Enzymes in Fish and Seafood Processing
- 1Department of Bioengineering, Institute for Biotechnology and Bioengineering, Instituto Superior Técnico, Universidade de Lisboa, Lisbon, Portugal
- 2Faculdade de Engenharia, Universidade Lusófona de Humanidades e Tecnologias, Lisbon, Portugal
Enzymes have been used for the production and processing of fish and seafood for several centuries in an empirical manner. In recent decades, a growing trend toward a rational and controlled application of enzymes for such goals has emerged. Underlying such pattern are, among others, the increasingly wider array of enzyme activities and enzyme sources, improved enzyme formulations, and enhanced requirements for cost-effective and environmentally friendly processes. The better use of enzyme action in fish- and seafood-related application has had a significant impact on fish-related industry. Thus, new products have surfaced, product quality has improved, more sustainable processes have been developed, and innovative and reliable analytical techniques have been implemented. Recent development in these fields are presented and discussed, and prospective developments are suggested.
Introduction
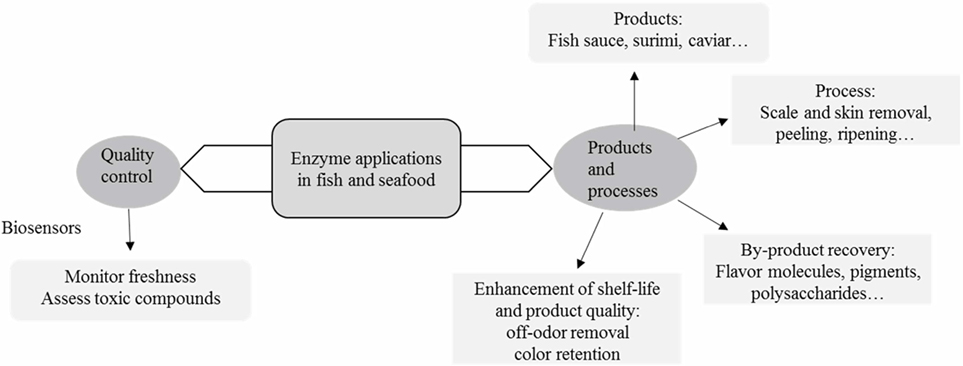
Enzymes are key tools in biotechnology and related areas because of their catalytic nature (Fraatz et al., 2014; Jemli et al., 2016). Accordingly, they have been extensively used in food production and processing for centuries, albeit in a rather empirical manner, which has been superseded by a rational approach in the last decades (Whitaker, 1994; Whitaker et al., 2002; Fraatz et al., 2014). In recent years, the focus has been on technical and scientific issues (enzyme formulations, molecular improvement of enzyme, screening for new/improved enzymes through traditional and metagenomics approaches, process improvement) as well as on legal and regulatory matters (definition of enzymes and technological purposes, procedures for safety assessment, harmonization of regulations, among others), all of these abridging the food industry (Fraatz et al., 2014; Li and Cirino, 2014; Alma’abadi et al., 2015; Jemli et al., 2016). Within this latter area, fish and seafood comprise a significant market (Morrissey and DeWitt, 2014), where enzyme action plays an effective role. In particular, and somehow not surprisingly, the use of enzymes from the marine environment has gradually been emerging as a relevant tool for fish and seafood processing (Diaz-López and García-Carreño, 2000; Shahidi and Janak Kamil, 2001; Venugopal, 2005; Sana, 2015), although this is sometimes overshadowed by other applications in food production and processing, e.g., bakery, beverages, and starch processing (Fraatz et al., 2014). This paper aims to provide an overview on the current status on the relevant uses of enzymes for fish and seafood processing and analysis. These are illustrated in Figure 1.
Enzyme Sources
When considering the enzymatic processing of fish and seafood, the role of both endogenous and added enzymes has to be considered. In the latter case, the enzymes used are from mammalian, plant, or microbial sources. Ease of manipulation and cultivation of the latter makes them the preferred source of enzymes. These are typically from terrestrial organisms, yet given the wide pool of marine microorganisms, the trend toward the use of these as enzyme sources has been increasing (Trincone, 2011, 2013). In particular, they are often adapted so as to display high activity at relatively low temperatures, unlike many of enzymes from terrestrial sources, thereby making them more effective in many processes that require often a low-temperature environment (Simpson, 2012).
Process Applications
Traditional use of enzymes in seafood processing involves the use of proteases, namely bacillolysin (Neutrase®), ficin, papain, pepsin, subtilisin (Alcalase®), trypsin, and a mixture of bacillolysin and subtilisin (Protamex®), of both endogenous and exogenous nature, for descaling and deskinning, peeling of shrimp, production of caviar and fish sauce, recovery of diverse molecules, and tenderization of squid, as reviewed by several authors (Haard and Simpson, 1994; Vilhelmsson, 1997; Diaz-López and García-Carreño, 2000; Suresh et al., 2015).
Proteases
Proteases are widely used in fish and seafood processing (Diaz-López and García-Carreño, 2000; Suresh et al., 2015), covering a wide array of applications.
Deskinning and Descaling
Deskinning involves the removal of fish skin without causing damage to the flesh, a process currently performed by rough mechanical procedures, imparting considerable risk of damaging the flesh and producing excessive waste. Moreover, enzymatic deskinning can improve the edible yield. Several specific applications have been implemented, specifically for processing herring, pollock, squid, skate, shrimp shells, and tuna, occasionally combined with physical treatment (Haard and Simpson, 1994; Rasika et al., 2013). Several of these methods involve the use of enzymes from marine organisms, for example, acid proteases from cod viscera for herring, protease extracts from minced arrowtooth flounder for pollock, and enzymes from squid for squid itself (Simpson, 2012). Recently, commercial proteases (Proleather FG-F® and Protease N®) and collagenase (CLS1®) were tested for the deskinning of catfish nuggets. Proleather FG-F® proved effective, and operational conditions (enzyme concentration, time, and temperature of incubation) were identified that optimized removal of the peritoneal membrane (Kim et al., 2014). Descaling can also be performed by mechanical methods, but again it is a harsh treatment and may result in tearing of the skin and lower filet yield. Thus, the milder enzyme approach is favored, particularly if mixtures of fish digestive proteases that enable operation at low temperatures, are used (Svenning et al., 1993; Gildberg et al., 2000; Gildberg, 2004). This approach has been assessed for scale removal of haddock and redfish (Haard and Simpson, 1994) in Japanese sashimi restaurants and fresh-fish markets (Simpson, 2012).
Proteases have also been used for the removal of raw meat from the head-shell of crustaceans, by immersing the latter in an enzyme solution (Gallant et al., 2001), although the reliability of the method has been questioned (Jabbour and Hognason, 2007). Early efforts for shrimp peeling and de-veining and for shucking clams have been reported, involving a mixture of ficin and amylase in the former case and ficin, amylases, and cellulases in the latter (Venugopal et al., 2000).
Fish Protein Hydrolyzate
One of the major established applications is the production of fish protein hydrolyzates (FPHs). FPH is the result of the enzymatic (endo- and/or exo-peptidases) or chemical hydrolysis of protein-rich byproduct waste of the fish processing industry, such as bones, head, liver, skin, trimmings, and viscera of fish flesh and of minces, leading to peptides with 2–20 amino acids, depending on the enzymes used, the fish used as source, the time of incubation, and concomitantly the degree of hydrolysis (Dh), defined as the ratio of the number of broken peptide bonds (p) to the total number of peptide bonds per mass unit (ptot).
Thus, a free α-amino group is formed for each hydrolyzed peptide bond (Nguyen et al., 2011; Chalamaiah et al., 2012; Benjakul et al., 2014; He et al., 2015; Suresh et al., 2015).
Traditional FPH hydrolysis was promoted either by acid or alkali action. Acid hydrolysis involves the use of concentrated hydrochloric acid, or occasionally sulfuric acid, operation at high temperatures and pressures, and neutralization of the hydrolyzate. Accordingly, the hydrolyzate contains large amounts of sodium chloride, which impairs its functionality. Moreover, tryptophan, a key amino acid, is destroyed in the process. Alkali hydrolysis also involves relatively high temperatures and concentrated sodium hydroxide. Moreover, during the process, several unwanted reactions occur, which lead to the formation of toxic compounds and impair the functionality of the hydrolyzate (Kristinsson and Rasco, 2000). The enzyme approach, although complex, occurs under mild conditions of temperature, pressure, and pH and involves the use of proteolytic enzymes, typically available at low cost, and deleterious reactions are virtually nonexistant. Hence, this approach is technically and economically attractive (He et al., 2013). FPHs display functional properties of interest for food formulation, namely emulsification and foam-forming ability, gelling activity, protein solubility, oil-binding capability, and water-holding capacity (Kristinsson and Rasco, 2000; Chalamaiah et al., 2010; He et al., 2015). FPHs compare favorably with poultry byproducts and protein hydrolyzate, both obtained by proteolysis with Alcalase, an outcome ascribed to the difference in amino acid composition (Taheri et al., 2013). Moreover, FPHs are envisaged as effective source of proteins for human nutrition, given their balanced amino acid composition and easier gastrointestinal adsorption when compared to free amino acids (Clemente, 2000). FPHs also exhibit antioxidant, antihypertensive, immunomodulatory, and antimicrobial activities, hence their incorporation in nutraceuticals and functional/health foods has recently emerged, as evidenced by the presence of several commercially available products (Chalamaiah et al., 2012; Hu et al., 2015). FPHs have a bitter taste, which is one of the key issues that prevents its dissemination in food products, East Asian condiments, and sauces (Kristinsson and Rasco, 2000). Moreover, excessive hydrolysis is likely to impair some functional properties or cause off-flavors in the final product (Balti et al., 2010). Several approaches for de-bittering have been tested, with different advantages and limitations. Among these, the enzymatic action of exopeptidases and concomitant removal of free amino acids has emerged as the most promising (Sujith and Hymavathi, 2011). Recently, the possibility of using FPHs as cryoprotective agents to preserve frozen fish, as an alternative to commonly used carbohydrate-based agents, was suggested, given the positive results obtained when hydrolyzates from Pacific hake (Merluccius productus) were obtained upon proteolysis using either Alcalase or Flavourzyme. FPHs were used for frozen storage of minced cod samples and displayed similar or better cryoprotective properties than a standard sucrose–sorbitol mixture (Cheung et al., 2009).
