Special Issue on Semantic Publishing with Formalization Papers1
1.Introduction
Considering the abundance of scientific articles that are published every day, keeping up with the latest research is becoming a significant challenge for researchers in many fields. This is at least partially due to the fact that we are still holding on to an archaic paradigm of scientific publishing: the canonical way to publish scientific results is by writing them up in long English texts called articles, which are in the best case easy to read by human experts but remain mostly inaccessible to automated approaches (except on a very superficial level with text mining techniques). These articles then undergo peer reviewing, which is typically done in a way that is secretive and not standardized, with the effect that the reviewing process may lack transparency and the valuable comments from the reviewers cannot be reused or build upon.
A range of approaches have been proposed to address some of these problems by making scientific texts machine-readable, allowing for automatic summarising, finding and retrieving information easier and even the ability to (partially) reason on the scientific texts themselves. Text mining approaches work reasonably well when it comes to simple entity extraction with techniques like named-entity recognition to extract the main concepts from a text (e.g. [1,14]), but accuracy dramatically drops with more complicated tasks like relation extraction or identifying links between entities [6,13,15].
The vast majority of existing approaches of making scientific texts machine-readable have one thing in common: they take the current paradigm of scientific articles for granted and therefore take them as their starting point to extract information. While it is important to try to process the vast amount of existing scientific literature that has the form of long English texts (and sometimes long texts in other languages), we should also think about how we can improve the way how we publish scientific insights in the first place. An important aspect of this is the vision of semantic publishing, which we mean here in the sense of genuine semantic publishing [9], where the machine-interpretable formal semantics cover the main scientific claims the work is making. Nanopublications [8], which are small RDF-based semantic packages, have emerged as a powerful concept and technology for enabling such genuine semantic publishing.
In the past, nanopublications were used to implement a semantic and fine-grained model for reviewing [2], and this was extended to semantically represent the full structure of (classical) scientific articles with their reviews and review responses as a single network of nanopublications [3]. In order to get closer to the vision of genuine semantic publishing, however, not just the structure but also the main content of these articles needs to be represented, most importantly their main scientific claims. To that aim, a semantic template called the super-pattern was proposed to represent the meaning of scientific claims in formal logic [4].
Taking an example from our previous study as illustration of the super-pattern, it has been stated in the scientific literature [7] that in particular kinds of cells in the rat brain (specifically, endothelial cells) some sort of stress called transient oxidative stress affects the expression of a protein called Pgp. The super-pattern consists of five slots that would in this example be filled in as follows:
Context class: rat brain endothelial cell
Subject class: transient oxidative stress
Qualifier: generally
Relation: affects
Object class: Pgp expression
Informally, we can read this in the following way: whenever there is an instance of transient oxidative stress in the context of an instance of a rat brain endothelial cell, then generally (meaning in at least 90% of the cases), that instance of stress has the relation of affecting an instance of Pgp expression. Formally, it directly maps to this logic formula:
In this special issue, we combine all these elements of previous research – namely semantic representation of reviews, scientific works as a networks of nanopublications, and representing the main claims with the super-pattern – in order to implement genuine semantic publishing and put it to the test. For practical reasons, we did not require the scientific claims to be novel ones, but they were selected from existing publications. This special issue consists of what we call formalization papers, which are nanopublication-based semantic publications whose novelty lies in the formalization of a previously published scientific claim.
2.Approach
For our approach, we committed to a number of features. First, we wanted the final contributions to be “real” papers in an established journal, which we achieved with the publication of this special issue. They should be fully semantically represented (in RDF) but also have classical views that makes them look like other articles. Like that, they should also seamlessly integrate with the existing bibliometric system and it should be straightforward to cite them in the classical way.
Second, we decided to fully focus on arguably the most interesting element of scientific articles, which happens to also be one of the most challenging to formally represent: the main scientific claims the article is making. Scientific articles have a large number of other interesting pieces of information, e.g. information about the used methods among many other things, but for the purpose of the study to be presented, we focus only on the main claims.
