Journals
Therapeutic Design of Peptide Modulators of the Interaction Between eNOS and p53 in Atherosclerosis
A B S T R A C T
Atherosclerosis is a cardiovascular disease featuring a chronic inflammation due to the accumulation of lipids within the tunica intima of arteries. The development of the disease depends on dynamic changes in the vascular biology. Immune system cells directly influence the pathogenesis of atherosclerosis during the inflammatory process. Currently, atherosclerosis diagnosis is performed by non-invasive or invasive methods depending on the type of arteries that are being investigated. New diagnostic and therapeutic procedures should improve the quality of life of patients. Some of the genes that could be biomarkers of cardiovascular diseases are TP53 and eNOS. The protein p53 is recognized as a tumor suppressor protein that controls DNA repair, cell cycle progression or arrest and apoptosis. These functions that p53 exerts are well known and some other functions are being investigated, such as its role in the cardiovascular system. The eNOS gene regulates the levels of nitric oxide, which is vital for several intracellular biological functions, such as vasodilation, vascular homeostasis, protection of arteries against injuries, cellular growth, signaling pathways and immune response among others. Here, we used an in-silico approach to predict four models of interaction between clinically important proteins (eNOS and p53), to predict the interface of interaction and to rationally design modulating peptides to be tested in vitro and in vivo and possibly used as a therapeutic agent.
K E Y W O R D S
eNOS, p53, atherosclerosis, PPI, interface of interaction, modulating peptides.
I N T R O D U C T I O N
Atherosclerosis is a chronic inflammatory disease that occurs by the accumulation of lipids in the innermost layer (tunica intima) of small, medium and large caliber arteries. The atheromatous plaque, together with platelet factors, stimulate the proliferation of muscle cells within this region. Thus, muscle cells, leukocytes and lipids remain stuck in this region leading to the narrowing of the arterial lumen. This intricate deposit might progress into fibrosis and the calcification of the atheromatous plaque. Its growth causes an obstruction of the artery and consequent local ischemia [1]. Moreover, the development of the disease depends on dynamic changes in the vascular biology [2]. The main etiopathogenic mechanism of cardiovascular diseases is the process of atherogenesis. Immune system cells play an important role in the pathogenesis of atherosclerosis during the inflammatory process that occur in the endothelium [3]. Atherosclerosis usually begins in childhood and progress silently over a long pre-clinical stage and eventually manifests clinically during the middle age of an individual [4]. Atherosclerotic cardiovascular diseases and its clinical manifestations, such as myocardial infarction and ischemic stroke, are the leading causes of morbidity and mortality worldwide [5]. Many factors have been reported to be associated with an increased risk of cardiovascular events [6]. The most widely studied factor is by far the low-density lipoprotein (LDL). Lipoproteins, such as LDL-cholesterol, containing apolipoprotein B, very low density lipoproteins (VLDL) and their remnants, intermediate density lipoproteins (IDL) and lipoprotein A directly influence the development of atherosclerosis [7].
Currently, atherosclerosis diagnosis is performed by non-invasive (echo Doppler) and invasive methods (angiotomography and catheterization), depending on the type of arteries that are being investigated. Even though the pathology has a genetic background, there is no genotypic technique efficient enough for a non-invasive and reliable diagnosis method. This is due to the complex genetic trait that characterizes atherosclerosis and the diversity of genes and polymorphisms that is related to the disease [8-10]. Several researches have been conducted in order to develop genetic and molecular tests to evaluate individuals at high risk of developing atherosclerosis [11-13].Some of the genes that could be biomarkers of cardiovascular diseases are TP53 (tumor protein 53) and eNOS (endothelial nitric oxide synthase). TP53 codes for the protein p53, which is known as the guardian of the genome due to its function related to genomic stability [14]. In addition, p53 is recognized as a tumor suppressor protein that controls DNA repair, cell cycle progression or arrest and apoptosis [15-17]. These functions that p53 exerts are well known and some other functions are being investigated, such as its role in the cardiovascular system. It is clear that p53 somehow influences cardiovascular homeostasis but details on how that is performed is not clear yet. Overexpression of p53 increases death rates in patients who suffered from myocardial infarction, decreases heart function, angiogenesis and distribution of oxygen [18-20]. Nitric oxide (NO) is a stress-signaling compound and increased level of NO can cause DNA damage, which activates p53 and reflects on vascular homeostasis and susceptibility to diseases [21, 22].
