Shigella Diversity and Changing Landscape: Insights for the Twenty-First Century
- 1Institut Pasteur, Unité de Pathogénie Microbienne Moléculaire, Paris, France
- 2Institut National de la Santé et de la Recherche Médicale, Unité 786, Paris, France
- 3Collège de France, Paris, France
Shigella is a pathovar of Escherichia coli comprising four groups, Shigella flexneri, Shigella sonnei, Shigella dysenteriae, and Shigella boydii, each of them, with the exception of S.sonnei, comprising several serotypes. Shigella accounts for the majority of dysentery causing infections occurring world-wide each year. Recent advancements in the Shigella field have led to a better understanding of the molecular mechanisms underlying host epithelial cell invasion and immune cell function manipulation, mainly using S. flexneri as a model. Host-cell invasion is the final step of the infection process, as Shigella's virulence strategy relies also on its ability to survive hostile conditions during its journey through the gastro-intestinal tract, to compete with the host microbiota and to cross the intestinal mucus layer. Hence, the diversity of the virulence strategies among the different Shigella species has not yet been deeply investigated, which might be an important step to understand the epidemiological spreading of Shigella species worldwide and a key aspect for the validation of novel vaccine candidates. The recent development of high-throughput screening and sequencing methods will facilitate these complex comparison studies. In this review we discuss several of the major avenues that the Shigella research field has taken over the past few years and hopefully gain some insights into the questions that remain surrounding this important human pathogen.
Introduction
Shigella are Gram-negative pathogenic enterobacteria and the etiological agent of bacillary dysentery or shigellosis. In humans, Shigella specifically invade and colonize the colonic mucosa leading to its disruption. Shigellosis is associated with fever and abdominal cramps; its diagnosis relies on the presence of erythrocytes, polymorphonuclear neutrophils (PMNs), and mucus in patient stools, which stand as diagnostic elements.
Shigella encompasses four subgroups (S. flexneri, S. sonnei, S, dysenteriae, and S. boydii), each composed of different serotypes, which are identified based on the structure of the lipopolysaccharide O-antigen repeats: S. dysenteriae encompasses 15 serotypes, S. flexneri, 14 serotypes, S. boydii, 20 serotypes, and S. sonnei a single serotype (Figure 1; reviewed in Marteyn et al., 2012). Chantemesse and Widal first described the bacillus causing non-amoebic dysentery in 1888, while Shiga first identified S. dysenteriae in 1898 (Shiga, 1898). Flexner first characterized S. flexneri in 1900 and S. sonnei was isolated and characterized by Sonne (1915). S. boydii was first described by Boyd in 1931.
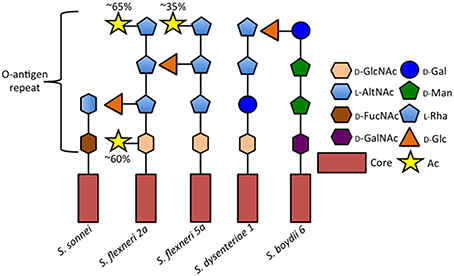
Figure 1. Comparison of selected Shigella O-antigen side chains. Schematic of repeated O-antigen side chains from serotypes S. sonnei (Gamian and Romanowska, 1982), S. flexneri 5a (Perepelov et al., 2010), S. dysenteriae 1 (Dmitriev et al., 1976), and S. boydii 6 (Senchenkova et al., 2005). Figure legend abreviations are as follows 2-Acetamido-2-deoxy-D-glucose (D-GlcNAc), 2-Acetamido-2-deoxy-L-altruronic acid (L-AltNAc), 2-Acetamido-4-amino-2, 4-dideoxy-D-fucose (D-FucNAc), 2-Acetamido-2-deoxy-D-galacturonic acid (D-GalNAc), D-Galactopyranose (D-Gal), D-Mannose (D-Man), L-Rhamnose (L-Rha), D-Glucose (D-Glc), O-acetyl (Ac).
Studies of host-pathogen interactions have mostly been restricted to the direct contact with and invasion of the host epithelial and immune cells. This is particularly true in the Shigella field. Its adhesion and invasion of host cells and its intracellular life have been characterized at a molecular level (reviewed in Tran Van Nhieu et al., 2000; Phalipon and Sansonetti, 2007). In this review, we envisage the host-pathogen interaction with a more global perspective, considering that prior to challenging epithelial or immune cells, Shigella must first interact and compete with the microbiome and subsequently face a thick mucus layer protecting the colonic epithelium. Shigella virulence mechanisms involved in these latter aspects have been under evaluated, although they represent key and essential features of a successful infection process.
Shigella virulence mechanisms have been mostly identified and characterized using S. flexneri as a model, while other groups show significantly high prevalence worldwide. In this review, we will emphasize the importance of performing comparative studies to better understand and characterize Shigella pathogenicity. The development of novel genetic tools and high-throughput screening and sequencing methods permits the analysis of Shigella virulence mechanism diversity.
Shigella Diversity: Epidemiology, Geographical Distribution, and Emerging Antibiotic Resistance
Shigella remains a leading cause of childhood morbidity and mortality. The recently conducted case-controlled Global Enteric Multicenter Study (GEMS) provided a solid update on the incidence of Shigella among severe forms of diarrhea, and convincingly demonstrated that in the sites considered (Sub-saharan Africa, and Asia), Shigella appeared amongst the top ranking pathogens identified (Kotloff et al., 2013).
Many observations have concluded that Shigella species are geographically stratified based on the level of economic development in a given country. S. flexneri is the primary infectious species in the developing world whereas S. sonnei rates increase with economic development. S. boydii is most commonly restricted to Bangladesh and South-East Asia and rarely occurs outside of these regions. S. dysenteriae type 1 (Sd1) occurs sporadically in outbreak settings with striking examples occurring in refugee camps during the civil war in Rwanda between November 1993 and February 1995 in which more than 180 thousand cases and significant mortality from Sd1 were recorded (Kernéis et al., 2009). The last major Sd1 outbreak occurred in 1999 during the civil war in Sierra Leone resulting in over 4000 cases (Guerin et al., 2004), and the cyclic occurrence of Sd1 in Bangladesh every 10 years is clearly discontinued for unknown reasons, illustrating major remaining uncertainties on changes of epidemiological patterns of these infections.