Third, in order to retain the flexibility and power of nanopublications, we decided to refrain from providing a custom-built and optimized user interface that hides the complexity and limits the flexibility. By using generic template-based nanopublication tools and by customizing them solely by providing the templates, we hoped to get a better understanding of how the nanopublication technology works for such kinds of content and workflows in general, and not just for our specific case. On the other hand, this also means that we were looking for a bit more technically minded authors who can handle interfaces that do not come with all the comfort of polished specific applications.
Fourth, we wanted to test a system that could be used to publish novel claims, but decided for practical reasons to focus on formalizing claims from previously published articles. Our approach is therefore based on what we call formalization papers that contribute novel formalizations of existing claims.
Finally, we wanted to cover not just these main claims, but the whole publishing workflow that involves the initial submission of contributions, their reviewing, the responses to the reviews, the updated versions, and the final decision, and represent these as independent but interlinked nanopublications.
3.Formalization papers
This special issue focuses on a new concept called “formalization papers”. A formalization paper contributes a semantic formalization of one of the main claims of an already published scientific article. Its novelty therefore lies solely in the formalization of a claim, not the claim itself. The authors of such formalization papers consequently take credit for the way how the formalization is done, but not for the original claim (unless that claim happens to come from the same authors).
Fig. 1.
Formalization paper template from Nanobench as used by the authors of the special issue.
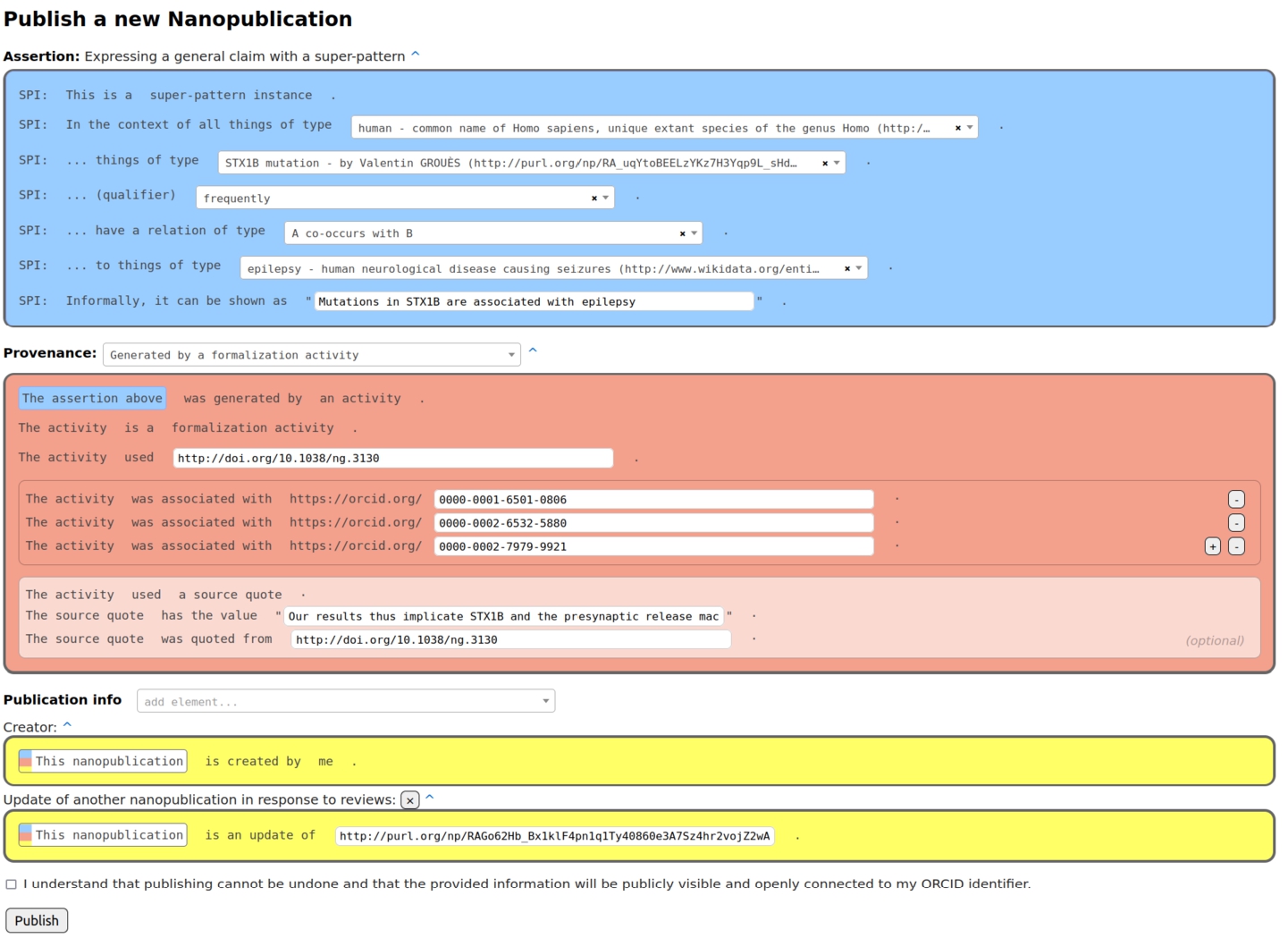