The endothelial dysfunction presented by atherosclerotic patients responds to several risk factors related to cardiovascular diseases. High cholesterol levels, hypertension, diabetes, smoking and other environmental factors lead to a severe pro-inflammatory and a pro-thrombotic endothelial state [23-28]. In addition, genetic polymorphisms influence endothelial dysfunction because several genes and the proteins they code for, exert crucial functions in regulating vascular endothelial stability [29-31]. The eNOS gene regulates the levels of nitric oxide, which is vital for several intracellular biological functions, such as vasodilation, vascular homeostasis, protection of arteries against injuries, cellular growth, signaling pathways and immune response among others [32-37]. The eNOS gene has been investigated as a possible biomarker for non-invasive diagnostic and more efficient treatment of cardiovascular diseases [31, 38-40].
Here, we used an in-silico approach to predict four models of interaction between clinically important proteins (eNOS and p53), to predict the interface of interaction and to rationally design modulating peptides to be tested in vitro and in vivo and possibly used as a therapeutic agent.
Materials and Methods
The three-dimensional structure of the proteins eNOS and p53 were modeled by the I-TASSER (Iterative Threading Assembly Refinement) server [41]. The modeling relies on templates based on homology from protein structures experimentally resolved and available in the PDB (protein databank). The predicted structure is assembled by fold recognition through Monte Carlo simulations. Briefly, the pipeline used for the prediction of the target protein structures consists of six basic steps. The prediction of the secondary structure by PSSpred (Protein Secondary Structure Prediction) and identification of templates by LOMETS (Local Meta-Threading-Server) [42]. Then, assembly of ranked fragments through Monte Carlo simulations; clusterization of structures according to conformation and energy using SPICKER in order to identify near native structures; molecular dynamics structure refinement and finally the prediction of biological function by COACH [42-45].
The domains of eNOS and p53 were identified by KBDOCK and InterPro [46, 47]. Protein-protein docking analyses were carried out by ClusPro, through clusterization and minimization of the predicted models [48]. The protein-protein interaction (PPI) and the interface of interaction between the target proteins are build based on three different coefficients considered individually (electrostatic-favored, hydrophobic-favored and Van der Waals-favored) or together (balanced-favored). The predicted PPIs are ranked according to energy scores based on those coefficients. The visualization software PyMol was used in order to analyze the PPI results, the interface of interaction between eNOS and p53, predicted hot spots, polymorphic residues and to design peptides able to modulate the interaction between those proteins. Amino acid residues that significantly contribute to the free-energy of binding and stability of PPI within the interface of interaction were recognized by KFC2 [49]. The basis for the identification of such amino acid residues is the structural and chemical analysis of the environment around residues. Moreover, hot spots experimentally determined by alanine scanning mutagenesis are taken into account for the prediction of hot spot present in the proteins under investigation. The hot spot prediction scores are based on conformation (scorea) and on biochemical properties (scoreb). Clinically important polymorphic residues for the eNOS and p53 proteins were identified through the dbSNP (database of single nucleotide polymorphism.