The production of FPHs typically requires the addition of enzymes from different sources, namely plants (papain, bromelain), microorganisms involving both commercial formulations (Alcalase, Flavourzyme, Neutrase, Protamex) and crude enzyme preparations (orientase, papain, trypsin, thermolysin), and fish digestive enzymes (pepsin, trypsin, chymotrypsin) (Kristinsson and Rasco, 2000; Benjakul et al., 2014; Jridi et al., 2014; Yarnpakdee et al., 2015). The production of FPHs using either endogenous enzymes such as cathepsin L in autocatalytic processes or endogenous enzymes combined with exogenous enzymes has also been performed (Kristinsson and Rasco, 2000; Samaranayaka and Li-Chan, 2008; Ovissipour et al., 2013). Still, processes based only on the use of endogenous enzymes lead to low protein content and recovery (Samaranayaka and Li-Chan, 2008; Ovissipour et al., 2013).
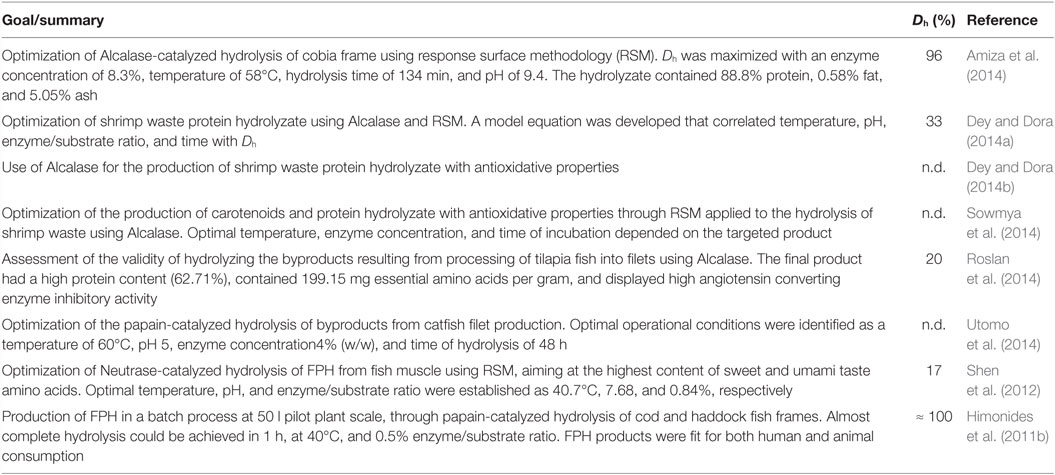
Efforts have been recently made to optimize the process of FPH production and to make better use of wastes in order and obtain products with relevant activity (Table 1). Further details on the use of enzymes for this application can be found in a recent review (Benjakul et al., 2014).
Fish Sauce
Fish sauce is the outcome of enzyme-solubilized and digested fish protein. The preparation is preserved in salt and used as an ingredient and condiment on vegetable dishes. Currently associated with Southeast Asia, fish sauce was quite popular in Roman culture, but since then it has almost vanished from Europe (Gildberg et al., 2000; Tanasupawat and Visessanguan, 2014). Protein hydrolysis takes place by autolysis, mainly involving trypsin and chymotrypsin, alongside cathepsins. As the pH of fish sauce decreases during fermentation, from around 7 to 5, the role of the enzymes in protein digestions is complementary because, while the two former ones are active at pH ~7, the latter are active in acidic environments (Lopetcharat et al., 2001; Turk et al., 2012). The extent of proteolysis is typically characterized by Dh.
The traditional process, which relies solely on autolysis involving fermentation and endogenous enzymes, is quite time consuming because it takes several months up to 3 years for full completion (Lopetcharat et al., 2001; Gildberg et al., 2007; Faisal et al., 2015; Lee et al., 2015). Therefore, the use of exogenous enzymes, such as bromelain, ficin, or papain, as well as the commercial preparations Protamex and particularly Protex 51FP and Neutrase, has been also shown to speed up fish fermentation (Beddows and Ardeshir, 1979; Ooshiro et al., 1981; Chuapoehuk and Raksakulthai, 1992; Aquerreta et al., 2001; Himonides et al., 2011a; Le et al., 2015). The use of exogenous enzymes can increase significantly the pace of fish fermentation (Aquerreta et al., 2001), yet their application should be carefully assessed in order not to tamper with the required functional and organoleptic properties and quality of the final product (Ooshiro et al., 1981; Himonides et al., 2011a; Ghaly et al., 2015). On the other hand, the use of exogenous enzymes can lead to a final product that fulfills the intended role and displays adequate features and even promising nutraceutical properties. This is due to the presence of polyunsaturated fatty acids, lower level of salt, and higher protein content than currently produced fish sauces in Southeast Asia. The preparation obtained with exogenous enzymes was also considered to be more similar to the fish sauce produced in classical Roman age (Aquerreta et al., 2001). Moreover, and besides process conditions, the functional and organoleptic properties and the nature of the final product are noticeably influenced by the starting material, as reported by Gildberg et al., who compared fish sauces obtained from several cold-water and tropical species. Thus, not only the overall content of proteins but also the profile of amino acids differs, and possibly that of fatty acids, which is likely the result of the diverse endogenous microbial and enzymatic activity (Gildberg et al., 2007). Different contents of protein and lipidic components were also observed when sardines and anchovy as raw materials were compared (Le et al., 2015). The ratio of salt to fish also conditions enzyme activity, with impact on the characteristics of the final product (Gildberg et al., 2007; Tanasupawat and Visessanguan, 2014; Le et al., 2015). Hence, the detailed characterization of catalytic activity requires chemical, physical, and sensorial evaluation, alongside microbiological analysis (Aquerreta et al., 2001; Gildberg et al., 2007; Tanasupawat and Visessanguan, 2014; Faisal et al., 2015).
Ripening
Proteases are also involved in the ripening of salted fish such as herring, anchovy, cod, and salmon, a complex biochemical process mostly characterized by the degradation of muscle proteins with endogenous enzymes, with increase of peptides and free amino acids. The activity of enzymes from the digestive tract, namely chymotrypsin and trypsin, is typically the most significant in the ripening process, although muscle proteases, namely cathepsins, also play a non-negligible role (Sikorski, 2007; Bjørkevoll et al., 2008; Rahaman, 2014).
Transglutaminase
Transglutaminases (TGAs), protein-glutamine γ-glutamyl-transferase (EC 2.3.2.13), promote acyl transfer reactions. These involve the γ-carboxyamide group of a peptide-bound glutamine residue as acyl donors and several primary amines as acyl acceptors, for example, the ɛ-amino group of lysine. Concomitantly, intra and intermolecular covalent bonds are formed, namely ɛ-(γ-glutamyl)–lysine, resulting in the cross-linking of peptides and proteins and polymerization. When primary amines are absent, water acts as acyl receptor, and the γ-carboxyamide groups of glutamine residues are deaminated into glutamic acid residues (Diaz-López and García-Carreño, 2000; Sikorski, 2007; Zilda, 2014). TGAs are available form mammalian, plant, and microbial sources, the two former being Ca2+ dependent (TGase), unlike the latter (microbial TGA; MTGase). This feature, along with the facile and more cost-effective production of enzymes from microbial sources, makes these the source of the diverse commercial formulations of TGAs (Sikorski, 2007; Serafini-Fracassini and Del Duca, 2008; Kieliszek and Misiewicz, 2014; Zilda, 2014).
Given their cross-linking ability, MTGases are used for the modification/improvement of the functional and mechanical properties of fish and seafood products, and therefore are used as binding ingredients for the restructuration of raw meats and in the production of surimi (Mariniello et al., 2008; Kieliszek and Misiewicz, 2014; Zilda, 2014). Also, TGAs are used in formulations of fish meat mince, modification of finfish texture, processing of shark fin, formation of collagen and gelatin bonds, and minimization of drip after thawing (Diaz-López and García-Carreño, 2000; Zilda, 2014; Suresh et al., 2015). Most of these processes involve the use of added MTGases; still, endogenous TGases account for ɛ-(γ-glutamyl)–lysine cross-linking in dried fish, frozen-stored surimi, and the polymerization of myosin heavy chains in the manufacture of kamaboko (Sikorski, 2007). The cost of enzymes may still limit the extention of this strategy to enhance the mechanical properties of these products. Still, their environmentally friendly nature, high activity, and specificity provide a promising alternative or complementary nature to the use of protease inhibitors (which can lead to unwanted changes in color and flavor), phosphates (which have a negative environmental impact), or oxidizing agents (Martín-Sánchez et al., 2009). The application of TGAs in fish and seafood has been reviewed recently (Kieliszek and Misiewicz, 2014; Zilda, 2014; Suresh et al., 2015), yet some illustrative examples of the mentioned applications are given in Table 2.
Lipases and Miscellaneous Process Applications
Lipases, triacylglycerol acylhydrolases (EC 3.1.1.3), promote the hydrolysis of tri-, di-, and monoglycerides to glycerol and fatty acids, in the presence of excess water, while in water-limiting conditions they promote ester synthesis. They often express other activities, namely phospholipase or other esterase type of activity, all of which have acknowledged industrial relevance (Venugopal et al., 2000; Verma et al., 2012). Lipases are of particular interest for the isolation of oil and fats from seafood byproducts as well as in the preparation of ω-3-poly-unsaturated fatty acids (ω-PUFAs) and enriched marine oils, given the nutritional value of these compounds (Chen et al., 2012; Walker et al., 2015). The enrichment performed chemically requires temperature and pH conditions harmful for the labile substrates, and hence the mild conditions required by lipases to promote transesterification are favored (Diaz-López and García-Carreño, 2000). Examples of applications can be found in a recent published review (Chaurasia et al., 2016).
Other applications of enzymes in fish and seafood processing include the production of caviar, the recovery of chitin, collagen, flavor molecules, minerals, and pigments from seafood byproducts, the removal of unwanted odors, and the improvement of shelf-life and color retention (Suresh et al., 2015).
Proteolytic enzymes, namely pepsins, have been used as alternative to mechanical and manual methods in the production of caviar. Extraction of caviar from roe sacs by enzymes minimizes damage to the eggs and results in a product free from connective tissue, as compared to the other methods, thereby leading to higher process yields (Martin et al., 2000; Venugopal, 2009).