Outside of outbreak settings, S. flexneri and S. sonnei account for the majority of Shigellosis cases. Recent epidemiological studies conducted around the world have discovered a rise in the proportion of S. sonnei isolates compared to S. flexneri. The expansion of S. sonnei can clearly be observed from clinical surveillance studies conducted in China which show the proportion of S. sonnei isolates increasing from 17.4% in 2003–2004 to 58.2% less than a decade later, closely following the rapid industrialization in China (Figure 2; Mao et al., 2013; Qiu et al., 2015). Noticeably, regions that had undergone significant industrialization reported decreases in S. flexneri and increasing cases of S. sonnei compared to under developed areas where flexneri levels remain high (Qiu et al., 2015). Rising cases of S. sonnei have also been detected in Bangladesh, which has historically been affected by all four species of Shigella. Between 2001 and 2011 the proportion of S. sonnei infections rose from 7 to 25% of reported cases in Bangladesh, which also corresponds with enhanced sanitation and clean water efforts throughout the country (Das et al., 2013; Hulland et al., 2013). The reasons for the counter-intuitive increase of S. sonnei in the face of better sanitation have not been determined, however several hypotheses have been put forward (Thompson et al., 2015). S. sonnei and Plesiomonas shigelloides share a common O-antigen that may lead to natural cross-protective immunity in populations that encounter high levels of P. shigelloides due to contaminated water supplies (Sack et al., 1994). Other avenues of interest include observations of increased survival and replication of S. sonnei in Acanthamoeba which may act as a reservoir and increased ability to acquire antibiotic resistances (Saeed et al., 2012). As more countries increase their level of development and sanitation it is likely that S. sonnei will become even more of a global public health concern which could have important impacts on vaccine development efforts.
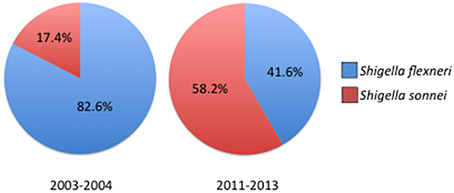
Figure 2. Prevelance of Shigella species across China 2003–2013. Comparison of reported Shigella species between 2003–2004 and 2011–2013 showing the increase in S. sonnei isolations. For years 2003–2004 n = 235 cases, for 2011–2013 n = 1049 cases. Prevelance data reported in Qiu et al. (2015).
A major concern surrounding Shigella is its capacity to rapidly acquire antibiotic resistances. Development of resistance to antibiotics is common in all Shigella species, particularly in S. sonnei, which can acquire resistance genes directly from E. coli through horizontal gene transfer. Several recent reports have suggested that S. sonnei is capable of sharing resistance plasmids through conjugation with commensal E. coli (Qu et al., 2014; Rashid and Rahman, 2015). Recently, S. sonnei resistance to the frontline antibiotic, ciprofloxacin, has been associated with travel to Asia and India and has been imported to the United States (Bowen et al., 2015; De Lappe et al., 2015; Kim et al., 2015). Preventing the global spread of resistant S. sonnei strains presents a significant public health challenge as intercontinental travel has become common.
Antibiotic resistance in S. flexneri is also well-documented with many studies finding high rates of resistance to at least one common antibiotic such as ampicillin, tetracycline, and chloramphenicol (Khaghani et al., 2014; Cui et al., 2015). These studies show that without access to proper testing facilities or multiple different antibiotics, patients affected by S. flexneri in developing countries have a high chance of not being successfully treated.
Two articles have shed some light on the expansion and spread of antibiotic resistance in Shigella. Sequencing and phylogenetic studies have pinpointed the emergence of S. sonnei from a single European clone that spread pandemically to multiple continents, diversifying and acquiring multiple antibiotic resistances along the way (Holt et al., 2013). The sequencing data also suggest that antibiotic resistant S. sonnei strains have been exported to new areas via infected travelers. In contrast, Connor et al. recently published results from a whole genome sequencing study of S. flexneri that found considerable independent acquisitions of antibiotic resistance yet little global spread of resistant flexneri strains, instead favoring a model of local resistance acquisition and persistence rather than pandemic spread (Connor et al., 2015). These complementary studies have yielded the clearest information to date on how Shigella has spread throughout the global community. Taken together a prediction can be made that efforts to combat the spread of antibiotic resistance should focus more heavily on S. sonnei surveillance while more localized efforts are needed to combat resistance in S. flexneri.
The changing dynamics of Shigella species, particularly trends like the disappearance of Sd1 and transition from S. flexneri to S. sonnei raise important unanswered questions. Thus, far, the molecular and cellular mechanisms of Shigella pathogenesis have largely considered the invasion step and likely do not account for the major differences among the four subgroups. Instead, research probing the early steps of establishment/colonization may provide new insights into explaining the epidemiological observations.
First Barriers to Shigella Infection: The Microbiota and the Mucus Layer
Shigella specifically invades the human colon, but not the small intestine. The molecular basis of this specificity remains unknown. Prior to reaching the epithelial lining, Shigella have to survive in the GI environment, to outcompete the colonic luminal commensal bacteria and to disrupt a thick protective mucus layer. For a long time, the mucus layer was considered as the first barrier to Shigella infection: it appears to be the second, the microbiota standing as the first. The inflammatory response induced by Shigella invasion might result in a perturbation of the composition and the function of these barriers, although these aspects are not yet well-described.
First Barrier: The Microbiota
The gut microbiome of healthy individuals comprises the most complex interface between microbes and human tissues in the body. Over 1000 different species of bacteria reside at varying amounts and locations throughout the GI tract and 70% of the host immune system is dedicated to maintaining its integrity (Vighi et al., 2008; Rajilić-Stojanović and de Vos, 2014). The colonic microbiota can vary significantly between individuals but is dominated at the phyla level by Bacteroidetes (~30%) and Firmicutes (~30%), with lower levels of Proteobacteria, Fusobacteria, Actinobacteria, Cyanobacteria, and Verrucomicrobia (Bäckhed et al., 2005; Andersson et al., 2008; Arumugam et al., 2011; Sankar et al., 2015; Figure 3). The microbiota is further stratified based on proximity to the mucosal lining of the colon with genera such as Coriobacteriaceae, Lachnospiraceae, and Ruminococcaceae showing greater abundance in the mucus layer whereas Bacteroidaceae is more prevalent in the lumen of the intestine (Lavelle et al., 2015). Significant amounts of research have worked to characterize the role of the gut microbiome in both health and disease states and a picture of this complex community is beginning to take shape.