Results and Discussion
The protein eNOS regulates the availability of NO, a lipophilic compound that takes part in several biological mechanism [50]. NO produced by the activity of eNOS modulate, beyond other functions, the degree of constriction experienced by a blood vessel, cell cycle progression, senescence or apoptosis, immune system cells activity and platelet aggregation [51-55]. Moreover, availability of NO influences cancer, genomic stability and cardiovascular diseases [31, 39, 56, 57]. Regarding the p53 protein, which is coded by the TP53 gene, is known as the guardian of the genome. The protein is a classical tumor suppressor protein related to cancer and several other diseases related to genomic instability, such as endometriosis, atherosclerosis and infertility [58-61]. Experimental approaches along with bioinformatic tools have contributed to increase our knowledge regarding diseases, development of new diagnosis and therapeutic strategies [62, 63]. The in silico prediction of hot spots within the interface of protein-protein complexes drive design of small peptides that can modulate PPIs, being a promisor technique for new treatment of diseases [63-65]. The basis for these approaches is the fact that certain amino acid residues are generally conserved among structure-related proteins and proteins with complementary functions. Variation on those conserved hot spots alter the conformational state of a protein and multi-protein complexes increasing the susceptibility to diseases to diseases through reduction, loss or gain of function [66]. It has been investigated the role of p53 in atherosclerosis and other cardiovascular diseases. NO has been implicated in p53 functions [21]. Since eNOS is responsible for NO synthesis and availability, we propose four different modes of interactions between p53 and eNOS according to energy parameters. Our approach led to the design of four peptides that could modulate the interaction between the target proteins and their function in atherosclerosis. To our knowledge, no study has aimed to propose such approach related to eNOS and p53.
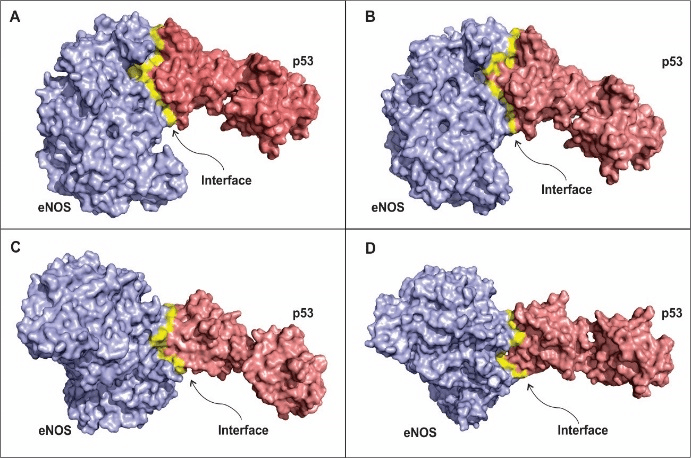
Figure 1 shows the four best stable modes of interaction between eNOS and p53 and the interface of interaction between the proteins under study. The modes of interaction are based on coefficients of energy, including electrostatic-favored (Figure 1B), hydrophobic-favored (Figure 1C), Van der Waals-favored (Figure 1D) and balanced coefficients of energy (Figure 1A). The latter was used to build a mode of interaction that considers all the other three types of coefficients. The mode of interaction for the proteins related to the balanced coefficients and that for the electrostatic-favored coefficients are very similar (Figures 1A and B). The main differences between these two predicted states are the hot spots residues identified for each situation, even though some of these hot spots repeat within the interface for both approaches (Tables 1 and 2). Several studies have shown how electrostatic forces contribute to PPIs, including those related to diseases development [67-69].
Figure 1C and 1D show a predicted mode of interaction between eNOS and p53 regarding hydrophobic-favored and Van der Waals-favored coefficients, respectively. The conformation of the complex for these two last coefficients are more similar to each other than the conformation predicted for electrostatic and hydrophobic forces. The hot spot residues that most contribute to the interaction and stabilization of the complex for the hydrophobic and Van der Waals forces are described in tables 3 and 4. Hydrophobic effect in PPIs are one of the main causes of hot spots clustering within the interface of interaction between proteins or protein and ligands [70].
Figure 1: Models for the eNOS and p53 interaction according to different chemical forces coefficients.
A – Model of interaction between eNOS and p53 taking into consideration all three energy coefficients used in the present study. B - Model for interaction between eNOS and p53 taking into consideration electrostatic-favored coefficients. C – Model for eNOS and p53 interaction regarding ahydrophobic-favored forces. D - Model for eNOS and p53 interaction regarding Van der Waals-favored coefficient. Blue: eNOS; red: p53 dimer; yellow: interface of interaction between the target proteins. Clusterization drives hot spot prediction and the design of modulating peptides in several approaches such the ones presented here. In fact, we found that our predicted hot spots in the interface of interaction between eNOS and p53 are near one to another (Table 3), forming clusters and contributing for the stability of the complex. Actually, clusters of hot spot residues were also predicted for the other coefficients of energy (Tables 2 and 4).