The content of a formalization paper is fully expressed in RDF in the form of nanopublications. Such a formalization paper can be shown in other formats to users, e.g. in HTML or PDF, but these are just views of the same underlying RDF content. The formalization papers consist of nanopublications in which the assertion contains the formalization of the scientific claim using the super-pattern [4], the provenance points to the original paper of the claim, and the publication information attributes the author of the formalization. Figure 1 shows an example of such a nanopublication in the interface the authors of the special issue used.
The instantiated super-pattern in the assertion part refers to a context class, a subject class, a qualifier, a relation type, and an object class according to the super-pattern ontology.22 In the process of coming up with such a formalization, one often realizes that for some of the class slots of the super-pattern (i.e. context, subject, and object class) the class that should be filled in to arrive at a correct formalization is not directly defined in any existing vocabulary or ontology and as such, this class might need to be minted as well. The provenance part of the nanopublication describes the “formalization activity” that was conducted in order arrive at this formalization from what is written in the source publication. The precise phrase from that source publication that was used can be quoted too.
4.Tools
In order to publish formalization papers, class definitions, and all the other kinds of nanopublications (submissions, reviews, responses to reviews, and decisions), Nanobench [10]33 was used. Figure 1 introduced above shows a screenshot of the publishing page of Nanobench. Publishing in Nanobench is based on templates, which are themselves expressed in nanopublications. The form shown in the screenshot is automatically generated based on the information found in several template nanopublications that we created and published for that purpose. All the application-specific behavior is therefore semantically represented in the templates, and Nanobench can flexibly be used for any other kind of data and workflow.
Fig. 2.
The Tapas interface listing submitted formalizations as the results of SPARQL queries over the nanopublication service network.