Chitin, a linear heterogeneous polysaccharide of N-acetyl-d-glucosamine and d-glucosamine, linked by β(1,4) glycosidic bonds, present in crustaceans and mollusks, is the most abundant and renewable polysaccharide on Earth, next to cellulose (Gortari and Hours, 2013). Overall, 10 billion tons of chitin is produced yearly (Zargar et al., 2015). Given its environmental friendliness, biocompatibility, biodegradability, and the relative ease of functionalization, chitin has a wide array of applications. These include food and feed, where chitin can be used as antioxidant, emulsifier, and thickening agent and for clarification of fruit juices, stabilization of color, reduction of lipid adsorption, and the preparation of dietary fibers. In addition, chitin is widely used in biomedical, pharmaceutical, tissue engineering, cosmetics, and wastewater sectors (Zargar et al., 2015). For this byproduct of the crustacean processing industry to be of use, the removal of minerals (demineralization), proteins (deproteinization), and pigments (decolorization) is required (Suresh et al., 2015; Zargar et al., 2015). The conventional process for chitin recovery and purification requires the use of strong alkali and acid solutions and relatively high temperatures. This involves an energy-consuming and environmentally hazardous process and leads to a product of relatively low quality. Moreover, the alkali-based deproteinization process prevents the use of the protein in animal feed (Synowiecki and Al-Khateeb, 2000; Gortari and Hours, 2013; Younes and Rinaudo, 2015; Zargar et al., 2015). Several proteolytic enzymes have been used for protein removal from chitin, such as Alcalase, pancreatine, papain, pepsin, and trypsin. Still, and despite enzyme screening and operational condition optimization, the enzymatic process is less effective than the conventional chemical method, as 5%–10% residual protein remains attached to the purified chitin when the former method is used (Younes and Rinaudo, 2015). In order to overcome this limitation, the use of a mild alkali treatment upon enzymatic proteolysis has been suggested (Younes and Rinaudo, 2015). Moreover, it has been reported that the order of demineralization and deproteinization is largely irrelevant in the yield and quality of chitin purification when chemical methods are used, whereas the presence of minerals may hamper the access of proteolytic enzymes to the substrate (Gortari and Hours, 2013; Younes and Rinaudo, 2015). This may be the reason underlying the data reported by Valdez-Peña and coworkers (Valdez-Pena et al., 2010). These authors reported an ecofriendly process for purifying chitin, with a sequential combination of enzymatic deproteinization and microwave irradiation for demineralization, where despite screening for enzyme activity, the residual protein was roughly half of the initial value (Valdez-Pena et al., 2010). Implementation of enzymatic deproteinization at the commercial scale is also limited by the cost of commercially available enzymes. An alternative approach, eventually more cost effective, involves the use of crude protease preparations from supernatants of microbial fermentations (Paul et al., 2015).
Collagen is a fibrous protein found in animal skin, bone, and connective tissue, accounting for about 30% of total protein content (Pal and Suresh, 2016). Collagen is widely used in the food and beverages industry as antioxidants, emulsifiers, thickeners, and preservatives, but also as edible films and coatings. Moreover, collagen is also used in biomedical, pharmaceutical, tissue engineering, and cosmetics areas (Benjakul et al., 2012; Hashim et al., 2015; Pal and Suresh, 2016). The outbreak of bovine spongiform encephalopathy and bird flu resulted in an increasing demand for collagen from fish, where it can be extracted from the skin, scale, swim bladder, fins, and bones (Benjakul et al., 2012; Pal and Suresh, 2016). Extraction of collagen must be performed at a temperature of ~ 4°C to minimize collagen degradation. Pepsin is the most common enzyme used for collagen extraction, occasionally used together with acetic acid. This enzymatic method displays particular features that are of interest, namely the hydrolysis of non-collagenous proteins, the hydrolysis of the telopeptides of collagen, enhancing its solubility in acid and concomitantly the extraction yield, and simultaneously reducing the antigenicity caused by telopeptides. Despite these advantages, acid extraction is by far the most widely used method for collagen extraction, possibly because of it low cost and ease of implementation (Benjakul et al., 2012; Hashim et al., 2015; Pal and Suresh, 2016).
The recovery of flavor molecules from seafood byproducts relies mostly on the use of commercial protease preparations, for example, Flavourzyme (leucyl aminopeptidase) and Protamex (Suresh and Prabhu, 2013). More recently, the effective use of bromelain has also been reported for the recovery of seafood-like flavor from byproducts of seaweed (Laohakunjit et al., 2014). Despite the limited availability of technical information, the selection of the most adequate enzyme for flavor recovery from seafood byproducts is largely casuistic and depends on the nature of the raw material (Suresh and Prabhu, 2013). Proteases are also used for the recovery of minerals form seafood byproducts, such as fishbone, which is rich in calcium and phosphorus (Suresh and Prabhu, 2013; Suresh et al., 2015).
Carotenoids and melanin are the major pigments found in the byproducts of seafood processing (Suresh and Prabhu, 2013). Carotenoids are used as additives in feed, to convey skin pigmentation in fish, suggestive of high quality and freshness to the consumer (Suresh and Prabhu, 2013; Malaweera and Wijesundara, 2014). The unstable carotenoids are typically extracted from crustaceous waste using organic solvents, which is environmentally hazardous and requires solvent recycling (Malaweera and Wijesundara, 2014). Enzymatic extraction relies on the use of proteases, mostly trypsin, to recover carotenoids in the form of carotenoproteins (Suresh and Prabhu, 2013). Recently, the use of crude protease extracts from the hepatopancreas of Pacific white shrimp allowed the extraction of carotenoproteins from shrimp waste rich in astaxanthin and displaying significant antioxidant activity. The crude nature of the active enzyme extract may underlie the development of a cost-effective methodology (Senphan et al., 2014).
The improvement of the shelf-life of fishery products depends on the development of strategies that prevent the action of deleterious agents such as endogenous enzymes, microbial contamination, and oxidation of lipid compounds. In alternative or alongside methodologies such as active packaging, controlled-atmosphere packaging, and natural preservatives such as plant materials, the use of glucose oxidase and catalase has been reported to prevent lipid oxidation (Campos et al., 2012; Erkmen, 2012; Siró, 2012; Khalafalla et al., 2015). Glucose oxidase has been also used for color retention in cooked shrimp and crab, as the enzyme prevents the oxidation of carotenoids (Venugopal et al., 2000).
Off-odors and fishy taste, mostly due to the presence of urea in the meat of sharks and rays, have been tackled by the use of materials rich in urease, such as soybean flour (Suresh et al., 2015).
Analytical Applications
Aquaculture and fish production has grown considerably in the last decades (FAO, 2014), but occasional toxic episodes, typically caused by toxins and involving shellfish, raises public health concerns and presents challenges for marketing those goods (FAO, 2011; Rodríguez et al., 2015). Several methods have therefore been developed for toxin screening, among which are enzyme-based methods such as enzyme-linked immunosorbent assays (ELISA). Briefly, this method involves the immobilization of the target antigen to a solid surface and its subsequent complexation with an antibody linked to an enzyme. The detection is carried out by incubating the enzyme, often peroxidase or alkaline phosphatase (Alp), in the presence of substrate, and assessing the formation of an easily measurable product (Rustad, 2010). Hence, commercially available ELISA kits have been successfully tested for the determination and quantification of antimicrobials (e.g., crystal violet, chloramphenicol, gentamicin, fluoroquinolone enrofloxacin, malachite green, metabolites of furaltadone and furazolidone) in fish from aquaculture, to assess illegal use of the compounds (Jester et al., 2014; Conti et al., 2015). These kits have also proved effective in the detection of toxins [e.g., paralytic shellfish poisoning (PSP), diarrhetic shellfish poisoning, neurotoxic shellfish poisoning, and amnesic shellfish] in shellfish and seafood (Garet et al., 2010; Huazhang et al., 2011; Eberhart et al., 2013; Turner and Goya, 2016) and pesticides in fish (Sapozhnikova et al., 2015). Still, when ELISA method was compared to a phosphatase 2A inhibition assay, the latter displayed more promising results as a screening tool for diarrhetic shellfish toxins, given the sensitivity and low level of false results (Eberhart et al., 2013).
Alongside commercial ELISA kits, researchers have developed setups based on ELISA methods anchored in horseradish peroxidase (Hrp). These have been assayed
(a) for the determination of anisakis larvae in seafood, with lower limits of detection within 5 to 250 parasites per kg of sample, depending on the specific features of the method and of the allergen targeted (Arilla et al., 2008; Xu et al., 2010);
(b) for the detection and quantification of malachite green, a dye with antimicrobial and antiparasitic properties, that has been illicitly used as an antifungal agent in aquaculture. The compound is rapidly metabolized to leucomalachite green, both compounds having putative carcinogenic activity. The setup developed allowed a limit of quantification (LOQ) and a limit of detection (LOD) for mixtures of the two compounds of 0.3 and 0.1 μg/kgfish, respectively, which are below the concentration of 2 μg/kgfish imposed by the EU as LOD and 1 μg/kgfish imposed by Canadian legislation as the limiting threshold above which fish is not allowed into the market (European Commission, 2004; Singh et al., 2011);
(c) for the detection of Vibrio parahaemolyticus, a pathogen that causes gastroenteritis, in seafood, resulting in a methodology that matched results obtained with a PCR (polymerase chain reaction)-based approach, while requiring less expertise and specialized and costly equipment (Kumar et al., 2011).
A peroxidase-based ELISA for the rapid and sensitive monitoring of PSP toxins in shellfish was recently presented. Data obtained with the new method correlated well with the reference mouse bioassay, but showed higher sensitivity, as the LOD was lower than the reference method. Moreover, when positive and negative results were compared based on the regulatory limit, the peroxidase-based ELISA method displayed a sensitivity of 100% and a specificity of 90% compared to the reference method (Kawatsu et al., 2014).
Once established that they allow for adequate response, LOD, and LOQ, the simplicity, high throughput capability, speed, and the relatively low cost of ELISA methods are competitive advantages when compared to costly, time- and man-power-demanding chromatographic or PCR-based methods. Still, ELISA methods depend heavily on the quality of antibodies, the preparation of which is time consuming. Alternatively, the use of aptamers has emerged. Aptamers are small-molecular-weight, single-stranded DNA or RNA molecules with high affinity and selectivity for proteins, which can be synthesized by chemical methods and are more stable than antibodies. An immunoassay based on aptamers, ELAA (enzyme-linked aptamer assay), was developed for the quantitative detection of Vibrio parahemolyticus, a pathogen related to seafood poisoning. The setup involves the use of Hrp immobilized onto gold nanoparticles. In the presence of the pathogen, down to 10 CFU/mL, the enzyme promotes a reaction involving 3,3′,5,5′-tetramethylbenzidine and H2O2, and an optical signal is produced in a linear manner in a logarithm plot within 10–106 CFU/mL (Wu et al., 2015). Also relying on the aptamer-based approach and on Hrp’s ability to catalyze the formation of colored products out of different substrates, an aptasensor for the detection and quantification of chloramphenicol in fish was developed. The sensor has a linear range 0.05–100 ngchloramphenicol/mL, which is within the range of the different methods already available, but displays an LOD of 0.015 ngchloramphenicol/mL, which is only surpassed by an electrochemical immunoassay. Moreover, when tested in real fish samples, the detection of chloramphenicol matched that obtained with a standard ELISA test (Miao et al., 2015).