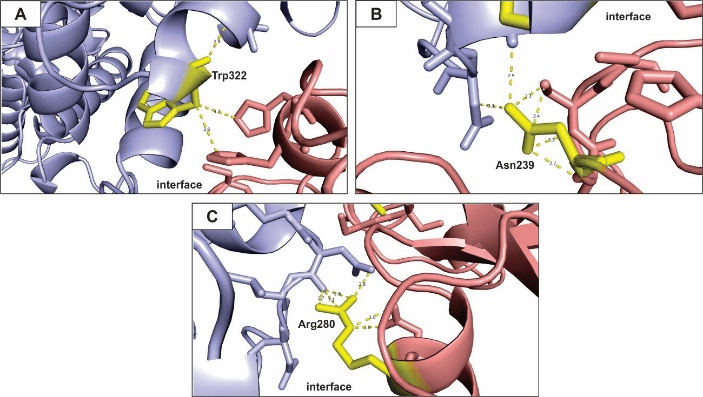
Next, we analyze the hot spot residues that most contribute for the stabilization of the complex in each model of eNOS and p53 interaction. We identified six mains hot spot residues for the balanced model of interaction (Figure 2). These amino acid residues establish polar contact with other hot spots present within the cluster and also with neighbor, less important, residues.
Table 1: Hotspot residues that significantly contribute to the free-energy of binding through balanced coefficients of energy
|
Residue |
Scorea |
Scoreb |
|
|
A |
Arg70 |
0.36 |
0.04 |
|
Trp244 |
1.38 |
0.29 |
|
|
Gln476 |
0.52 |
0.06 |
|
|
B |
Asp478 |
1.37 |
0.01 |
|
Trp480 |
1.28 |
0.29 |
|
|
His178 |
1.21 |
0.04 |
|
|
a |
Met243 |
1.14 |
0.22 |
|
Arg280 |
0.47 |
0.07 |
Scorea – Score based on conformation
Scoreb – Score based on biochemical properties
Table 2: Hotspot residues that significantly contribute to the free-energy of binding through electrostatic-favored coefficient of energy.
|
Residue |
Scorea |
Scoreb |
|
|
A |
Phe105 |
0.63 |
0.04 |
|
Trp244 |
1.40 |
0.32 |
|
|
A |
Arg474 |
0.99 |
0.14 |
|
Gln476 |
0.55 |
0.02 |
|
|
Trp480 |
1.66 |
0.31 |
|
|
B |
Arg70 |
0.80 |
0.16 |
|
a |
Arg175 |
0.78 |
0.36 |
|
His178 |
1.20 |
0.17 |
|
|
His179 |
1.46 |
0.29 |
|
|
a |
Arg181 |
0.72 |
0.06 |
|
a |
Asn239 |
1.18 |
0.01 |
Scorea – Score based on conformation
Scoreb – Score based on biochemical properties
Table 3: Hotspot residues that significantly contribute to the free-energy of binding through hydrophobic-favored coefficients of energy
|
Residue |
Scorea |
Scoreb |
|
|
Trp322 |
1.46 |
0.25 |
|
|
A |
Leu326 |
0.60 |
0.14 |
|
His178 |
1.64 |
0.26 |
|
|
His179 |
0.52 |
0.18 |
Scorea – Score based on conformation
Scoreb – Score based on biochemical properties
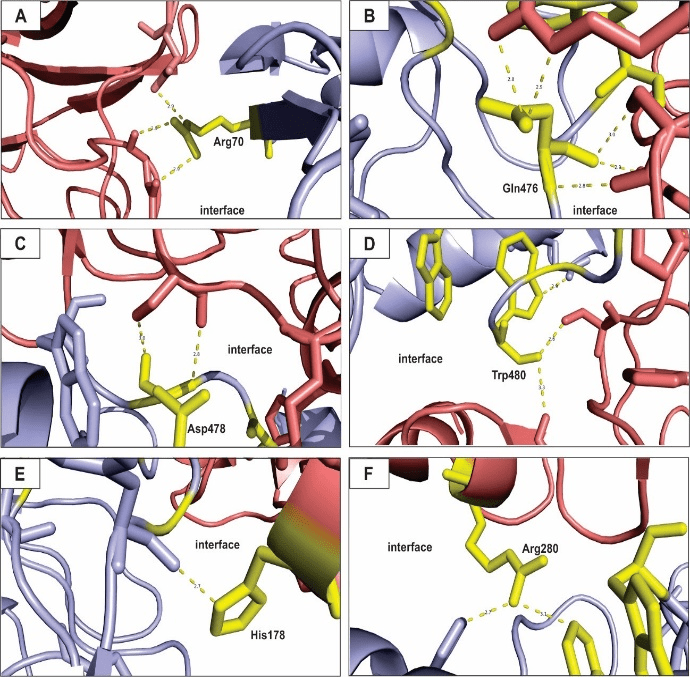