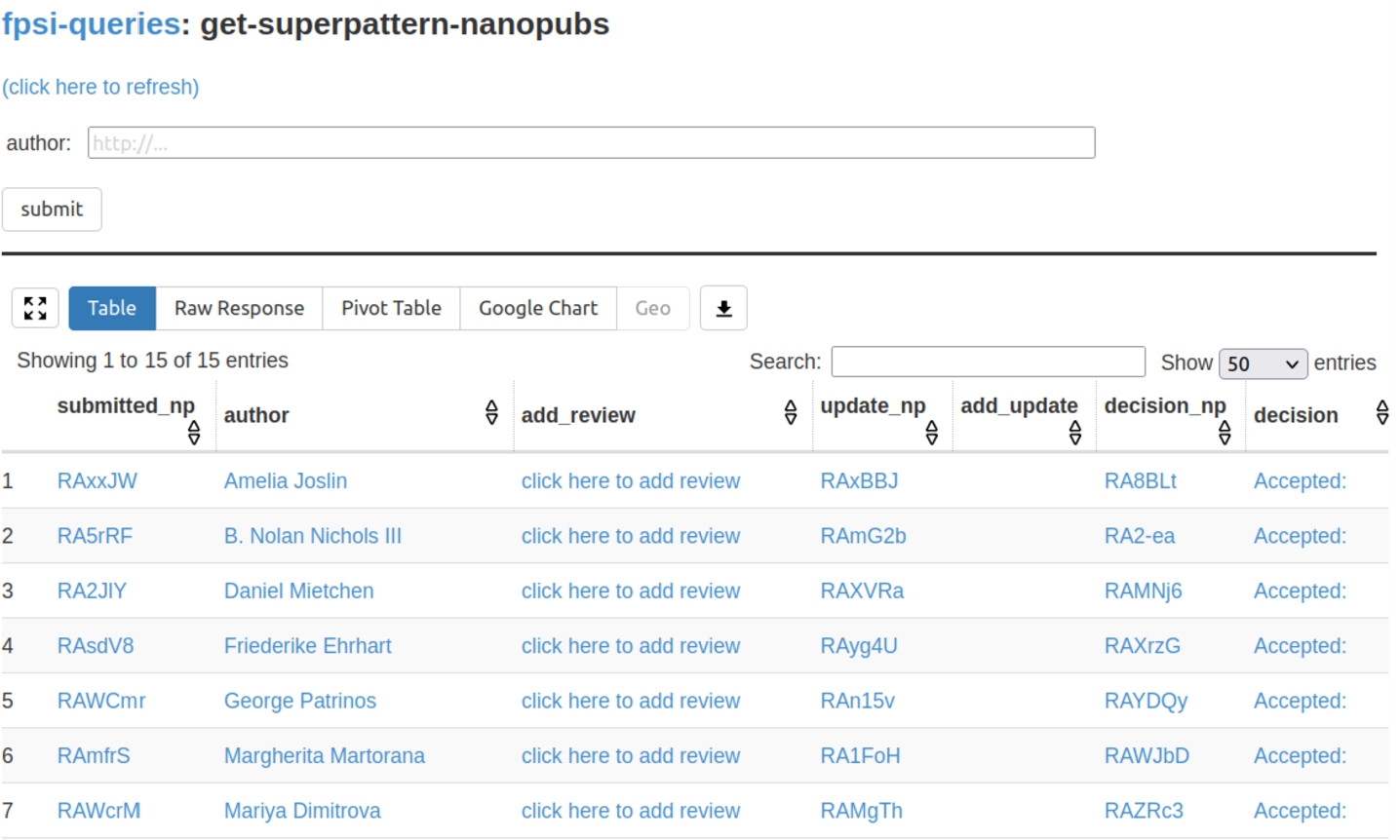
The second tool that was used, Tapas [11],44 is equally generic. It is a simple user interface component built on top of grlc [12] that allows to run template-based SPARQL queries on RDF triple stores. In our case, it was run on SPARQL endpoints provided by the nanopublication service network [10]. Tapas was used to show aggregations and overviews of submissions and reviews. Figure 2 shows a screenshot of the main submission overview. Tapas by itself is read-only, but it was connected to the Nanobench tool with links that lead to partially filled-in forms (e.g. “click here to add review” in the screenshot).
5.Design and planning of this special issue
Interested authors could submit formalization papers, which upon acceptance were published in this special issue. The goal of this was to demonstrate for the first time that scientific articles can be formalized and therefore machine-interpretable including the main scientific claims. Additionally, we also wanted to see whether we can also represent the scientific publishing workflow that lead to the publication of such claims in a machine-friendly way using nanopublications.
Because the user interfaces we had at our disposal were still quite rough and technical, we restricted the set of possible authors and sent the call for papers on a by-invitation basis to selected groups of researchers who have previously worked or had experience with technologies like RDF and semantics. We expect to be able to build more accessible user interfaces in the future that can show the inherent complexity in a way that does not require technical skills, but how this can be achieved was out of scope for this work.