Enzymatic methods have also been used to establish the freshness of fish and seafood, based on the concentration of nucleotides present (Aristoy et al., 2010). Briefly, once death occurs, adenosine triphosphate (ATP) ceases to be synthesized/regenerated, and it is rapidly decomposed to adenosine monophosphate (AMP) and then to inosine monophosphate (IMP), the former accumulating mostly in crustaceans while the later in fish, where it is responsible for conveying the pleasant fresh flavor (Luong and Male, 1992). IMP spontaneously degrades slowly to inosine (INO), a process slowed by cold environments, and INO is converted to hypoxanthine (HX), which conveys a bitter taste in the presence of either nucleoside phosphorylase (Np) or inosine nucleosidase (In). HX is sequentially oxidized to xanthine (XA) and to uric acid by xanthine oxidase (Xo), with release of hydrogen peroxide in both steps (Nielsen and Nielsen, 2012; Kostić et al., 2015). The key steps of this pathway are illustrated in Figure 2. INO and HX are typically used as indicators of freshness, but given that the variability in the degradation of mononucleotides depends on several factors, e.g., source of material and physical methods of processing, often multiparametric indicators are advised, such as
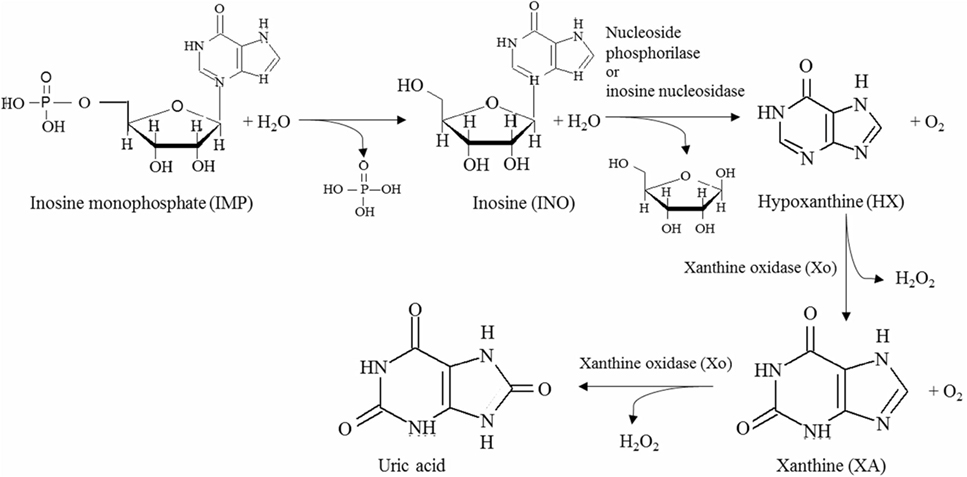
Figure 2. Relevant steps in the main pathway for nucleotide degradation in fish leading to the production of spoilage indicators, namely inosine and hypoxanthine.
In any case, the higher the concentration of IMP, the fresher the fish. Enzymatic analysis can be performed with one or more enzymes either in solution or immobilized, in the latter case typically as a biosensor, where the biological component is connected to a physical-chemical transducer and an optoelectronic interface (Aristoy et al., 2010; Thakur and Ragavan, 2013). A review on the materials and principles underlying biosensor assembly and operation targeted for food analysis was published recently (Thakur and Ragavan, 2013).
Biogenic amines (BAs) are nonvolatile, low-molecular-weight organic bases, such as histamine, cadaverine, putrescine, and tyramine, that are formed in foods as the outcome of microbial decarboxylation of the corresponding amino acids or of transamination of aldehydes and ketones by amino acid transaminases (Prester, 2011; Visciano et al., 2012; Zhai et al., 2012). Since BAs are produced by spoilage bacteria, their level can represent the quality of food (Hosseini et al., 2013). The consumption of high amounts of BAs, particularly histamine, can result in food poisoning; hence the maximum content in histamine is regulated. In Europe, these are within 100–200 mg/kg for fish species, and within 200–400 mg/kg for enzyme-processed foods. ELISA kits and histamine-specific enzyme kits, based on the oxidation of histamine by histamine dehydrogenase, are commercially available (Köse et al., 2011; Hungerford and Wu, 2012; Visciano et al., 2012).
The use of free enzymes, preferentially for the quantification of IMP, INO, and HX, involves the sequential use of either 5′-nucleotidase (Nt) or adenosinedeaminase (Ad), Np, and Xo, ultimately resulting in the formation of uric acid and hydrogen peroxide (Luong et al., 1989; Luong and Male, 1992; Cho et al., 1999, 2000). Measurements can be carried out by polarography, as the response of a Clark hydrogen peroxide probe electrode to uric acid and hydrogen peroxide is additive (Luong et al., 1989), or by spectrophotometry (Luong and Male, 1992; Cho et al., 1999, 2000). The combination of Alp, Np, and Xo, conjugated with WST-8, a color developing agent that reacts with hydrogen peroxide allowing spectrophotometric readings, was used for the colorimetric-based quantification of IMP, INO, and HX. Amorphous freeze-dried enzyme formulations in the presence of gelatin and sucrose were prepared, which allowed promising shelf-life, as the Ki values determined after 6 months of storage at 40°C were not significantly altered when compared to those of newly prepared formulation. Moreover, the use of additives enhanced the enzyme activity (Srirangsan et al., 2010). Developments making the methodology amenable to implementation in microtiter plates and spectrophotometric quantification, thus allowing for high throughput, have been also presented (Goodrich and Balakireva, 2015).
Assessment of fish and seafood freshness through the use of immobilized enzymes has relied on both multi-enzyme and single-enzyme systems, the former mostly aiming to determine several metabolites of ATP degradation and Ki values (Watanabe et al., 1984, 1986; Luong et al., 1989). Some of these multi-enzymes involved the flow injection analysis approach (Carsol and Mascini, 1998; Okuma and Watanabe, 2002). Still, enzymatic biosensors often rely on the quantification of HX using immobilized Xo, although detection of BAs has also been considered (Lawal and Adeloju, 2012a; Visciano et al., 2012). Immobilization strategies have been selected both aiming to allow for activity retention upon immobilization and enzyme stability and also to remove interferences form hydrogen peroxide, uric acid, or ascorbic acid that can be present in the sample for analysis (Aristoy et al., 2010; Lawal and Adeloju, 2012a). Specific details on the methods of immobilization and operation of recently developed biosensors are given in Table 3 and in Figures 3 and 4.
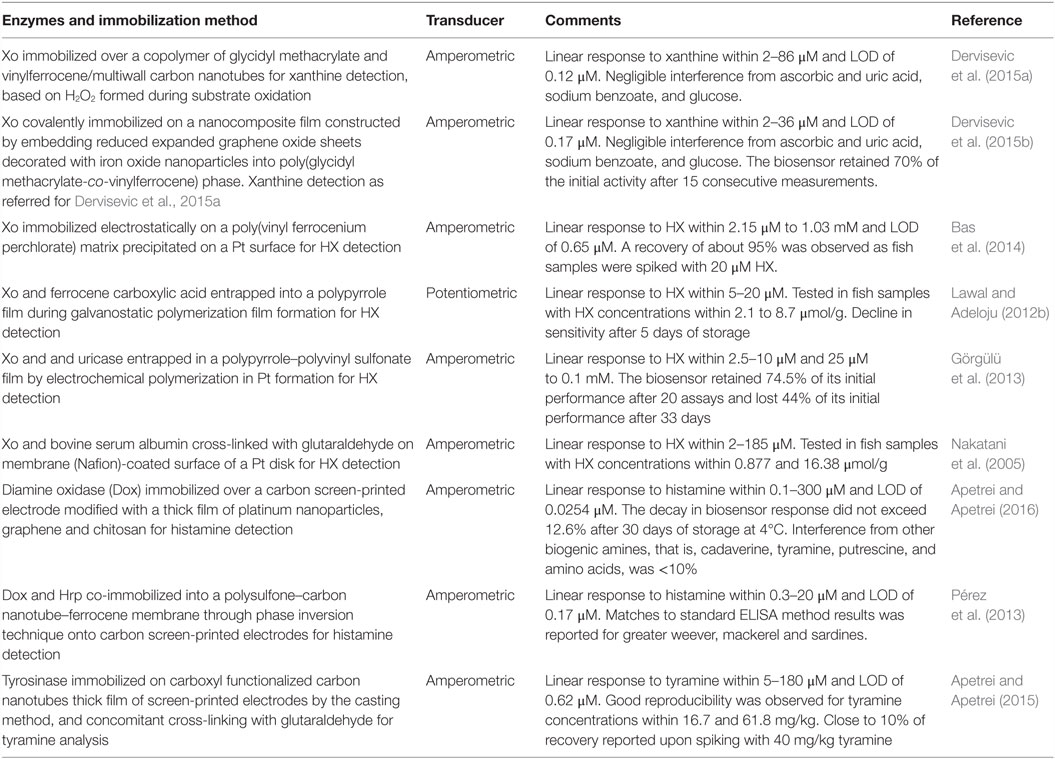
Table 3. Recent examples of enzymatic biosensors developed for assessment of fish and seafood quality and freshness.
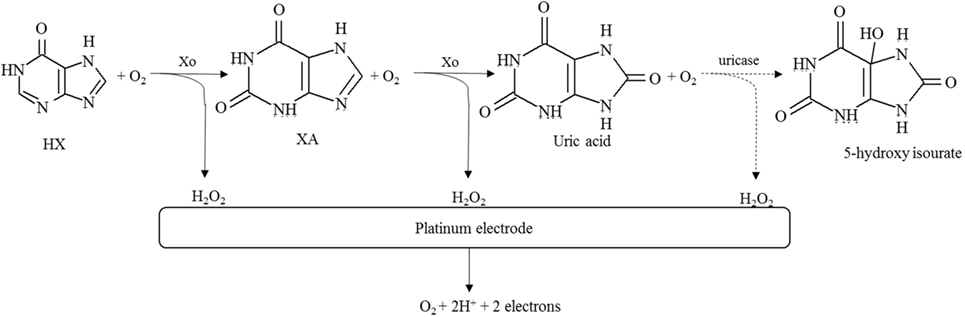
Figure 3. Schematic diagram of the reactions involved in the quantification of hypoxanthine as an indicator of fish freshness. The methodologies rely on the determination of hydrogen peroxide formed in consecutive reactions catalyzed by xanthine oxidase (solid lines) and eventually also uricase (dashed lines), combined with a platinum-based electrode. Further information and references as detailed in Table 3.