The residue Arg70 (Figure 2A), present in the eNOS structure, interacts with two residues of the p53 polypeptide chain, while Arg280 (Figure 2F) interacts with two other residues. The Arg side chain is amphipathic and the amino acid is usually found on the surface of proteins, with its hydrophilic part interacting with other polar residues of partner proteins or interacting with the environment surrounding it [71]. The residues Gln 476 (Figure 1B), Asp478 (Figure 1C) and Trp480 (Figure 1D) are present in the p53 structure and they contribute significantly to the stability of the complex, binding to residues from the eNOS chain within the interface of interaction. Finally, His178 belong to a cluster of hot spots on the eNOS structure and significantly contribute to the free-energy of the binding proteins, which is clearly important for the biological function of the complex [72, 73].
Table 4: Hotspot residues that significantly contribute to the free-energy of binding through Van Der Waals-favored coefficients of energy
|
Residue |
Scorea |
Scoreb |
|
|
Trp322 |
0.47 |
0.10 |
|
|
a |
Asn 239 |
0.67 |
0.06 |
|
Arg280 |
0.52 |
0.04 |
Scorea – Score based on conformation
Scoreb – Score based on biochemical properties
Figure 2: Hot spots prediction for balanced coefficients model of eNOS and p53 interaction.
All figures presented here show polar interactions for the amino acid residues classified as hot spots (Table 1). A – Arg70; B – Glu476; C – Asp478; D – Trp480; E – His178; F – Arg280. Blue: eNOS; red: p53; yellow: hot spot residues.
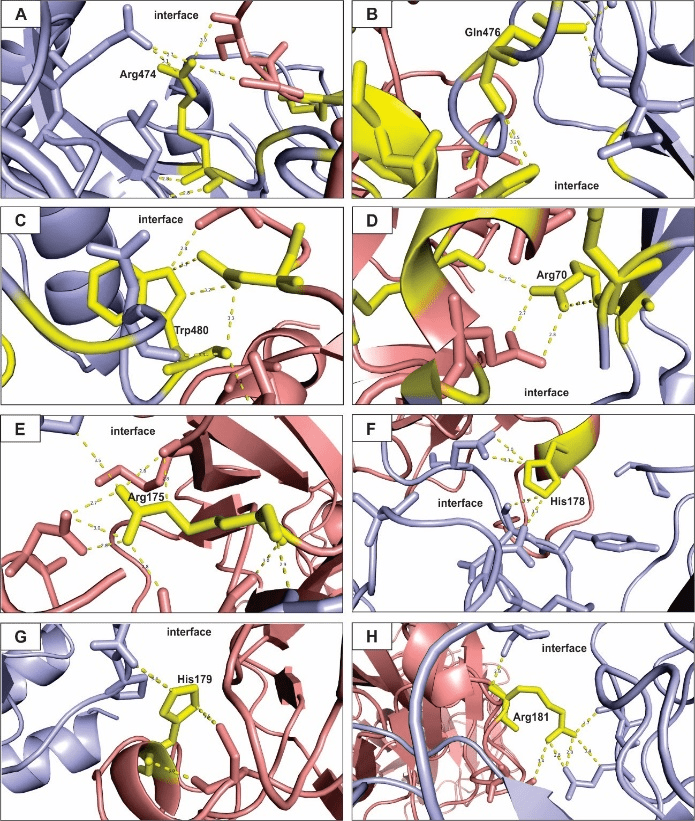
Figure 3 shows the hot spots prediction for the electrostatic-favored model of interaction between eNOS and p53. Interestingly, more Arg residues (Figure 3A, D, E and I) participate in polar interactions in the interface between the proteins target of the present approach. Arg residues have been shown to contribute significantly to the binding of toxin proteins and ion channel proteins through electrostatic forces and they can act as electrostatic adhesive forces among biomolecules [74, 75]. Here, Arg hot spot residues develop polar interactions with neighbor amino acids from the same polypeptide chain and with amino acids from the polypeptide chain of the interacting protein. Thus, it greatly influences the conformation stability of the eNOS-p53 complex.