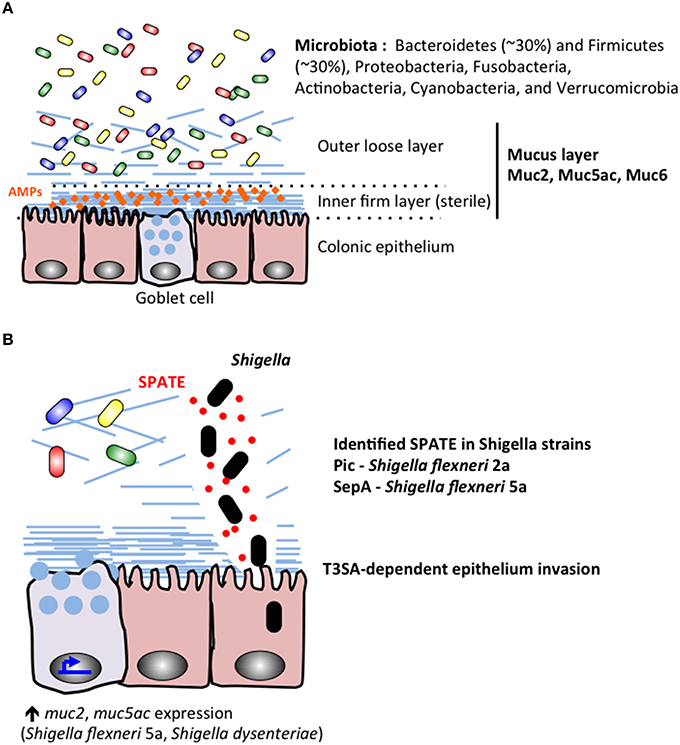
Figure 3. During Shigella infection, the colonic microbiota and mucus layers stand as the first barriers protecting the epithelium lineage. (A) The microbiota colonizes the colonic lumenal compartment, and the outer mucus layer. The inner mucus sterile remains sterile probaly due to its mechanical properties and accumulation of secreted antimicrobial peptides (AMPs). (B) Shigella secrete proteins belonging to the SPATE family, including Pic characterized in S. flexneri 2a (Henderson et al., 1999; Gutiérrez-Jiménez et al., 2008; Navarro-Garcia et al., 2010; Ruiz-Perez et al., 2011) and SepA identified in S. flexneri 5a (Benjelloun-Touimi et al., 1995). Shigella SPATE proteins cleave mucins to allow the bacteria to reach the epithelial layer. Shigella Type III secretion apparatus (T3SA) is required for epithelium invasion.
The study of how pathogens interact with and supplant host commensal bacteria is fast becoming an area of intense research. These interactions generally occur at the earliest stages of disease and set up the environment for a pathogen to gain entry into host tissues. Currently, very few studies have investigated the role of the microbiota in Shigella infections. The infectious dose of Shigella is thought to be quite low, (~10) organisms, so it is likely that Shigella has evolved some mechanism(s) to survive while being vastly outnumbered.
Early works assessed the role of individual commensal organisms in preventing the colonization of Shigella. Germ free mice and Guinea pigs monocolonized with E. coli were shown to block the establishment of S. flexneri whereas monocolonization with Bacteroides, Bifidobacterium, or Streptococcus had no effect on Shigella levels (Formal et al., 1961; Maier and Hentges, 1972). Other studies have shown treatment with streptomycin produces an environment that supports Shigella colonization in mice (Pongpech et al., 1989; Martino et al., 2005). Together these studies suggest a complex interplay between Shigella and commensal bacteria that are not easily reconciled with the low infectious dose observed in humans. It is possible that the overwhelming inflammatory environment produced during human disease masks more subtle and complex early interactions between Shigella and commensal bacteria. Revisiting these issues with newer techniques will be important in deciphering the role of host microbiota in Shigellosis.
The production and resistance to secreted small molecule inhibitors such as colicins mediate one common form of competition between enterobacteraceae. Colicins are small, secreted proteins that can have various cytotoxic properties against phylogenetically related bacteria and are are almost always carried on plasmids that encode both the colicin and its cognate immunity factor, reviewed in Cascales et al. (2007). Strains that are capable of producing a colicin are better able to compete against colicin sensitive strains over ecological niches. In the case of S. sonnei, it has been reported that up to 93% of tested strains in Kolkata, India, harbor at least one colicin-encoding plasmid which were shown to target closely related susceptible E. coli strains, which as previously mentioned, were important for restricting Shigella colonization (Calcuttawala et al., 2015). Meanwhile, the chromosomally encoded O-antigen chain length regulator WzzB was recently reported to contribute to colicin E2 resistance in S. flexneri (Tran et al., 2014). Regulation of the O-antigen chain length has an important impact during the intracellular phase of Shigella infection, but Tran et al. suggest the exact number of repeat units has evolved to help mediate colicin resistance as well (Morona et al., 2003; Tran et al., 2014). The production and resistance to colicins by Shigella may have an important role in its initial competition with the host microbiota.
A growing number of reports suggest that members of the lactobacilli genus may contribute to colonization resistance against Shigella strains by inducing an anti-inflammatory state or by blocking adherence to epithelial cells (Tien et al., 2006; Moorthy et al., 2010; Zhang et al., 2012). Specifically, proteins found in the S-layers of gram positive Lactobacillus species have been found to inhibit adhesion of several different pathogens including Salmonella enteritidis, EHEC O157:H7, and S. sonnei to tissue culture cell lines although the exact mechanism(s) remains to be discovered (Golowczyc et al., 2007; Johnson-Henry et al., 2007; Zhang et al., 2012). Further assessments of the interaction of commensal bacteria with Shigella will be important to gain a more complete picture of how beneficial bacteria aid in host defenses against pathogens. Similarly, studies probing how Shigella overcomes the resident microbiota during successful infection would be of great interest.
Second Barrier: The Mucus Layer
A complex layer of mucus naturally protects human epithelium in the gastrointestinal tracts (stomach, small intestine, colon; Ambort et al., 2012). The role of the mucus layer is to prevent direct interaction between commensal bacteria and epithelial cells (Johansson et al., 2011), avoiding further stimulation of the immune system (Pelaseyed et al., 2014; Figure 3 upper panel). In addition to the barrier effect created by the mucus layer, antimicrobial peptides (AMPs) secreted by epithelial and phagocytic cells accumulate in the mucus layer and act as a further inhibitory blockade against microbial species (Meyer-Hoffert et al., 2008). Mucus is mainly composed of mucins (20 identified mucins in humans) reviewed in McGuckin et al. (2011), which are high-molecular weight (1–10 million Da) glycoproteins linked with each other through intermolecular disulfide-bonds between cysteine-rich D domains located in mucins amino and carboxyl termini. Mucins are either oligomeric, soluble and secreted (MUC2, MUC5ac, MUC5b, MUC6, and Muc7) or are membrane tethered, which might be associated with different functions. Mucins are naturally produced and released on epithelial surfaces: mainly by goblet cells but also (in a lesser extent) by epithelial cells. It has been recently shown that the surface colonic goblet cells secrete continuously, while the secretion of goblet cells located in the small intestine or colonic crypts are inducible (Birchenough et al., 2015). The thickness of the mucus layer varies along the GI tract in humans and various animal models (Varum et al., 2012). While the ileal mucus layer is thin (< 300 μm), the thickness of the colon mucus layer can reach 1 mm in humans. The colonic mucus layer is organized in two layers: the inner layer (50 μm) is firmly attached to the epithelium, the outer loose layer (>100 μm) is unattached (Figure 3). Several studies have described the ability of commensal bacteria to colonize and adhere to the abundant O-glycans present in the outer mucus layer, while the inner mucus layer remains sterile, due to its mechanical properties or capacity to retain secreted antimicrobial compounds, avoiding further stimulation of epithelial cells by commensal bacteria (Johansson et al., 2011). The sterility of the inner mucus layer might be altered by impairment of AMP production leading to colitis or other inflammatory bowel conditions (Kocsis et al., 2008; Johansson et al., 2014). The colonic mucus layer backbone is mainly composed of MUC2 (Karlsson et al., 1996), associated with MUC5AC and MUC6 (reviewed in McGuckin et al., 2011; Figure 3). Differential MUC2 glycosylation profiles are described in the ileum and in the colon, being enriched in either sialilated or sulfated oligosaccharide species respectively (Karlsson et al., 1996; Robbe et al., 2003). It has been shown that S. dysenteriae 1 adheres to human colonic mucins, not to small intestine mucins (Sudha et al., 2001), although no specific adhesion molecule has been identified.