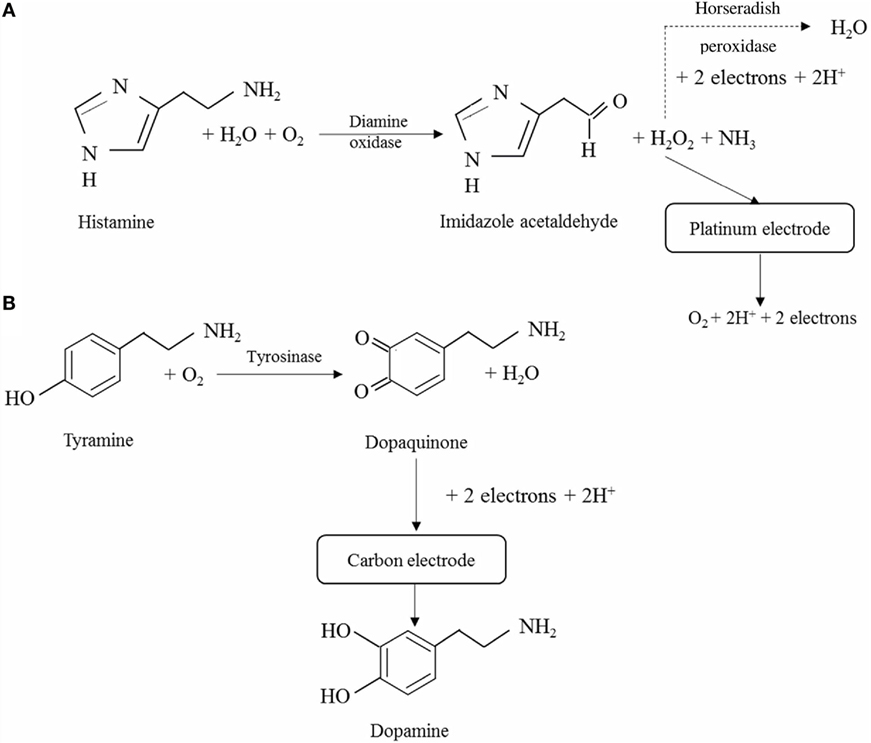
Figure 4. Schematic diagrams of the reactions involved in the quantification of biogenic amines. (A) Quantification of histamine based on hydrogen peroxide formation and either direct assessment of the latter in a platinum electrode (solid line, Apetrei and Apetrei, 2016) or through the use of horseradish peroxidase (dashed line, Pérez et al., 2013). (B) Quantification of tyrosine through oxidation with tyrosinase to dopaquinone and reduction of the latter to dopamine on the surface of a carbon electrode (Apetrei and Apetrei, 2015).
Conclusion
Fish and seafood industries have a key role as providers of healthy food. As an outcome of the increasing public awareness of the significance of a balanced diet to health, the demand for fish and seafood concomitantly grows, a trend that is foreseen to continue in the near future. Given the perishable nature of the products and their complexity, their effective processing and monitoring is a challenging task. Physical and chemical processes have been often the mainstay, with a minor contribution of endogenous enzymes. Advances in enzyme technology are turning the tide, as a result of a growing insight into the mechanisms of enzyme action, access to marine sources of enzymes, and improvements in heterologous expression, coupled with the need to minimize wastes and add value to byproducts that typically are often discarded. Within this, the enzymatic approach contributes significantly to overcome the environmental impact of traditional processes, thereby contributing to the implementation of sustainable and cost-effective processes. Moreover, enzyme technology can also contribute to the quality control of fish and seafood goods. It is a fact that the role of enzymes in fish and seafood industries lags behind other fields in food processing. Still, the combined developments in several complementary fields, such as heterologous expression of enzymes that can particularly allow for cost-effective production of some key enzymes, computational methods for predictive models, the ability to use the marine microbiota to provide new/improved enzymatic activities, enzyme formulations and immobilization methods, and materials science for enhanced operational stability or delivery systems, suggest that significant developments can be expected for enzyme applications in fish and seafood industries.
Author Contributions
As a single author mauscript, all the work (concept, selection of papers/patents/thesis, design of the review, writing) is accounted to the one author.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding
This work was funded by the European Union’s Horizon 2020 Research and Innovation Programme under grant agreement no. 634486 –“INMARE Industrial Applications of Marine Enzymes: Innovative screening and expression platforms to discover and use the functional protein diversity from the sea.”
References
Alma’abadi, A. D., Gojobori, T., and Mineta, K. (2015). Marine metagenome as a resource for novel enzymes. Genomics Proteomics Bioinformatics 13, 290–295. doi: 10.1016/j.gpb.2015.10.001
Amiza, M. A., Mohamad, J., and Hasan, R. (2014). Optimization of enzymatic protein hydrolysis from cobia (Rachycentron canadum) frame using Alcalase®. J. Aquatic Food Prod. Technol. 23, 303–312. doi:10.1080/10498850.2012.717593
Apetrei, I. M., and Apetrei, C. (2015). The biocomposite screen-printed biosensor based on immobilization of tyrosinase onto the carboxyl functionalised carbon nanotube for assaying tyramine in fish products. J. Food Eng. 149, 1–8. doi:10.1016/j.jfoodeng.2014.09.036
Apetrei, I. M., and Apetrei, C. (2016). Amperometric biosensor based on diamine oxidase/platinum nanoparticles/graphene/chitosan modified screen-printed carbon electrode for histamine detection. Sensors 16, 422. doi:10.3390/s16040422
Aquerreta, Y., Astiasarán, I., and Bello, J. (2001). Use of exogenous enzymes to elaborate the Roman fish sauce ‘garum’. J. Sci. Food Agric. 82, 107–112. doi:10.1002/jsfa.1013
Arilla, M. C., Ibarrola, I., Martinez, A., Monteseirin, J., Conde, J., and Asturias, J. A. (2008). An antibody-based ELISA for quantification of Ani s 1, a major allergen from Anisakis simplex. Parasitology 135, 735–740. doi:10.1017/S0031182008004332
Aristoy, M. C., Mora, L., Hernández-Cázares, A. S., and Toldrá, F. (2010). “Nucleotides and nucleosides,” in Handbook of Seafood and Seafood Products Analysis, eds L. M. L. Nollet and F. Toldrá (Boca Raton, FL: CRC Press), 58–68.
Balti, R., Bougatef, A., El-Hadj, A. N., Zekri, D., Barkia, A., and Nasri, M. (2010). Influence of degree of hydrolysis on functional properties and angiotensin I-converting enzyme-inhibitory activity of protein hydrolysates from cuttlefish (Sepia officinalis) by-products. J. Sci. Food Agric. 90, 2006–2014. doi:10.1002/jsfa.4045
Bas, S. K., Gülce, H., and Yildiz, S. (2014). Hypoxanthine biosensor based on immobilization of xanthine oxidase on modified Pt electrode and its application for fish meat. Int. J. Polym. Mat. Polym. Biomat. 63, 476–485. doi:10.1080/00914037.2013.854215
Beddows, C. G., and Ardeshir, A. G. (1979). The production of soluble fish protein solution for use in fish sauce manufacture. I. The use of added enzymes. J. Food. Technol. 14, 603–612. doi:10.1111/j.1365-2621.1979.tb00907.x
Benjakul, S., Nalinanon, S., and Shahidi, F. (2012). “Fish collagen,” in Food Biochemistry and Food Processing, 2nd Edn, eds B. K. Simpson, L. M. L. Nollet, F. Toldra, S. Benjakul, G. Paliyath and Y. H. Hui (Chichester: John Wiley & Sons, Inc), 365–387.
Benjakul, S., Yarnpakdee, S., Senphan, T., Halldorsdottir, S. M., and Kristinsson, H. G. (2014). “Fish protein hydrolysates: production, bioactivities, and applications,” in Antioxidants and Functional Components in Aquatic Foods, eds H. G. Kristinsson and S. Raghavan (Chichester, West Sussex: John Wiley & Sons, Ltd.), 237–281.
Bjørkevoll, I., Lauritzsen, K., Gundersen, B., Dahl, R. W., Eilertsen, G., Sivertsen, A. H., et al. (2008). Ripening of Salted Cod. Tromsø: Nofima AS.
Campos, C. A., Castro, M. P., Aubourg, S. P., and Velázquez, J. B. (2012). “Use of natural preservatives in seafood,” in Novel Technologies in Food Science, eds A. McElhatton and P. J. A. Sobral (New York, NY: Springer), 325–360.
Carsol, M. A., and Mascini, M. (1998). Development of a system with enzyme reactors for the determination of fish freshness. Talanta 47, 335–342. doi:10.1016/S0039-9140(98)00129-5
Chalamaiah, M., Dinesh Kumar, B., Hemalatha, R., and Jyothirmayi, T. (2012). Fish protein hydrolysates: proximate composition, amino acid composition, antioxidant activities and applications: a review. Food Chem. 135, 3020–3038. doi:10.1016/j.foodchem.2012.06.100
Chalamaiah, M., Narsing Rao, G., Rao, D. G., and Jyothirmayi, T. (2010). Protein hydrolysates from meriga (Cirrhinus mrigala) egg and evaluation of their functional properties. Food Chem. 120, 652–657. doi:10.1016/j.foodchem.2009.10.057
Chaurasia, S. P., Bhandaria, K., Sharmaa, A., and Dalai, A. K. (2016). A review on lipase catalysed synthesis of DHA rich glyceride from fish oils. Int. J. Res. Sci. Innov. 3, 9–19.
Chen, B., McClements, D. J., and Decker, E. A. (2012). Design of foods with bioactive lipids for improved health. Annu. Rev. Food Sci. Technol. 4, 35–56. doi:10.1146/annurev-food-032112-135808
Cheung, I. W., Liceaga, A. M., and Li-Chan, E. C. (2009). Pacific hake (Merluccius productus) hydrolysates as cryoprotective agents in frozen pacific cod fillet mince. J. Food Sci. 74, C588–C594. doi:10.1111/j.1750-3841.2009.01307.x
Cho, Y. J., Im, Y. S., Kim, S. M., and Choi, Y. J. (1999). Enzymatic method for measuring ATP related compounds in fish sauces. J. Korean Fish. Soc. 32, 385–390.
Cho, Y. J., Im, Y. S., Seo, D. H., Kim, T. J., Min, J. G., and Choi, Y. J. (2000). Enzymatic method for measuring ATP related compounds in Jeotkals. J. Korean Fish. Soc 33, 16–19.
Chuapoehuk, P., and Raksakulthai, N. (1992). Use of papain and bromelain in the production of oyster sauce. Asean Food J. 7, 199–203.