The other amino acid residues that contribute to the free-energy of binding through the electrostatic-favored coefficient are Gln476, Trp480, His178 and His 179. The former has been related to play important roles on the intermolecular association and aggregation of proteins through polar bonds [76]. In addition, Gln influences formation of macromolecular complexes formed by proteins and RNAs [77]. Trp has also been identified as an important component of protein-ligand interfaces, playing anchoring roles among structural binding proteins and stabilizing binding sites of proteins [78, 79]. The latter residues in this hot spot cluster, His178 and His179, have multiple roles in the molecular interactions due to the properties showed by the structure of histidine. Special interest is directed to His residues duet to its ability of modulating electrostatic interactions of charged residues. A feature that is promising for the regulation of the stability of protein complex and the design of modulating small organic molecules in therapeutics approaches [80, 81].
Figure 3: Hot spots prediction for the electrostatic-favored coefficients model of eNOS and p53 interaction.
All figures presented here show polar interactions for the amino acid residues predicted as hot spots within the interface of interaction of the proteins under study (Table 2). A – Arg474; B – Gln476; C – Trp480; D – Arg70; E – Arg175; F – His178; G – His179; H – Arg181. Blue: eNOS; red: p53; yellow: hot spot residues.
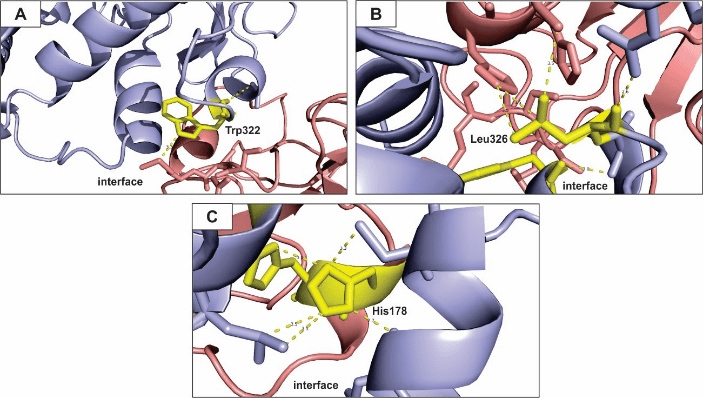
Figure 4: Hot spots prediction for the hydrophobic-favored coefficients model of eNOS and p53 interaction.
All figures presented here show polar interactions for the amino acid residues predicted as hot spots within the interface of interaction of the proteins under study (Table 3). A – Trp322; B – Leu326; C – His178. Blue: eNOS; red: p53; yellow: hot spot residues.
Figure 5: Hot spots prediction for the Van der Waals-favored coefficient model of eNOS and p53 interaction.
All figures presented here show polar interactions for the amino acid residues predicted as hot spots within the interface of interaction of the proteins under study (Table 3). A – Trp322; B – Asn239; C – Arg280. Blue: eNOS; red: p53; yellow: hot spot residues.