The authors of formalization papers formalized their own previously published claim, or a claim from a paper published by others. In the latter case, the formalization paper authors take credit for the formalization of the claim but not for the claim itself. All submissions to this special issue were peer-reviewed (also as nanopublications) using a previously proposed reviewing ontology [2]. Upon acceptance, these formalization papers were published in this special issue, thereby giving them the same bibliometric status as other scientific articles, which leads to regular indexing in scientific article databases, counting of citations, and so on.
Fig. 3.
Timeline publication for the special issue with formalization papers at the data science journal.
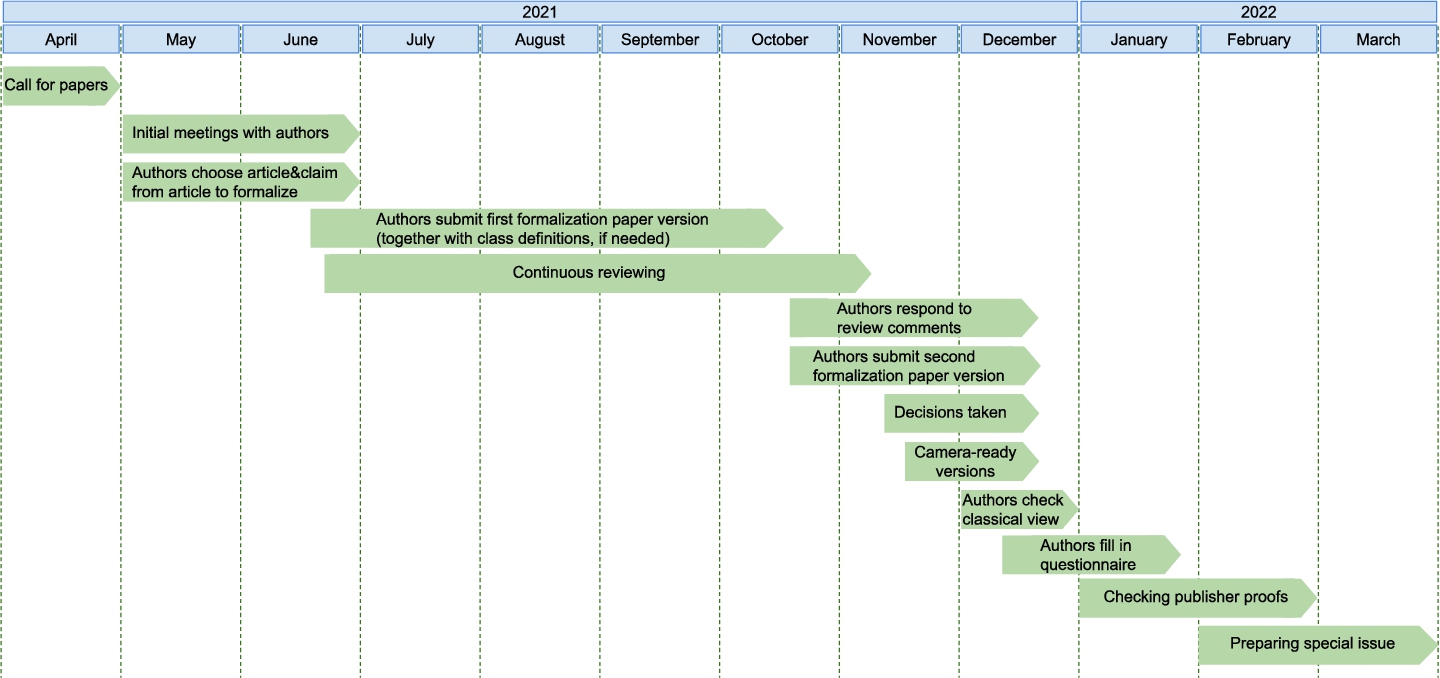
The whole timeline of the special issue can be seen in Figure 3. The authors received close guidance on how to represent a claim of their choosing in RDF using the super-pattern and nanopublications, and on the various stages of the publication process. Authors took part in several information sessions and discussion meetings and were provided at each step with helper materials, videos, and even direct assistance if needed. In total, 24 such individual sessions were organized from May to December 2021.
