.2018.00225
46. Wu HB, Huang SS, Lu CG, Tian SD, Chen M. CircAPLP2 regulates the proliferation and metastasis of colorectal cancer by targeting miR-101-3p to activate the Notch signalling pathway. Am J Transl Res (2020) 12(6):2554–69.
47. Haugen BR. American Thyroid Association Management guidelines for adult patients with thyroid nodules and differentiated thyroid cancer: what is new and what has changed? Cancer (2015) 123:372–81. doi: 10.1002/cncr.30360
48. He T, Wang H, Sun J, Wu J, Gong F, Li B, et al. Altered expression of DLG1-AS1 distinguished papillary thyroid carcinoma from benign thyroid nodules[J]. BMC Endocr Disord (2019) 19(1):122. doi: 10.1186/s12902-019-0440-x
49. Fu XM, Guo W, Li N, Liu HZ, Liu J, Qiu SQ, et al. The expression and function of long noncoding RNA lncRNA-ATB in papillary thyroid cancer[J]. Eur Rev Med Pharmacol Sci (2017) 21(14):3239–46.
50. Bahn JH, Zhang Q, Li F, Chan TM, Lin X, Kimi Y, et al. The landscape of microRNA, Piwi-interacting RNA, and circular RNA in human saliva. Clin Chem (2015) 61(1):221–30. doi: 10.1373/clinchem.2014.230433
Keywords: papillary thyroid carcinoma, circRNA, characteristics, ceRNA, signaling pathway, diagnosis, prognosis
Citation: Xu X and Jing J (2021) Advances on circRNAs Contribute to Carcinogenesis and Progression in Papillary Thyroid Carcinoma. Front. Endocrinol. 11:555243. doi: 10.3389/fendo.2020.555243
Received: 24 April 2020; Accepted: 13 November 2020;
Published: 21 January 2021.
Edited by:
Christoph Reiners, University Hospital Würzburg, GermanyReviewed by:
Jason David Prescott, Johns Hopkins Medicine, United StatesMarialuisa Appetecchia, Istituti Fisioterapici Ospitalieri (IRCCS), Italy
Copyright © 2021 Xu and Jing. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiexian Jing, 2912972872@qq.com
 Xiaoqin Xu
Xiaoqin Xu Jiexian Jing
Jiexian Jing