We identified three important hot spot residues that contribute to the free-energy of binding in the eNOS-p53 complex regarding the hydrophobic-favored coefficient (Table 3). Trp residues and their interactions with neighbor residues drive the protein complex folding due to its hydrophobic nature and tendency to be located inside the protein structure [82].
Figure 4A shows a Trp hot spot residue on the eNOS polypeptide chain, its structure does not project into the interface of interaction, it is rather buried within eNOS structure. Even so, it is able to establish polar interactions with a residue from the p53 polypeptide chain and contribute to the free-energy of binding of the complex. The Trp residue interact with intra-chain residues and contribute to the stability of the conformation of eNOS structure as it binds to p53. In addition, Trp residues have been related to influence refolding and stability of beta‐sheets [83, 84]. Here, Trp 322 belongs to a beta‐sheet chain and we hypothesize that it drives the conformation of eNOS differently when the protein is bind or free from the p53 partner. Polymorphic Trp 322 has significant clinical relevance (dbSNP short genetic variations) as it may increase susceptibility to diseases, such as atherosclerosis and cancer, due to differences in eNOS and eNOS-p53 folding and refolding mechanisms.
Figure 4B shows a hydrophobic Leu residue interacting with neighboring residues within the interface of interaction and contributing to the stability of the complex. It has been show that Leu residues and other hydrophobic residues (such as Trp) largely contribute to the stability of protein complex through polar interaction within interfaces, a feature also shared by Hys residues [80, 81, 85, 86]. Figure 4C shows a residue of His in a hot spot cluster interacting with residues located in intra and inter polypeptide chains in the interface of interaction of eNOS and p53.
Van de Waals force are weak, individually, but they greatly contribute to the free-energy of binding when the whole structure of the interface of interaction is taken into account [87]. Out approach predicted three amino acid residues within the eNOS-p53 interface of interaction (Table 4). Trp and other sulfur-containing amino acids govern complex stabilization through Van der Waals interaction and hydrogen bonding in certain proteins [88]. Asn residues have been shown to contribute to the interface of interaction in an antigen-antibody complex and to drive amide orientation through interaction with other neighboring amino acids [89, 90]. Finally, Arg residues contribute to nucleosome structure due to Van der Waals forces between histones and DNA [91]. Here, these amino acids (Trp, Asn and Arg) contribute significantly for the stability of the eNOS-p53 complex (Figure 5).
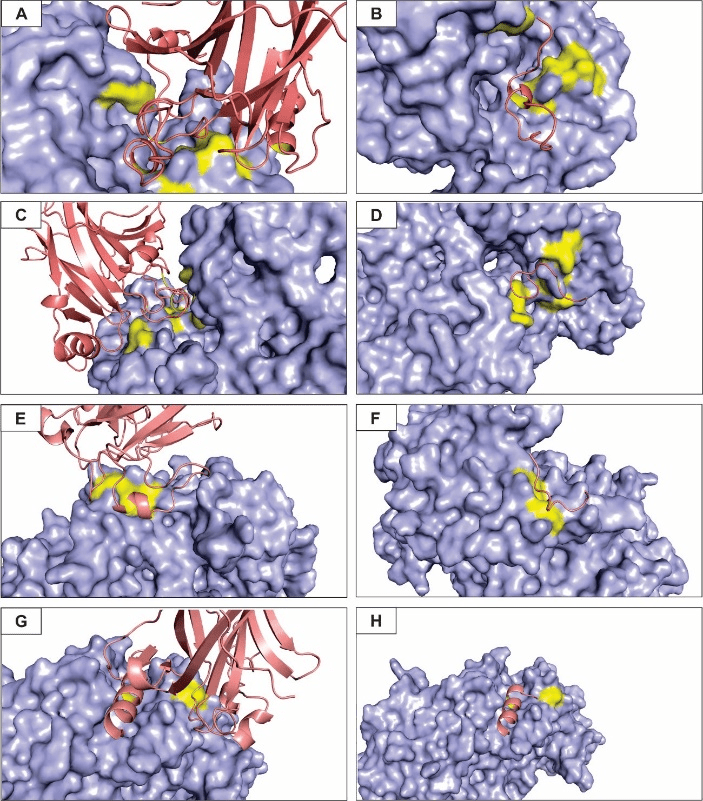
A – Interface of interaction between eNOS and p53 for a balanced coefficient of energy. B – Peptide rationally designed to modulate the eNOS-p53 mode of interaction based on balanced coefficients. C – Interface of interaction for the electrostatic-favored coefficient. D – Peptide rationally designed based on the electrostatic-favored coefficient. E – Interface of interaction for the hydrophobic-favored coefficient. F – Peptide rationally designed based on the hydrophobic-favored coefficient. G – Interface of interaction for the Van der Waals-favored coefficient. H – Peptide rationally designed action based on the Van der Waals-favored coefficient