Clemente, A. (2000). Enzymatic protein hydrolysates in human nutrition. Trends Food Sci. Technol. 11, 254–262. doi:10.1016/S0924-2244(01)00007-3
Conti, G. O., Copat, C., Wang, Z., D’Agati, P., Cristaldi, A., and Ferrante, M. (2015). Determination of illegal antimicrobials in aquaculture feed and fish: an ELISA study. Food Control 50, 937–941. doi:10.1016/j.foodcont.2014.10.050
Dervisevic, M., Custiuc, E., Çevik, E., and Şenel, M. (2015a). Construction of novel xanthine biosensor by using polymeric mediator/MWCNT nanocomposite layer for fish freshness detection. Food Chem. 181, 277–283. doi:10.1016/j.foodchem.2015.02.104
Dervisevic, M., Custiuc, E., Çevik, E., Durmuş, Z., Şenel, M., and Durmuş, A. (2015b). Electrochemical biosensor based on REGO/Fe3O4 bionanocomposite interface for xanthine detection in fish sample. Food Control 57, 402–410. doi:10.1016/j.foodcont.2015.05.001
Dey, S. S., and Dora, K. C. (2014a). Optimization of the production of shrimp waste protein hydrolysate using microbial proteases adopting response surface methodology. J. Food Sci. Technol. 51, 16–24. doi:10.1007/s13197-011-0455-4
Dey, S. S., and Dora, K. C. (2014b). Antioxidative activity of protein hydrolysate produced by alcalase hydrolysis from shrimp waste (Penaeus monodon and Penaeus indicus). J. Food Sci. Technol. 51, 449–457. doi:10.1007/s13197-011-0512-z
Diaz-López, M., and García-Carreño, F. L. (2000). “Applications of fish and shellfish enzymes in food and feed products,” in Seafood Enzymes, eds N. F. Haard and B. K. Simpson (New York, NY: Marcel Dekker), 571–618.
Eberhart, B. T., Moore, L. K., Harrington, N., Adams, N. G., Borchert, J., and Trainer, V. L. (2013). Screening tests for the rapid detection of diarrhetic shellfish toxins in Washington State. Mar. Drugs 11, 3718–3734. doi:10.3390/md11103718
Erkmen, O. (2012). “Modified-atmosphere storage of foods,” in Progress in Food Preservation, eds R. Bath and G. Paliyath (Oxford: John Wiley & Sons Ltd.), 49–66.
European Commission. (2004). Commission decision of 22 December 2003 amending Decision 2002/657/EC as regards the setting of minimum required performance limits(MRPLs) for certain residues in food of animal origin. Off J. Eur. Communities. L6, 38–39.
Faisal, M., Islami, S. N. E., Islam, M. N., Kamal, M., and Khan, M. N. A. (2015). Study on microbial and physical changes in fish sauce during fermentation. Res. Agric. Livest. Fish 2, 375–383. doi:10.3329/ralf.v2i2.25024
FAO. (2011). Assessment and Management of Biotoxin Risks in Bivalve Molluscs. Rome: FAO Fisheries and Aqualculture.
FAO. (2014). The State of the World Fisheries and Aquaculture. Rome: Food and Agriculture Organization of the United Nations.
Fraatz, M. A., Rühl, M., and Zorn, H. (2014). Food and feed enzymes. Adv. Biochem. Eng. Biotechnol. 143, 229–256. doi:10.1007/10_2013_235
Gallant, C. G., Hong, L., and Ablett, R. (2001). Methods for Recovering Raw Meat from the Head-Shells of Crustaceans, e.g. Lobsters and Use Thereof for Stuffed Crustaceans, e.g. Lobster and Stuffing So-Provided. U.S. Patent No 6,235,338. Washington, DC: U.S. Patent and Trademark Office.
Garet, E., González-Fernández, A., Lago, J., Vieites, J. M., and Cabado, A. G. (2010). Comparative evaluation of enzyme-linked immunoassay and reference methods for the detection of shellfish hydrophilic toxins in several presentations of seafood. J. Agric. Food Chem. 58, 1410–1415. doi:10.1021/jf904448z
Ghaly, A. E., Ramakrishnan, V. V., Brooks, M. S., Budge, S. M., and Dave, D. (2015). Fish processing wastes as a potential source of proteins, amino acids and oils: a critical review. J. Microb. Biochem. Technol. 5, 4. doi:10.4172/1948-5948.1000110
Gildberg, A. (2004). Digestive enzyme activities in starved pre-slaughter farmed and wild–captured, Atlantic cod (Gadus morhua). Aquaculture 238, 343–353. doi:10.1016/j.aquaculture.2004.03.021
Gildberg, A., Simpson, B. K., and Haard, N. F. (2000). “Uses of enzymes from marine organisms,” in Seafood Enzymes: utilization and influence in postharvest seafood quality, eds N. F. Haard and B. K. Simpson (New York: Marcel Dekker), 619–640.
Gildberg, A., Wichaphon, J., Lertsiri, S., Assavanig, A., Sørensen, N. K., and Thongthai, C. (2007). Chemical and organoleptic comparison of fish sauce made from cold water species and typical Thai fish sauce. J. Aquatic Food Prod. Technol. 16, 31–42. doi:10.1300/J030v16n03_04
Gonçalves, A. A., and Passos, M. G. (2010). Restructured fish product from white croacker (Micropogonias furnieri) mince using microbial transglutaminase. Braz. Arch. Biol. Technol. 53, 987–995. doi:10.1590/S1516-89132010000400030
Goodrich, W., and Balakireva, L. (2015). A freshness assay for seafood. Food Qual. Saf. 42–43. Available at: http://www.foodqualityandsafety.com/article/a-freshness-assay-for-seafood/
Görgülü, M., Çete, S., Arslan, H., and Yaşar, A. (2013). Preparing a new biosensor for hypoxanthine determination by immobilization of xanthine oxidase and uricase in polypyrrole-polyvinyl sulphonate film. Artif. Cells Nanomed. Biotechnol 41, 327–331. doi:10.3109/21691401.2012.744993
Gortari, M. C., and Hours, R. A. (2013). Biotechnological processes for chitin recovery out of crustacean waste: a mini-review. Electronic J. Biotechnol. 16, 3. doi:10.2225/vol16-issue3-fulltext-10
Haard, N. F., and Simpson, B. K. (1994). “Proteases from aquatic organisms and their uses in the seafood industry,” in Fisheries Processing – Biotechnological Applications, ed. A. M. Martin (Dordrecht: Springer Science+Business Media), 132–154.
Hashim, P., Mohd Ridzwan, M. S., Bakar, J., and Mat Hashim, D. (2015). Collagen in food and beverage industries. Int. Food Res. J. 22, 1–8.
He, S., Franco, C., and Zhang, W. (2013). Functions, applications and production of protein hydrolysates from fish processing co-products (FPCP). Food Res. Int. 50, 289–297. doi:10.1016/j.foodres.2012.10.031
He, S., Franco, C., and Zhang, W. (2015). Fish protein hydrolysates: application in deep-fried food and food safety analysis. J. Food Sci. 80, E108–E115. doi:10.1111/1750-3841.12684
Himonides, A. T., Taylor, A. K. D., and Morris, A. J. (2011a). A study of the enzymatic hydrolysis of fish frames using model systems. Food Nutr. Sci. 2, 575–585. doi:10.4236/fns.2011.26081
Himonides, A., Taylor, A., and Morris, A. (2011b). Enzymatic hydrolysis of fish frames using pilot plant scale systems. Food Nutr. Sci. 2, 586–593. doi:10.4236/fns.2011.26082
Hosseini, S. V., Hamzeh, A., Moslemi, M., Lashkan, A. B., Iglesias, A., and Feás, X. (2013). Effect of delayed icing on biogenic amines formation and bacterial contribution of iced common carp (Cyprinus carpio). Molecules 18, 15464–15473. doi:10.3390/molecules181215464
Hu, F.-Y., Chi, C.-F., Wang, B., and Deng, S.-G. (2015). Two novel antioxidant nonapeptides from protein hydrolysate of skate (Raja porosa) muscle. Mar. Drugs 13, 1993–2009. doi:10.3390/md13041993
Huazhang, L., Wen-xue, L., Quan-xin, Z., Guang-yu, Y., Jun-tao, L., Li, M., et al. (2011). Detection of shellfish toxins from scallops in Guangzhou seafood market. J. Venom. Anim. Toxins Incl. Trop. Dis. 17, 74–84. doi:10.1590/S1678-91992011000100010
Hungerford, J., and Wu, W. H. (2012). Comparison study of three rapid test kits for histamine in fish: biooscientific MaxSignal enzymatic assay neogen veratox ELISA, and the neogen reveal histamine screening test. Food Control 25, 448–457. doi:10.1016/j.foodcont.2011.11.007
Jabbour, T., and Hognason, G. (2007) Method for Shucking Lobster, Crab or Shrimp. Patent Application US20070254569.
Jemli, S., Ayadi-Zouari, D., Hlima, H. B., and Bejar, S. (2016). Biocatalysts: application and engineering for industrial purposes. Crit. Rev. Biotechnol. 36, 246–258. doi:10.3109/07388551.2014.950550
Jester, E. L., Abraham, A., Wang, Y., El Said, K. R., and Plakas, S. M. (2014). Performance evaluation of commercial ELISA kits for screening of furazolidone and furaltadone residues in fish. Food Chem. 145, 593–598. doi:10.1016/j.foodchem.2013.08.090
Jridi, M., Lassoued, I., Nasri, R., Ayadi, M. A., Nasri, M., and Souissi, N. (2014). Characterization and potential use of cuttlefish skin gelatin hydrolysates prepared by different microbial proteases. Biomed. Res. 2014, 461728. doi:10.1155/2014/461728
Kaewudom, P., Benjakuln, S., and Kijroongrojana, K. (2013). Properties of surimi gel as influenced by fish gelatin and microbial transglutaminase. Food Biosci. 1, 39–47. doi:10.1016/j.fbio.2013.03.001
Kawatsu, K., Kanki, M., Harada, T., and Kumeda, Y. (2014). A highly rapid and simple competitive enzyme-linked immunosorbent assay for monitoring paralytic shellfish poisoning toxins in shellfish. Food Chem. 162, 94–98. doi:10.1016/j.foodchem.2014.04.038
Khalafalla, F. A., Ali, F. H. M., and Hassan, A.-R. H. A. (2015). Quality improvement and shelf-life extension of refrigerated Nile tilapia (Oreochromis niloticus) fillets using natural herbs. Beni-Suef Univ. J. Basic Appl. Sci. 4, 33–40. doi:10.1016/j.bjbas.2015.02.005
Kieliszek, M., and Misiewicz, A. (2014). Microbial transglutaminase and its application in the food industry. A review. Folia Microbiol. 59, 241–250. doi:10.1007/s12223-013-0287-x
Kim, T., Silva, J. L., Parakulsuksatid, P., and Wang, D. S. (2014). Optimization of enzymatic treatments for deskinning of catfish nuggets. J. Aquatic Food Prod. Technol. 23, 385–393. doi:10.1080/10498850.2012.720651
Köse, S., Kaklikkaya, N., Koral, S., Tufan, B., Buruk, C. K., and Aydin, F. (2011). Commercial test kits and the determination of histamine in traditional (ethnic) fish products – evaluation against an EU accepted HPLC method. Food Chem. 125, 1490–1497. doi:10.1016/j.foodchem.2010.10.069
Kostić, D. A., Dimitrijević, D. S., Stojanović, G. S., Palić, I. R., Đrdević, A. S., and Ickovski, J. D. (2015). Xanthine oxidase: isolation, assays of activity, and inhibition. J. Chem. 2015, 8. doi:10.1155/2015/294858
Kristinsson, H. G., and Rasco, B. A. (2000). Fish protein hydrolysates: production, biochemical, and functional properties. Crit. Rev. Food Sci. Nutr. 40, 43–81. doi:10.1080/10408690091189266
Kumar, B. K., Raghunah, P., Devegowda, D., Deekshit, K., Venugopal, N. V., Karunasagar, I., et al. (2011). Development of monoclonal antibody based sandwich ELISA for the rapid detection of pathogenic Vibrio parahaemolyticus in seafood. Intl. J. Food Microbiol. 145, 244–249. doi:10.1016/j.ijfoodmicro.2010.12.030
Laohakunjit, N., Selamassakul, O., and Kerdchoechuen, O. (2014). Seafood-like flavour obtained from the enzymatic hydrolysis of the protein by-products of seaweed (Gracilaria sp.). Food Chem. 158, 162–170. doi:10.1016/j.foodchem.2014.02.101
Lawal, A. T., and Adeloju, S. B. (2012a). Progress and recent advances in fabrication and utilization of hypoxanthine biosensors for meat and fish quality assessment: a review. Talanta 100, 217–228. doi:10.1016/j.talanta.2012.07.085
Lawal, A. T., and Adeloju, S. B. (2012b). Mediated xanthine oxidase potentiometric biosensors for hypoxanthine based on ferrocene carboxylic acid modified electrode. Food. Chem. 135, 2982–2987. doi:10.1016/j.foodchem.2012.07.052
Le, C. M., Donnay-Moreno, C., Bruzac, S., Baron, R., Nguyen, H. T. M., and Bergé, J. P. (2015). Proteolysis of sardine (Sardina pilchardus) and anchovy (Stolephorus commersonii) by commercial enzymes in saline solutions. Food Technol. Biotechnol. 53, 87–90. doi:10.17113/ftb.53.01.15.3893
Lee, H. W., Oh, J. Y., and Kang, D. I. (2015). Method for Manufacturing a Fish Sauce and a Fish Sauce Prepared By the Same. Patent Application US 20150296854 A1.