Fig. 4.
The “classical view” of a formalization paper.

In order to create the appearance of the accepted papers in the special issue as classical papers, “classical views” were created semi-automatically in the form of HTML and PDF versions from the corresponding nanopublications. Also, this view allowed the formalization papers to be integrated in the publisher’s content management system, and to make them connect to the existing bibliometric system, as can be seen in Figure 4.
Table 1
Instantiated super-patterns accepted for publication in the special issue. Submissions marked with ◇ are formalizations in which authors extracted a scientific claim from their own previously published article; classes minted using Nanobench are marked with *, while newly minted Wikidata classes are marked with **
6.Overview of the papers in this special issue
In total, we had an initial number of 20 people that replied to our call for papers from 12 different institutions from the United States of America, Germany, Luxembourg, Bulgaria, and The Netherlands from fields like biomedicine, bioinformatics, health sciences, ecology, data science, and computer science. After an initial information session, out of the 20 authors that responded to the call for papers, 18 decided to continue their participation. All these 18 authors that responded to the call for formalization papers managed in the end to publish (upon acceptance) their articles in this special issue.
We had a total of 15 formalization paper submissions, 13 with individual authors and 2 with joint authorship. Out of the total of 18 authors, two of these have both an individual submission and a joint-authorship one. The super-pattern instantiations of the final accepted formalization paper submissions can be seen in Table 1. Here, the classes used to instantiate the super-patterns that comprise the formalizations are given for each submission: the context, subject and object classes for each submission are listed, together with the qualifier and relations selected from the SuperPattern ontology [4].
Looking at Table 1, we see that the super-pattern instances exhibit quite a broad variety of scientific fields (bioinformatics, biomedicine, pharmacology, data science, computer science) mostly linked to the life sciences. 7 out of the 15 submissions contain a formalization in which authors extracted a scientific claim from their own previously published article (submission number marked with ◇). Additionally, out of the total 44 classes used in the formalizations, 22 new classes were minted using Nanobench (marked with *), while 4 were newly minted Wikidata classes (marked with **). 13 already-existing classes were reused from Wikidata (their Wikidata identifier is specified next to the class name) and 4 classes were referenced from other ontologies.
7.Conclusion
The publication of the special issue with formalization papers at the Data Science journal shows not only that nanopublications and the super-pattern can be used to implement the basic steps and entities of a journal workflow, but also that authors of such formalization papers can be taught to use these in order to publish in a novel journal publication workflow as the publication of the special issue demonstrates. We found that the super-pattern can be well understood conceptually, and that its application in a practical setting is feasible. With the publication of this special issue we demonstrate for the first time that the content of a scientific journal can be made fully machine-interpretable.
In the future, we can take the next logical step by publishing novel claims in this way from the start, and not depend on claims from already-published papers. These contributions will then also have to be accompanied by statements about the methods, equipment, and all other relevant scientific concepts, and can include not just the high-level claim but more lower-level ones, possibly all the way down to the raw data. This representation would then ideally cover the entire scientific workflow, starting from a motivation, leading to the design and execution of a study, and ending in new scientific insights. Such fully formalized scientific contributions can be seen as a major step – even a breakthrough – for the Semantic Web and Open Science movements and will bring us closer to a world where machines can interpret scientific knowledge and help us organize and understand it in a reliable and transparent manner.