Shigella efficiently invade the colon, which is protected by the thickest layer of mucus in the GI tract. In addition, it has been shown that S. dysenteriae infection induces the expression of MUC2 and MUC5AC in vitro in an IL-1β dependent-manner (Prakash et al., 2011; Raja et al., 2012), but also in vivo in a rat ileal loop model (Gopal et al., 2014). Similarly, S. flexneri induces MUC2, MUC5AC, MUC4, and MUC15 expression in vitro (HT29-MTX cells; Sperandio et al., 2013). Together with the fact that Shigella are devoid of flagella (which promote the mobility of various pathogens, such as Helicobacter pylori (Ottemann and Lowenthal, 2002), it suggests that Shigella express and releases mucolytic molecules allowing its progression through the mucus layer.
It has long been known that the degradation of the mucus outer layer and the release of mucin glycans, by glycoside hydrolase-expressing bacteria composing the microbiota, were used as a source of nutrients and energy (Deplancke et al., 2002; Sonnenburg et al., 2005). So far, no report describes similar adaptation of Shigella in the colonic lumen to promote its proliferation.
Mucus integrity can be altered by the several classes of enzymes secreted by enterobacteria (Figure 3 lower panel). It includes mucinases, glycosidases, sulfatases, sialidases (or neuraminidases), and sialilate O-acetylesterases (SIAE; Corfield et al., 1992; reviewed in McGuckin et al., 2011; Corfield, 2015). It was shown by Haider and colleagues that all four Shigella species were able to produce mucinases and neuraminidases (Haider et al., 1993), although since this initial observation no comparative study was performed. Pic, a S. flexneri 2A (4257T) autosecreted protein belonging to the SPATE (Serine Protease Autotransporters of Enterobacteriaceae) was characterized as a mucinase (Henderson et al., 1999; Gutiérrez-Jiménez et al., 2008). It was shown that the deletion of pic in S. flexneri 2a clinical isolates reduced its pathogenicity (Navarro-Garcia et al., 2010; Zhang et al., 2013). Pic homologous protein is expressed by S. sonnei, not in S. flexneri 5a, which instead secretes another protein belonging to the SPATE family (SepA), whose function remains unknown (Benjelloun-Touimi et al., 1995, 1998). S. dysenteriae, S. flexneri 2a, S. sonnei and S. boydii express homologous proteins to Enterotoxigenic E. coli (ETEC) EatA, belonging to the SPATE family, which has been shown to degrade MUC2 (Kumar et al., 2013). Until now, the role of EatA in Shigella virulence mechanisms has not yet been characterized.
After traversing the mucus layer, Shigella contact and inject epithelial cells with effector molecules that down-regulate production of AMPs, supporting an environment that is more conducive to further penetration into the tissue and providing evidence that AMPs are an important host defense factor against Shigella (Sperandio et al., 2008; Gudmundsson et al., 2010).
Altogether, these results suggest that Shigella potentially express and secrete mucinases, although the specificity of these enzymes will have to be characterized and their cytotoxicity further studied. It would also be of great interest to study the consequences of disrupting of the mucus layer on local AMP concentrations and inhibition of invading Shigella bacteria. Disruption of the mucus layer and dispersion of AMPs could play a significant role in Shigella survival in addition to aiding colonization.
Global Tools to Decipher Shigella Virulence Mechanism Diversity
The advent of affordable, high-throughput sequencing technologies is allowing the field of microbiology to design powerful experiments that have previously been impossible to perform due to time, money, or computing restraints. With this technology however has come the very real problem of how to adequately mine the vast quantities of data generated for important information. In the Shigella field there has been an explosion in the number of sequenced strains and phylogenetic trees establishing a detailed history of the evolution of this pathogen. The most sophisticated sequencing analysis of S. sonnei to date comprised of 132 strains spread across four continents has yielded very interesting information on its origins and recent global spread (Holt et al., 2012). In this paper the authors estimate the most recent common ancestor to S. sonnei arose in Europe less than 500 years ago between 1554 and 1763 A.D (Holt et al., 2012). Additionally, it was found that the global spread of S. sonnei has only begun quite recently, since the mid twentieth century (Holt et al., 2012). A subsequent massive sequencing study of 263 S. sonnei strains, isolated over 15 years in Vietnam, has been used to track its evolution and to date its first arrival in the country to the 1980's (Holt et al., 2013). The result is a fascinating “high resolution” view of local adaptations and evolutionary changes that have occurred over the past 15 years from what started as a single imported clone (Holt et al., 2013). Sequencing studies like these are providing invaluable information about how pathogens evolve, expand, and become fixed in a population. However, the massive amount of data generated from sequencing so many isolates can present a daunting challenge to mine for insights into the functional outcomes of microbial evolution. Holt et al.'s (2012) paper alone reported detecting 10,111 SNP mutations in the sequenced strains making it difficult to investigate individual mutations for their impact on the fitness of Shigella. Ideally, it would be intriguing to assess Shigella's pathogenic traits over time based upon its evolution.
Alternatively, high-throughput sequencing has found another important use in new generations of mutant library design and screening applications. In Vibrio cholerae, pooled transposon mutant libraries have been screened in rich media and under in vivo conditions, coupled with next generation transposon sequencing (TN-seq) resulting in a global picture of genes essential for survival in a rabbit animal model (Kamp et al., 2013). This global approach allowed over 100,000 individual mutants to be screened simultaneously in a single model system, creating a nearly complete picture of the genes required for V. cholerae to survive in a host (Kamp et al., 2013). This technique has also successfully been demonstrated in the gram positive pathogen Staphylococcus aureus, where over 71,000 transposon mutants were screened in vivo (Valentino et al., 2014). Adapting this protocol to Shigella, coupled with the newly developed Guinea pig colonic infection model could give significant insights into the repertoire of genes needed to survive and replicate in the colon during infection (Arena et al., 2015). Furthermore, performing such experiments in representative S. sonnei and S. flexneri strains could allow for an interesting comparison study, highlighting similarities, and differences in growth and survival strategies between Shigella species which up until now have been impossible to perform.
Conclusions
For several decades, S. flexneri strains (2a and 5a) were used as model organisms to characterize Shigella virulence mechanisms. However, each Shigella groups and subgroups exploit various virulence mechanisms and as a consequence deserved a specific analysis. The development of high-throughput technologies represents a key opportunity to address this fastidious but essential question without bias. It will allow comparative studies and a better comprehension of various Shigella group specific virulence features. This aspect is well-illustrated by the diversity of SPATE proteins expressed and secreted by different groups (see Section First Barriers to Shigella Infection: The Microbiota and the Mucus Layer), coming studies will probably highlight additional specificities.