Li, Y., and Cirino, P. C. (2014). Recent advances in engineering proteins for biocatalysis. Biotechnol. Bioeng. 111, 1273–1287. doi:10.1002/bit.25240
Liu, Z., Lu, Y., Ge, X., and Zeng, M. (2011). Effects of transglutaminase on rheological and film forming properties of fish gelatin. Adv. Mat. Res. 236-238, 2877–2880. doi:10.4028/www.scientific.net/AMR.236-238.2877
Lopetcharat, K., Choi, Y. J., Park, J. D., and Daeschel, M. A. (2001). Fish sauce products and manufacturing: a review. Food Rev. Int. 17, 65–88. doi:10.1081/FRI-100000515
Luong, J. H. T., and Male, K. B. (1992). Development of a new biosensor system for the determination of the hypoxanthine ratio, an indicator of fish freshness. Enzyme Microb. Technol. 14, 125–130. doi:10.1016/0141-0229(92)90169-O
Luong, J. H. T., Male, K. B., and Nguyen, A. L. (1989). Application of polarography for monitoring the fish post-mortem metabolite transformation. Enzyme Microb. Technol. 11, 277–282. doi:10.1016/0141-0229(89)90042-2
Malaweera, B. O., and Wijesundara, W. M. N. M. (2014). “Use of seafood processing by-products in the animal feed industry,” in Seafood Processing By-Products – Trends and Applications, ed. S.-K. Kim (New York: Springer), 315–339.
Mariniello, L., Di Pierro, P., Giosafatto, C. V. L., Sorrentino, A., and Porta, R. (2008). “Transglutaminase in food biotechnology,” in Recent Research Developments in Food Biotechnology. Enzymes as Additives or Processing Aids, eds R. Porta, P. Di Pierro and L. Mariniello (Kerala, India: Research Signpost), 105–211.
Martin, R. E., Fernandes, C. E., and Learson, R. L. (2000). “Optional processing methods,” in Marine and Freshwater Products Handbook, eds R. E. Martin, E. P. Carter, E. P. Carter, G. J. Flick Jr and L. M. Davis (Lancaster, PA: Technomic Publishing Company, Inc.), 343–354.
Martín-Sánchez, A. M., Navarro, C., Pérez-Álvarez, J. A., and Kuri, V. (2009). Alternatives for efficient and sustainable production of surimi: a review. Compr. Rev. Food Sci. Food Saf. 8, 359–374. doi:10.1111/j.1541-4337.2009.00087.x
Miao, Y., Gan, N., Li, T., Zhang, H., Cao, Y., and Jiangba, Q. (2015). A colorimetric aptasensor for chloramphenicol in fish based on double-stranded DNA antibody labeled enzyme-linked polymer nanotracers for signal amplification. Sens. Actuators B 220, 679–687. doi:10.1016/j.snb.2015.05.106
Moreno, H., Carballo, J., and Borderías, J. (2013). Raw-appearing restructured fish models made with sodium alginate or microbial transglutaminase and effect of chilled storage. Food Sci. Technol. (Campinas) 33, 137–145. doi:10.1590/S0101-20612013005000004
Morrissey, M., and DeWitt, C. (2014). “Value-added seafood,” in Seafood Processing: Technology, Quality and Safety, ed. I. S. Boziaris (Chichester: John Wiley & Sons, Ltd), 343–358.
Nakatani, H. S., dos Santos, L. V., Pelegrine, C. P., Gomes, S. T. M., Matsushita, M., de Souza, N. E., et al. (2005). Biosensor based on xanthine oxidase for monitoring hypoxanthine in fish meat. Am. J. Biochem. Biotechnol. 1, 85–89. doi:10.3844/ajbbsp.2005.85.89
Nguyen, H. T. M., Sylla, K. S. B., Randriamahatody, Z., Donnay-Moreno, C., Moreau, J., Tran, L. T., et al. (2011). Enzymatic hydrolysis of yellowfin tuna (Thunnus albacares) by-products using protamex protease. Food Technol. Biotechnol. 49, 48–55.
Nielsen, M. K., and Nielsen, H. H. (2012). “Seafood enzymes,” in Food Biochemistry and Food Processing, 2nd Edn, eds B. K. Simpson, L. M. L. Nollet, F. Toldra, S. Benjakul, G. Paliyath and Y. H. Hui (Chichester: John Wiley & Sons, Inc), 247–262.
Oh, J.-H. (2012). Characterization of edible film fabricated with channel catfish Ictalurus punctatus gelatin by cross-linking with transglutaminase. Fish Aquat. Sci. 15, 9–14. doi:10.5657/FAS.2012.0009
Okuma, H., and Watanabe, E. (2002). Flow system for fish freshness determination based on double multi-enzyme reactor electrodes. Biosens. Bioelectron. 17, 367–372. doi:10.1016/S0956-5663(01)00309-8
Ooshiro, Z., Ok, T., Une, H., Hayashi, S., and Itakura, T. (1981). Study on use of commercial proteolytic enzymes in production of fish sauce. Mem. Fac. Fish. Kagoshima Univ. 30, 383–394.
Ovissipour, M., Rasco, B., Shiroodi, S. G., Modanlow, M., Gholami, S., and Nemati, M. (2013). Antioxidant activity of protein hydrolysates from whole anchovy sprat (Clupeonella engrauliformis) prepared using endogenous enzymes and commercial proteases. J. Sci. Food Agric. 93, 1718–1726. doi:10.1002/jsfa.5957
Pal, G. K., and Suresh, P. V. (2016). Sustainable valorisation of seafood by-products: recovery of collagen and development of collagen-based novel functional food ingredients. Innov. Food Sci. Emerg. Technol doi:10.1016/j.ifset.2016.03.015
Paul, T., Halder, S. K., Das, A., Ghosh, K., Mandal, A., Payra, P., et al. (2015). Production of chitin and bioactive materials from Black tiger shrimp (Penaeus monodon) shell waste by the treatment of bacterial protease cocktail. 3 Biotech 5, 483–493. doi:10.1007/s13205-014-0245-6
Pérez, S., Bartrolí, J., and Fàbregas, E. (2013). Amperometric biosensor for the determination of histamine in fish samples. Food Chem. 141, 4066–4072. doi:10.1016/j.foodchem.2013.06.125
Prester, L. (2011). Biogenic amines in fish, fish products and shellfish: a review. Food Addit. Contam. Part A Chem. Anal. Control Expo. Risk. Assess. 28, 1547–1560. doi:10.1080/19440049.2011.600728
Rahaman, M. A. (2014). Effect of Processing Parameters on Salting of Herring. Ph.D. thesis, Norwegian University of Science and Technology, Uppsala, Sweden. Available at: http://www.diva-portal.org/smash/get/diva2:743707/FULLTEXT01.pdf
Rasika, D. M. D., Ranadheera, C. S., and Vidanarachchi, J. K. (2013). “Applications of marine derived peptides and proteins in the food industry,” in Marine Proteins and Peptides: Biological Activities and Applications, ed. S.-K. Kim (Chichester: John Wiley & Sons, Inc), 545–587.
Rodríguez, L. P., González, V., Martínez, A., Paz, B., Lago, J., Cordeiro, V., et al. (2015). Occurrence of lipophilic marine toxins in shellfish from Galicia (NW of Spain) and synergies among them. Mar. Drugs 13, 1666–1687. doi:10.3390/md13041666
Roslan, J., Yunos, K. F. M., Abdullah, N., and Kamal, S. M. M. (2014). Characterization of fish protein hydrolysate from tilapia (Oreochromis niloticus) by-product. Agric. Agric. Sci. Procedia 2, 312–319. doi:10.1016/j.aaspro.2014.11.044
Rustad, T. (2010). “Peptides and proteins,” in Handbook of Seafood and Seafood Products Analysis, eds L. M. L. Nollet and F. Toldrá (Boca Raton, FL: CRC Press), 11–19.
Samaranayaka, A. G. P., and Li-Chan, E. C. Y. (2008). Autolysis-assisted production of fish protein hydrolysates with antioxidant properties from Pacific hake (Merluccius productus). Food Chem. 107, 768–776. doi:10.1016/j.foodchem.2007.08.076
Sana, B. (2015). “Marine microbial enzymes: current status and future prospects,” in Springer Handbook of Marine Biotechnology, ed. S.-K. Kim (Berlin: Springer-Verlag), 905–917.