Notes
References
[1] | T. Al-Moslmi, M.G. Ocaña, A.L. Opdahl and C. Veres, Named entity extraction for knowledge graphs: A literature overview, IEEE Access 8: ((2020) ), 32862–32881. doi:10.1109/ACCESS.2020.2973928. |
[2] | C.-I. Bucur, T. Kuhn and D. Ceolin, Peer reviewing revisited: Assessing research with interlinked semantic comments, in: K-CAP 2019: Proceedings of the 10th International Conference on Knowledge Capture, (2019) , pp. 179–187. doi:10.1145/3360901.3364434. |
[3] | C.-I. Bucur, T. Kuhn, D. Ceolin and J. van Ossenbruggen, A unified nanopublication model for effective and user-friendly access to the elements of scientific publishing, in: EKAW2020, (2020) . doi:10.1007/978-3-030-61244-3_7. |
[4] | C.-I. Bucur, T. Kuhn, D. Ceolin and J. van Ossenbruggen, Expressing high-level scientific claims with formal semantics, in: Proceedings of the 11th on Knowledge Capture Conference, K-CAP’21, Association for Computing Machinery, New York, NY, USA, (2021) , pp. 233–240. ISBN 9781450384575. doi:10.1145/3460210.3493561. |
[5] | C.-I. Bucur, T. Kuhn, D. Ceolin and J. van Ossenbruggen, Nanopublication-Based Semantic Publishing and Reviewing: A Field Study with Formalization Papers, 2022, available at https://arxiv.org/abs/2203.01608. |
[6] | O. Etzioni, M. Cafarella, D. Downey, A.-M. Popescu, T. Shaked, S. Soderland, D.S. Weld and A. Yates, Unsupervised named-entity extraction from the web: An experimental study, Artificial Intelligence 165: (1) ((2005) ), 91–134. doi:10.1016/j.artint.2005.03.001. |
[7] | R.A. Felix and M.A. Barrand, P-glycoprotein expression in rat brain endothelial cells: Evidence for regulation by transient oxidative stress, Journal of Neurochemistry 80: ((2002) ), 64–72. doi:10.1046/j.0022-3042.2001.00660.x. |
[8] | P. Groth, A. Gibson and J. Velterop, The anatomy of a nanopublication, Inf. Serv. Use 30: ((2010) ), 51–56. doi:10.3233/ISU-2010-0613. |
[9] | T. Kuhn and M. Dumontier, Genuine semantic publishing, Data Sci. 1: ((2017) ), 139–154. doi:10.3233/DS-170010. |
[10] | T. Kuhn, R. Taelman, V. Emonet, H. Antonatos, S. Soiland-Reyes and M. Dumontier, Semantic micro-contributions with decentralized nanopublication services, PeerJ Computer Science 7: ((2021) ), e387. doi:10.7717/peerj-cs.387. |
[11] | P. Lisena, A. Meroño-Peñuela, T. Kuhn and R. Troncy, Easy web API development with SPARQL transformer, in: The Semantic Web – ISWC 2019, C. Ghidini et al., eds, Lecture Notes in Computer Science, Vol. 11779: , Springer, Cham, (2019) . doi:10.1007/978-3-030-30796-7_28. |
[12] | A. Meroño-Pe nuela and R. Hoekstra, grlc makes GitHub taste like linked data APIs, in: European Semantic Web Conference, Springer, (2016) , pp. 342–353. doi:10.1007/978-3-319-47602-5_48. |
[13] | Y. Xu, L. Mou, G. Li, Y. Chen, H. Peng and Z. Jin, Classifying relations via long short term memory networks along shortest dependency paths, in: Proceedings of the 2015 Conference on Empirical Methods in Natural Language Processing, Association for Computational Linguistics, Lisbon, Portugal, (2015) , pp. 1785–1794. doi:10.18653/v1/D15-1206. |
[14] | V. Yadav and S. Bethard, A survey on recent advances in named entity recognition from deep learning models, in: COLING, (2018) , available at https://aclanthology.org/C18-1182.pdf. |
[15] | D. Zeng, K. Liu, S. Lai, G. Zhou and J. Zhao, Relation classification via convolutional deep neural network, in: COLING, (2014) , available at https://aclanthology.org/C14-1220.pdf. |