The diversity of Shigella groups identified worldwide has changed over the last decades. Efforts to improve water sanitation have lead to a decrease in S. flexneri infections however S. sonnei rapidly moves in replacing S. flexneri. For this reason improving water supplies alone is not sufficient to eliminate the threat of Shigella. Vaccines targeting S. sonnei and remaining S. flexneri serotypes (i.e., 2a, 3a, and 6) +/− S. dysenteriae 1 must be part of a long-term strategy that takes in account the serotype-specificity of immune protection against Shigella, to reduce the global dysentery burden. However, the development of live, rationally-attenuated, oral vaccine candidates against Shigellosis has been handicapped by two major issues. (i) The difficulty to assemble a multivalent set of these epidemiologically-relevant serotypes, in the absence of a definitely-established cross-protective antigen (Levine et al., 2007). (ii) A clear discrepancy in the colonization and potential protective capacity of the so-far tested candidates when tested in western volunteers and in individuals living in endemic areas (Coster et al., 1999; Rahman et al., 2011).
Clearly, more basic research aimed at better understanding the basic mechanisms of Shigella colonization is warranted to master this key step and improve the design of future live-attenuated orally-administered vaccines.
Author Contributions
All authors listed, have made substantial, direct and intellectual contribution to the work, and approved it for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Ambort, D., Johansson, M. E. V., Gustafsson, J. K., Ermund, A., and Hansson, G. C. (2012). Perspectives on mucus properties and formation–lessons from the biochemical world. Cold Spring Harb. Perspect. Med. 2:a014159. doi: 10.1101/cshperspect.a014159
Andersson, A. F., Lindberg, M., Jakobsson, H., Bäckhed, F., Nyrén, P., and Engstrand, L. (2008). Comparative analysis of human gut microbiota by barcoded pyrosequencing. PLoS ONE 3:e2836. doi: 10.1371/journal.pone.0002836
Arena, E. T., Campbell-Valois, F.-X., Tinevez, J.-Y., Nigro, G., Sachse, M., Moya-Nilges, M., et al. (2015). Bioimage analysis of Shigella infection reveals targeting of colonic crypts. Proc. Natl. Acad. Sci. U.S.A. 112, E3282–E3290. doi: 10.1073/pnas.1509091112
Arumugam, M., Raes, J., Pelletier, E., Le Paslier, D., Yamada, T., Mende, D. R., et al. (2011). Enterotypes of the human gut microbiome. Nature 473, 174–180. doi: 10.1038/nature09944
Bäckhed, F., Ley, R. E., Sonnenburg, J. L., Peterson, D. A., and Gordon, J. I. (2005). Host-bacterial mutualism in the human intestine. Science 307, 1915–1920. doi: 10.1126/science.1104816
Benjelloun-Touimi, Z., Sansonetti, P. J., and Parsot, C. (1995). SepA, the major extracellular protein of Shigella flexneri: autonomous secretion and involvement in tissue invasion. Mol. Microbiol. 17, 123–135. doi: 10.1111/j.1365-2958.1995.mmi_17010123.x
Benjelloun-Touimi, Z., Si Tahar, M., Montecucco, C., Sansonetti, P. J., and Parsot, C. (1998). SepA, the 110 kDa protein secreted by Shigella flexneri: two-domain structure and proteolytic activity. Microbiology 144(Pt 7), 1815–1822. doi: 10.1099/00221287-144-7-1815
Birchenough, G. M. H., Johansson, M. E., Gustafsson, J. K., Bergström, J. H., and Hansson, G. C. (2015). New developments in goblet cell mucus secretion and function. Mucosal Immunol. 8, 712–719. doi: 10.1038/mi.2015.32
Bowen, A., Hurd, J., Hoover, C., Khachadourian, Y., Traphagen, E., Harvey, E., Libby, T., et al. (2015). Importation and domestic transmission of Shigella sonnei resistant to ciprofloxacin - United States, May 2014-February 2015. Morb. Mortal. Wkly. Rep. 64, 318–320.
Calcuttawala, F., Hariharan, C., Pazhani, G. P., Ghosh, S., and Ramamurthy, T. (2015). Activity spectrum of colicins produced by Shigella sonnei and genetic mechanism of colicin resistance in conspecific S. sonnei strains and Escherichia coli. Antimicrob. Agents Chemother. 59, 152–158. doi: 10.1128/AAC.04122-14
Cascales, E., Buchanan, S. K., Duche, D., Kleanthous, C., Lloubes, R., Postle, K., et al. (2007). Colicin biology. Microbiol. Mol. Biol. Rev. 71, 158–229. doi: 10.1128/MMBR.00036-06
Connor, T. R., Barker, C. R., Baker, K. S., Weill, F. X., Talukder, K. A., Smith, A. M., et al. (2015). Species-wide whole genome sequencing reveals historical global spread and recent local persistence in Shigella flexneri. Elife 4:e07335. doi: 10.7554/eLife.07335
Corfield, A. P., Wagner, S. A., Clamp, J. R., Kriaris, M. S., and Hoskins, L. C. (1992). Mucin degradation in the human colon: production of sialidase, sialate O-acetylesterase, N-acetylneuraminate lyase, arylesterase, and glycosulfatase activities by strains of fecal bacteria. Infect. Immun. 60, 3971–3978.
Corfield, A. P. (2015). Mucins: a biologically relevant glycan barrier in mucosal protection. Biochim. Biophys. Acta 1850, 236–252. doi: 10.1016/j.bbagen.2014.05.003
Coster, T. S., Hoge, C. W., VanDeVerg, L. L., Hartman, A. B., Oaks, E. V., Venkatesan, M. M., et al. (1999). Vaccination against shigellosis with attenuated Shigella flexneri 2a strain SC602. Infect Immun 67, 3437–3443.
Cui, X., Wang, J., Yang, C., Liang, B., Ma, Q., Yi, S., et al. (2015). Prevalence and antimicrobial resistance of Shigella flexneri serotype 2 variant in China. Front Microbiol 6:435. doi: 10.3389/fmicb.2015.00435
Das, S. K., Ahmed, S., Ferdous, F., Farzana, F. D., Chisti, M. J., Latham, J. R., et al. (2013). Etiological diversity of diarrhoeal disease in Bangladesh. J. Infect. Dev. Ctries 7, 900–909. doi: 10.3855/jidc.3003
De Lappe, N., O'Connor, J., Garvey, P., McKeown, P. J., and Cormican, M. (2015). Ciprofloxacin-resistant Shigella sonnei associated with travel to India. Emerg. Infect. Dis. 21, 894–896. doi: 10.3201/eid2105.141184
Deplancke, B., Vidal, O., Ganessunker, D., Donovan, S. M., Mackie, R. I., and Gaskins, H. R. (2002). Selective growth of mucolytic bacteria including Clostridium perfringens in a neonatal piglet model of total parenteral nutrition. Am. J. Clin. Nutr. 76, 1117–1125.