Sapozhnikova, Y., Simons, T., and Lehotay, S. J. (2015). Evaluation of a fast and simple sample preparation method for polybrominated diphenyl ether (PBDE) flame retardants and dichlorodiphenyltrichloroethane (DDT) pesticides in fish for analysis by ELISA compared with GC-MS/MS. J. Agric. Food Chem. 63, 4429–4434. doi:10.1021/jf505651g
Senphan, T., Benjakul, S., and Kishimura, H. (2014). Characteristics and antioxidative activity of carotenoprotein from shells of Pacific white shrimp extracted using hepatopancreas proteases. Food Biosci. 5, 54–63. doi:10.1016/j.fbio.2013.11.004
Serafini-Fracassini, D., and Del Duca, S. (2008). Transglutaminases: widespread cross-linking enzymes in plants. Ann. Bot 102, 145–152. doi:10.1093/aob/mcn075
Shahidi, F., and Janak Kamil, Y. V. A. (2001). Enzymes from fish and aquatic invertebrates and their application in the food industry. Trends Food Sci. Technol. 12, 435–464. doi:10.1016/S0924-2244(02)00021-3
Shen, Q., Guo, R., Dai, Z., and Zhang, Y. (2012). Investigation of enzymatic hydrolysis conditions on the properties of protein hydrolysate from fish muscle (Collichthys niveatus) and evaluation of its functional properties. J. Agric. Food Chem. 60, 5192–5198. doi:10.1021/jf205258f
Sikorski, Z. E. (2007). “The role of proteins in food,” in Chemical and Functional Properties of Food Components, ed. Z. E. Sikorski (Boca Raton, FL: CRC Press), 129–176.
Singh, G., Koerner, T., Gelinas, J. M., Abbott, M., Brady, B., Huet, A. C., et al. (2011). Design and characterization of a direct ELISA for the detection and quantification of leucomalachite green. Food Addit. Contam. Part A Chem. Anal. Control Expo. Risk Assess. 28, 731–739. doi:10.1080/19440049.2011.567360
Siró, I. (2012). “Active and intelligent packaging of food,” in Progress in Food Preservation, eds R. Bath and G. Paliyath (Oxford: John Wiley & Sons Ltd.), 23–48.
Sowmya, R., Ravikumar, T. M., Vivek, R., Rathinaraj, K., and Sachindra, N. M. (2014). Optimization of enzymatic hydrolysis of shrimp waste for recovery of antioxidant activity rich protein isolate. J. Food Sci. Technol. 51, 3199–3207. doi:10.1007/s13197-012-0815-8
Srirangsan, P., Hamada-Sato, N., Kawai, K., Watanabe, M., and Suzuki, T. (2010). Improvement of fish freshness determination method by the application of amorphous freeze-dried enzymes. J. Agric. Food Chem. 58, 12456–12461. doi:10.1021/jf102363a
Sujith, P. A., and Hymavathi, T. V. (2011). Recent developments with debittering of protein hydrolysates. As. J. Food Ag-Ind. 4, 365–381.
Suresh, P. V., Nidheesh, T., and Pal, G. V. (2015). “Enzymes in seafood processing,” in Enzymes in Food and Beverage Processing, ed. M. Chandrasekaran (Boca Raton, FL: CRC Press), 354–377.
Suresh, P. V., and Prabhu, G. N. (2013). “Seafood,” in Valorization of Food Processing Byproducts, ed. M. Chandrasekaran (Boca Raton, FL: CRC Press), 685–736.
Svenning, R., Stenberg, E., Gildberg, A., and Nilsen, K. (1993). Biotechnological descaling of fish. Infofish Int. 6, 30–31.
Synowiecki, J., and Al-Khateeb, N. A. A. Q. (2000). The recovery of protein hydrolysate during enzymatic isolation of chitin from shrimp Crangon crangon processing discards. Food Chem. 68, 147–152. doi:10.1016/S0308-8146(99)00165-X
Taheri, A., Anvar, S. A. A., Ahari, H., and Fogliano, V. (2013). Comparison the functional properties of protein hydrolysates from poultry byproducts and rainbow trout (Onchorhynchus mykiss) viscera. Iran J. Fish. Sci. 12, 154–169.
Tanasupawat, S., and Visessanguan, W. (2014). “Fish fermentation,” in Seafood Processing: Technology, Quality and Safety, ed. I. S. Boziaris (Chichester: John Wiley & Sons, Ltd), 177–207.
Thakur, M. S., and Ragavan, K. V. (2013). Biosensors in food processing. J. Food Sci. Technol. 50, 625–641. doi:10.1007/s13197-012-0783-z
Trincone, A. (2011). Marine biocatalysts: enzymatic features and applications. Mar. Drugs 9, 478–499. doi:10.3390/md9040478
Turk, V., Stoka, V., Vasiljeva, O., Renko, M., Sun, T., Turk, B., et al. (2012). Cysteine cathepsins: from structure, function and regulation to new frontiers. Biochim. Biophys. Acta 1824, 68–88. doi:10.1016/j.bbapap.2011.10.002
Turner, A. D., and Goya, A. B. (2016). Comparison of four rapid test kits for the detection of Okadaic acid-group toxins in bivalve shellfish from Argentina. Food Control 59, 829–840. doi:10.1016/j.foodcont.2015.07.005
Utomo, B. S. B., Suryaningrum, T. D., and Harianto, H. R. (2014). Optimization of enzymatic hydrolysis of fish protein hydrolysate (FPH) processing from waste of catfish fillet production. Squalen Bull. Mar. Fish. Postharvest Biotechnol. 9, 115–126. doi:10.15578/squalen.v9i3.79
Valdez-Pena, A. U., Espinoza-Perez, J. D., Sandoval-Fabian, G. C., Balagurusamy, N., Hernandez-Rivera, A., De-la-Garza-Rodriguez, I. M., et al. (2010). Screening of industrial enzymes for deproteinization of shrimp head for chitin recovery. Food Sci. Biotechnol. 19, 553–557. doi:10.1007/s10068-010-0077-z
Venugopal, V. (2005). “Applications of enzymes in fish processing and quality,” in Seafood Processing – Adding Value Through Quick Freezing, Retortable Packaging and Cook-Chilling, ed. V. Venugopal (Boca Raton, FL: CRC Press), 401–424.
Venugopal, V., Lakshmanan, R., Doke, S. N., and Bongirwar, D. R. (2000). Enzymes in fish processing, biosensors and quality control. Food Biotechnol. 14, 21–27. doi:10.1080/08905430009549980
Verma, N., Thakur, S., and Bhatt, A. K. (2012). Microbial lipases: industrial applications and properties (a review). Int. Res. J. Biol. Sci. 1, 88–92.
Vilhelmsson, O. (1997). The state of enzyme biotechnology in the fish processing industry. Trends Food Sci. Technol. 8, 266–270. doi:10.1016/S0924-2244(97)01057-1
Visciano, P., Schirone, M., Tofalo, R., and Suzzi, G. (2012). Biogenic amines in raw and processed seafood. Front. Microbiol. 3:188. doi:10.3389/fmicb.2012.00188
Walker, R., Decker, E. A., and McClements, D. J. (2015). Development of food-grade nanoemulsions and emulsions for delivery of omega-3 fatty acids: opportunities and obstacles in the food industry. Food Funct. 6, 42–55. doi:10.1039/c4fo00723a
Watanabe, E., Endo, H., Takeuchi, N., Hayashi, T., and Toyama, K. (1986). Determination of fish freshness with a multielectrode enzyme sensor system. Bull. Jpn. Soc. Sci. Fish. 52, 489–495. doi:10.2331/suisan.52.489
Watanabe, E., Tokimatsu, S., and Toyama, K. (1984). Simultaneous determination of hypoxanthine, inosine-5′-phosphate and adenosine 5′-phosphate with a multielectrode enzyme sensor. Anal. Chim. Acta 164, 139–146. doi:10.1016/S0003-2670(00)85625-1
Whitaker, J. R. (1994). Principles of Enzymology for the Food Science, 2nd Edn. New York, NY: Marcel Dekker.
Whitaker, J. R., Voragen, A. G. J., and Wong, D. S. W. (eds) (2002). Handbook of Food Enzymology. Boca Raton, FL: CRC Press.
Wolska, J., Jonkers, J., Holst, O., and Adlercreutz, P. (2015). The addition of transglutaminase improves the physical quality of extruded fish feed. Biotechnol. Lett. 37, 2265–2270. doi:10.1007/s10529-015-1911-4
Wu, S., Wang, Y., Duan, N., Ma, H., and Wang, Z. (2015). Colorimetric aptasensor based on enzyme for the detection of Vibrio parahemolyticus. J. Agric. Food Chem. 63, 7849–7854. doi:10.1021/acs.jafc.5b03224
Xu, X., Sui, J., Cao, L., and Lin, H. J. (2010). Direct competitive enzyme-linked immunosorbent assay (ELISA) for rapid screening of anisakids larvae in seafood. Sci. Food Agric. 90, 877–881. doi:10.1002/jsfa.3898
Yarnpakdee, S., Benjakul, S., Kristinsson, H. G., and Kishimura, H. (2015). Antioxidant and sensory properties of protein hydrolysate derived from Nile tilapia (Oreochromis niloticus) by one- and two-step hydrolysis. J. Food Sci. Technol. 52, 3336–3349. doi:10.1007/s13197-014-1394-7
Yin, T., and Park, J. W. (2015). Optimum processing conditions for slowly heated surimi seafood using protease-laden Pacific whiting surimi. LWT Food Sci. Technol. 63, 490–496. doi:10.1016/j.lwt.2015.02.033
Younes, I., and Rinaudo, M. (2015). Chitin and chitosan preparation from marine sources. structure, properties and applications. Mar. Drugs 3, 1133–1174. doi:10.3390/md13031133
Zargar, V., Asghari, M., and Dashti, A. (2015). A review on chitin and chitosan polymers: structure, chemistry, solubility, derivatives, and applications. ChemBioEng Rev. 2, 204–226. doi:10.1002/cben.201400025
Zhai, H., Yang, X., Li, L., Xia, G., Cen, J., Huang, H., et al. (2012). Biogenic amines in commercial fish and fish products sold in southern China. Food Control 25, 303–308. doi:10.1016/j.foodcont.2011.10.057
Keywords: biocatalysis, biosensors, proteases, transglutaminases, lipases, oxidases
Citation: Fernandes P (2016) Enzymes in Fish and Seafood Processing. Front. Bioeng. Biotechnol. 4:59. doi: 10.3389/fbioe.2016.00059
Received: 18 April 2016; Accepted: 24 June 2016;
Published: 07 July 2016
Edited by:
Antonio Trincone, Istituto di Chimica Biomolecolare, Consiglio Nazionale delle Ricerche, ItalyReviewed by:
Tania Primavera Pozzo Mendoza, University of California Davis, USAHelena M. Moreno Conde, Institute of Food Science, Technology and Nutrition (ICTAN), Spain
Copyright: © 2016 Fernandes. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pedro Fernandes, pedro.fernandes@tecnico.ulisboa.pt
 Pedro Fernandes
Pedro Fernandes