Dmitriev, B. A., Knirel, Y. A., Kochetkov, N. K., and Hofman, I. L. (1976). Somatic antigens of Shigella. Eur. J. Biochem. 66, 559–566. doi: 10.1111/j.1432-1033.1976.tb10582.x
Formal, S. B., Dammin, G., Sprinz, H., Kundel, D., Schneider, H., Horowitz, R. E., et al. (1961). Experimental Shigella infections. V. Studies in germ-free guinea pigs. J. Bacteriol. 82, 284–287.
Gamian, A., and Romanowska, E. (1982). The core structure of Shigella sonnei lipopolysaccharide and the linkage between O-specific polysaccharide and the core region. Eur. J. Biochem. 129, 105–109. doi: 10.1111/j.1432-1033.1982.tb07027.x
Golowczyc, M. A., Mobili, P., Garrote, G. L., Abraham, A. G., and De Antoni, G. L. (2007). Protective action of Lactobacillus kefir carrying S-layer protein against Salmonella enterica serovar Enteritidis. Int. J. Food Microbiol. 118, 264–273. doi: 10.1016/j.ijfoodmicro.2007.07.042
Gopal, A., Iyer, S. C., Gopal, U., Devaraj, N., and Halagowder, D. (2014). Shigella dysenteriae modulates BMP pathway to induce mucin gene expression in vivo and in vitro. PLoS ONE 9:e111408. doi: 10.1371/journal.pone.0111408
Gudmundsson, G. H., Bergman, P., Andersson, J., Raqib, R., and Agerberth, B. (2010). Battle and balance at mucosal surfaces–the story of Shigella and antimicrobial peptides. Biochem. Biophys. Res. Commun. 396, 116–119. doi: 10.1016/j.bbrc.2010.03.081
Guerin, P. J., Brasher, C., Baron, E., Mic, D., Grimont, F., Ryan, M., et al. (2004). Case management of a multidrug-resistant Shigella dysenteriae serotype 1 outbreak in a crisis context in Sierra Leone, 1999-2000. Trans. R. Soc. Trop. Med. Hyg. 98, 635–643. doi: 10.1016/j.trstmh.2004.01.005
Gutiérrez-Jiménez, J., Arciniega, I., and Navarro-Garcia, F. (2008). The serine protease motif of Pic mediates a dose-dependent mucolytic activity after binding to sugar constituents of the mucin substrate. Microb Pathog. 45, 115–123. doi: 10.1016/j.micpath.2008.04.006
Haider, K., Hossain, A., Wanke, C., Qadri, F., Ali, S., and Nahar, S. (1993). Production of mucinase and neuraminidase and binding of Shigella to intestinal mucin. J. Diarrhoeal. Dis. Res. 11, 88–92.
Henderson, I. R., Czeczulin, J., Eslava, C., Noriega, F., and Nataro, J. P. (1999). Characterization of pic, a secreted protease of Shigella flexneri and enteroaggregative Escherichia coli. Infect. Immun. 67, 5587–5596.
Holt, K. E., Baker, S., Weill, F.-X., Holmes, E. C., Kitchen, A., Yu, J., et al. (2012). Shigella sonnei genome sequencing and phylogenetic analysis indicate recent global dissemination from Europe. Nat. Genet. 44, 1056–1059. doi: 10.1038/ng.2369
Holt, K. E., Thieu Nga, T. V., Thanh, D. P., Vinh, H., Kim, D. W., Vu Tra, M. P., et al. (2013). Tracking the establishment of local endemic populations of an emergent enteric pathogen. Proc. Natl. Acad. Sci. U.S.A. 110, 17522–17527. doi: 10.1073/pnas.1308632110
Hulland, K. R. S., Leontsini, E., Dreibelbis, R., Unicomb, L., Afroz, A., Dutta, N. C., et al. (2013). Designing a handwashing station for infrastructure-restricted communities in Bangladesh using the integrated behavioural model for water, sanitation and hygiene interventions (IBM-WASH). BMC Public Health 13:877. doi: 10.1186/1471-2458-13-877
Johansson, M. E. V., Gustafsson, J. K., Holmén-Larsson, J., Jabbar, K. S., Xia, L., Xu, H., et al. (2014). Bacteria penetrate the normally impenetrable inner colon mucus layer in both murine colitis models and patients with ulcerative colitis. Gut 63, 281–291. doi: 10.1136/gutjnl-2012-303207
Johansson, M. E. V., Larsson, J. M. H., and Hansson, G. C. (2011). The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions. Proc. Natl. Acad. Sci. U.S.A. 108(Suppl. 1), 4659–4665. doi: 10.1073/pnas.1006451107
Johnson-Henry, K. C., Morona, R., Hagen, K. E., Daniels, C., Gordonpour, M., Van Den Bosch, L., et al. (2007). Surface-layer protein extracts from Lactobacillus helveticus inhibit enterohaemorrhagic Escherichia coli O157:H7 adhesion to epithelial cells. Cell Microbiol. 9, 356–367. doi: 10.1111/j.1462-5822.2006.00791.x
Kamp, H. D., Patimalla-Dipali, B., Lazinski, D. W., Wallace-Gadsden, F., and Camilli, A. (2013). Gene fitness landscapes of Vibrio cholerae at important stages of its life cycle. PLoS Pathog. 9, e1003800. doi: 10.1371/journal.ppat.1003800
Karlsson, N. G., Johansson, M. E., Asker, N., Karlsson, H., Gendler, S. J., Carlstedt, I., et al. (1996). Molecular characterization of the large heavily glycosylated domain glycopeptide from the rat small intestinal Muc2 mucin. Glycoconj. J. 13, 823–831. doi: 10.1007/BF00702346
Kernéis, S., Guerin, P. J., Seidlein von, L., Legros, D., and Grais, R. F. (2009). A look back at an ongoing problem: Shigella dysenteriae type 1 epidemics in refugee settings in Central Africa (1993-1995). PLoS ONE 4:e4494. doi: 10.1371/journal.pone.0004494
Khaghani, S., Shamsizadeh, A., Nikfar, R., and Hesami, A. (2014). Shigella flexneri: a three-year antimicrobial resistance monitoring of isolates in a Children Hospital, Ahvaz, Iran. Iran J. Microbiol. 6, 225–229.
Kim, J. S., Kim, J. J., Kim, S. J., Jeon, S.-E., Seo, K. Y., Choi, J.-K., et al. (2015). Outbreak of Ciprofloxacin-resistant Shigella sonnei associated with travel to Vietnam, Republic of Korea. Emerg. Infect. Dis. 21, 1247–1250. doi: 10.3201/eid2107.150363
Kocsis, A. K., Lakatos, P. L., Somogyvári, F., Fuszek, P., Papp, J., Fischer, S., et al. (2008). Association of beta-defensin 1 single nucleotide polymorphisms with Crohn's disease. Scand. J. Gastroenterol. 43, 299–307. doi: 10.1080/00365520701682615
Kotloff, K. L., Nataro, J. P., Blackwelder, W. C., Nasrin, D., Farag, T. H., Panchalingam, S., et al. (2013). Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet 382, 209–222. doi: 10.1016/S0140-6736(13)60844-2
Kumar, P., Luo, Q., Vickers, T. J., Sheikh, A., Lewis, W. G., and Fleckenstein, J. M. (2013). EatA, an immununogenic protective antigen of Enterotoxigenic Escherichia coli degrades intestinal mucin. Infect. Immun. 82, 500–508. doi: 10.1128/IAI.01106-13
Lavelle, A., Lennon, G., O'Sullivan, O., Docherty, N., Balfe, A., Maguire, A., et al. (2015). Spatial variation of the colonic microbiota in patients with ulcerative colitis and control volunteers. Gut 64, 1553–1561. doi: 10.1136/gutjnl-2014-307873
Levine, M. M., Kotloff, K. L., Barry, E. M., Pasetti, M. F., and Sztein, M. B. (2007). Clinical trials of Shigella vaccines: two steps forward and one step back on a long, hard road. Nat. Rev. Microbiol. 5, 540–553. doi: 10.1038/nrmicro1662
Maier, B. R., and Hentges, D. J. (1972). Experimental Shigella infections in laboratory animals. I. Antagonism by human normal flora components in gnotobiotic mice. Infect. Immun. 6, 168–173.
Mao, Y., Fernandez, M.-I., Cui, E., Regnault, B., Bao, C., Mulet, C., et al. (2013). Changing trends and serotype distribution of Shigella species in Beijing from 1994 to 2010. J. Immunol. 5, 21–4930. doi: 10.1186/1757-4749-5-21
Marteyn, B., Gazi, A., and Sansonetti, P. (2012). Shigella: a model of virulence regulation in vivo. Gut Microbes 3, 104–120. doi: 10.4161/gmic.19325
Martino, M. C., Rossi, G., Martini, I., Tattoli, I., Chiavolini, D., Phalipon, A., et al. (2005). Mucosal lymphoid infiltrate dominates colonic pathological changes in murine experimental shigellosis. J. Infect. Dis. 192, 136–148. doi: 10.1086/430740
McGuckin, M. A., Lindén, S. K., Sutton, P., and Florin, T. H. (2011). Mucin dynamics and enteric pathogens. Nat. Rev. Microbiol. 9, 265–278. doi: 10.1038/nrmicro2538
Meyer-Hoffert, U., Hornef, M. W., Henriques-Normark, B., Axelsson, L.-G., Midtvedt, T., Putsep, K., et al. (2008). Secreted enteric antimicrobial activity localises to the mucus surface layer. Gut 57, 764–771. doi: 10.1136/gut.2007.141481
Moorthy, G., Murali, M. R., and Niranjali Devaraj, S. (2010). Lactobacilli inhibit Shigella dysenteriae 1 induced pro-inflammatory response and cytotoxicity in host cells via impediment of Shigella-host interactions. Dig. Liver Dis. 42, 33–39. doi: 10.1016/j.dld.2009.04.021
Morona, R., Daniels, C., and Van Den Bosch, L. (2003). Genetic modulation of Shigella flexneri 2a lipopolysaccharide O antigen modal chain length reveals that it has been optimized for virulence. Microbiology 149(Pt 4), 925–939. doi: 10.1099/mic.0.26141-0
Navarro-Garcia, F., Gutierrez-Jimenez, J., Garcia-Tovar, C., Castro, L. A., Salazar-Gonzalez, H., and Cordova, V. (2010). Pic, an autotransporter protein secreted by different pathogens in the Enterobacteriaceae family, is a potent mucus secretagogue. Infect. Immun. 78, 4101–4109. doi: 10.1128/IAI.00523-10
Ottemann, K. M., and Lowenthal, A. C. (2002). Helicobacter pylori uses motility for initial colonization and to attain robust infection. Infect. Immun. 70, 1984–1990. doi: 10.1128/IAI.70.4.1984-1990.2002
Pelaseyed, T., Bergström, J. H., Gustafsson, J. K., Ermund, A., Birchenough, G. M. H., Schütte, A., et al. (2014). The mucus and mucins of the goblet cells and enterocytes provide the first defense line of the gastrointestinal tract and interact with the immune system. Immunol. Rev. 260, 8–20. doi: 10.1111/imr.12182
Perepelov, A. V., Shevelev, S. D., Liu, B., Senchenkova, S. N., Shashkov, A. S., Feng, L., et al. (2010). Structures of the O-antigens of Escherichia coli O13, O129, and O135 related to the O-antigens of Shigella flexneri. Carbohydr. Res. 345, 1594–1599. doi: 10.1016/j.carres.2010.04.023
Phalipon, A., and Sansonetti, P. J. (2007). Shigella's ways of manipulating the host intestinal innate and adaptive immune system: a tool box for survival? Immunol. Cell Biol. 85, 119–129. doi: 10.1038/sj.icb7100025
Pongpech, P., Hentges, D. J., Marsh, W. W., and Eberle, M. E. (1989). Effect of streptomycin administration on association of enteric pathogens with cecal tissue of mice. Infect. Immun. 57, 2092–2097.
Prakash, R., Bharathi Raja, S., Devaraj, H., and Devaraj, S. N. (2011). Up-regulation of MUC2 and IL-1β expression in human colonic epithelial cells by Shigella and its interaction with mucins. PLoS ONE 6:e27046. doi: 10.1371/journal.pone.0027046
Qiu, S., Xu, X., Yang, C., Wang, J., Liang, B., Li, P., et al. (2015). Shift in serotype distribution of Shigella species in China, 2003-2013. Clin. Microbiol. Infect. 21, 252.e5–e8. doi: 10.1016/j.cmi.2014.10.019
Qu, F., Ying, Z., Zhang, C., Chen, Z., Chen, S., Cui, E., et al. (2014). Plasmid-encoding extended-spectrum beta-lactamase CTX-M-55 in a clinical Shigella sonnei strain, China. Future Microbiol. 9, 1143–1150. doi: 10.2217/fmb.14.53
Rahman, K. M., Arifeen, S. E., Zaman, K., Rahman, M., Raqib, R., Yunus, M., et al. (2011). Safety, dose, immunogenicity, and transmissibility of an oral live attenuated Shigella flexneri 2a vaccine candidate (SC602) among healthy adults and school children in Matlab, Bangladesh. Vaccine 29, 1347–1354. doi: 10.1016/j.vaccine.2010.10.035
Raja, S. B., Murali, M. R., Devaraj, H., and Devaraj, S. N. (2012). Differential expression of gastric MUC5AC in colonic epithelial cells: TFF3-wired IL1 β/Akt crosstalk-induced mucosal immune response against Shigella dysenteriae infection. J. Cell Sci. 125, 703–713. doi: 10.1242/jcs.092148
Rajilić-Stojanović, M., and de Vos, W. M. (2014). The first 1000 cultured species of the human gastrointestinal microbiota. FEMS Microbiol. Rev. 38, 996–1047. doi: 10.1111/1574-6976.12075
Rashid, H., and Rahman, M. (2015). Possible transfer of plasmid mediated third generation cephalosporin resistance between Escherichia coli and Shigella sonnei in the human gut. Infect. Genet. Evol. 30, 15–18. doi: 10.1016/j.meegid.2014.11.023
Robbe, C., Capon, C., Maes, E., Rousset, M., Zweibaum, A., Zanetta, J. P., et al. (2003). Evidence of regio-specific glycosylation in human intestinal mucins: presence of an acidic gradient along the intestinal tract. J. Biol. Chem. 278, 46337–46348. doi: 10.1074/jbc.M302529200
Ruiz-Perez, F., Wahid, R., Faherty, C. S., Kolappaswamy, K., Rodriguez, L., Santiago, A., et al. (2011). Serine protease autotransporters from Shigella flexneri and pathogenic Escherichia coli target a broad range of leukocyte glycoproteins. Proc. Natl. Acad. Sci. U.S.A. 108, 12881–12886. doi: 10.1073/pnas.1101006108
Sack, D. A., Hoque, A. T., Huq, A., and Etheridge, M. (1994). Is protection against shigellosis induced by natural infection with Plesiomonas shigelloides? Lancet 343, 1413–1415. doi: 10.1016/S0140-6736(94)92531-3
Saeed, A., Johansson, D., Sandström, G., and Abd, H. (2012). Temperature depended role of Shigella flexneri invasion plasmid on the interaction with Acanthamoeba castellanii. Int. J. Microbiol. 2012, 917031. doi: 10.1155/2012/917031
Sankar, S. A., Lagier, J.-C., Pontarotti, P., Raoult, D., and Fournier, P.-E. (2015). The human gut microbiome, a taxonomic conundrum. Syst. Appl. Microbiol. 38, 276–286. doi: 10.1016/j.syapm.2015.03.004
Senchenkova, S. N., Feng, L., Yang, J., Shashkov, A. S., Cheng, J., Liu, D., et al. (2005). Structural and genetic characterization of the Shigella boydii type 10 and type 6 O antigens. J. Bacteriol. 187, 2551–2554. doi: 10.1128/JB.187.7.2551-2554.2005
Shiga, K. (1898). Ueber den erreger der Dysenterie in Japan. Vorlaufige Mitteilung Zentralbal Bakteriol. Microbiol. Hyg. 23, 599–600.
Sonne, C. (1915). Ueber die Bakteriologie der giftarmen Dysenteriebacillen (Para-dysenteriebacillen). Zentr. Bakt. Parsitenk. Orig. 76, 408–456.
Sonnenburg, J. L., Xu, J., Leip, D. D., Chen, C.-H., Westover, B. P., Weatherford, J., et al. (2005). Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science 307, 1955–1959. doi: 10.1126/science.1109051
Sperandio, B., Fischer, N., Joncquel Chevalier-Curt, M., Rossez, Y., Roux, P., Robbe Masselot, C., et al. (2013). Virulent Shigella flexneri affects secretion, expression, and glycosylation of gel-forming mucins in mucus-producing cells. Infect. Immun. 81, 3632–3643. doi: 10.1128/IAI.00551-13
Sperandio, B., Regnault, B., Guo, J., Zhang, Z., Stanley, S. L. Jr. Sansonetti, P. J., et al. (2008). Virulent Shigella flexneri subverts the host innate immune response through manipulation of antimicrobial peptide gene expression. J. Exp. Med. 205, 1121–1132. doi: 10.1084/jem.20071698
Sudha, P. S., Devaraj, H., and Devaraj, N. (2001). Adherence of Shigella dysenteriae 1 to human colonic mucin. Curr. Microbiol. 42, 381–387. doi: 10.1007/s002840010234
Thompson, C. N., Duy, P. T., and Baker, S. (2015). The rising dominance of Shigella sonnei: an intercontinental shift in the etiology of bacillary dysentery. PLoS Negl. Trop. Dis. 9:e0003708. doi: 10.1371/journal.pntd.0003708
Tien, M.-T., Girardin, S. E., Regnault, B., Le Bourhis, L., Dillies, M. A., Coppée, J. Y., et al. (2006). Anti-inflammatory effect of Lactobacillus casei on Shigella-infected human intestinal epithelial cells. J. Immunol. 176, 1228–1237. doi: 10.4049/jimmunol.176.2.1228
Tran, E. N. H., Papadopoulos, M., and Morona, R. (2014). Relationship between O-antigen chain length and resistance to colicin E2 in Shigella flexneri. Microbiology 160, 589–601. doi: 10.1099/mic.0.074955-0
Tran Van Nhieu, G., Bourdet-Sicard, R., Duménil, G., Blocker, A., and Sansonetti, P. J. (2000). Bacterial signals and cell responses during Shigella entry into epithelial cells. Cell Microbiol. 2, 187–193. doi: 10.1046/j.1462-5822.2000.00046.x
Valentino, M. D., Foulston, L., Sadaka, A., Kos, V. N., Villet, R. A., Santa Maria, J. Jr., et al. (2014). Genes contributing to Staphylococcus aureus fitness in abscess- and infection-related ecologies. MBio 5, e01729–e01714. doi: 10.1128/mBio.01729-14
Varum, F. J. O., Veiga, F., Sousa, J. S., and Basit, A. W. (2012). Mucus thickness in the gastrointestinal tract of laboratory animals. J. Pharm. Pharmacol. 64, 218–227. doi: 10.1111/j.2042-7158.2011.01399.x
Vighi, G., Marcucci, F., Sensi, L., Di Cara, G., and Frati, F. (2008). Allergy and the gastrointestinal system. Clin. Exp. Immunol. 153(Suppl. 1), 3–6. doi: 10.1111/j.1365-2249.2008.03713.x
Zhang, J., Qian, L., Wu, Y., Cai, X., Li, X., Cheng, X., et al. (2013). Deletion of pic results in decreased virulence for a clinical isolate of Shigella flexneri 2a from China. BMC Microbiol. 13:31. doi: 10.1186/1471-2180-13-31
Keywords: Shigella, diversity, virulence, microbiota, mucus
Citation: Anderson M, Sansonetti PJ and Marteyn BS (2016) Shigella Diversity and Changing Landscape: Insights for the Twenty-First Century. Front. Cell. Infect. Microbiol. 6:45. doi: 10.3389/fcimb.2016.00045
Received: 07 January 2016; Accepted: 28 March 2016;
Published: 19 April 2016.
Edited by:
William D. Picking, University of Kansas, USAReviewed by:
V. K. Viswanathan, University of Arizona, USASuzanne Mariane Janete Fleiszig, University of California, Berkeley, USA
Copyright © 2016 Anderson, Sansonetti and Marteyn. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Benoit S. Marteyn, marteyn@pasteur.fr
 Mark Anderson1,2
Mark Anderson1,2  Benoit S. Marteyn
Benoit S. Marteyn