Based on the predicted hot spots and the interface of interaction between the complex formed by eNOS and p53 according to specific energy coefficients, we rationally designed modulating peptides for each model (Figure 6). To our knowledge, no study aimed at the design of small molecules for the interaction of such proteins, although several other studies have been trying to find efficient designed peptides that could modulate eNOS and p53 activities individually or when interacting with other target proteins [92, 93].
Figure 6A shows the surface of the eNOS protein, the hot spots residues within the interface of interaction and a secondary structure of p53 monomer representation in order to highlight how they interact according to balanced coefficients of energy. Figure 6B shows the designed peptide (the sequence of the peptides is not shown) anchored on eNOS surface. Regarding electrostatic-favored forces clusters of hot spot residues on the p53 surface form loops that fit in clefts present on the surface of eNOS (Figure 6C). Although the predicted complex structure and interface of interaction between eNOS and p53 are very similar for the balanced coefficients and the electrostatic-favored coefficient, the peptide designed for the latter is rather smaller, but with an energy score similar to the former (Figure 6D).
The prediction of the interface of interaction for the hydrophobic-favored and Van der Waals-favored coefficients was in a quite different region of the eNOS protein when compared to the other coefficients. A smaller interface of interaction was identified (Figure 6E and G) as the best score and a smaller number of hot spots was found for the hydrophobic-favored and Van der Waals-favored (Tables 3 and 4). Figures 6F and H shows the rationally designed peptides for the latter coefficients, respectively. Interestingly, the structure of the peptide predicted for the Van der Waals coefficient is a beta-sheet and fits perfectly in a cleft present on the eNOS surface.
Concluding Remarks
Cardiovascular diseases are the leading cause of deaths worldwide. Genetic and environmental factors increase the susceptibility to such diseases. Recently, research has focused on the prediction of proteins structure, interaction and other properties that could enhance diagnostic and therapeutic procedures. Bioinformatic tools have become a promisor way to achieve such goals and several different approaches have been proposed with promising results. Here, we used an in-silico approach to predict four models of interaction between clinically important proteins (eNOS and p53), to predict the interface of interaction and to rationally design modulating peptides to be tested in vitro and in vivo and possibly used as a therapeutic agent.
A – Interface of interaction between eNOS and p53 for a balanced coefficient of energy. B – Peptide rationally designed to modulate the eNOS-p53 mode of interaction based on balanced coefficients. C – Interface of interaction for the electrostatic-favored coefficient. D – Peptide rationally designed based on the electrostatic-favored coefficient. E – Interface of interaction for the hydrophobic-favored coefficient. F – Peptide rationally designed based on the hydrophobic-favored coefficient. G – Interface of interaction for the Van der Waals-favored coefficient. H – Peptide rationally designed action based on the Van der Waals-favored coefficient
Article Info
Article Type
Research ArticlePublication history
Received: Fri 08, Mar 2019Accepted: Mon 15, Apr 2019
Published: Fri 10, May 2019
Copyright
© 2023 Silva KSF. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. Hosting by Science Repository.DOI: 10.31487/j.JICOA.2019.02.01
Author Info
Araújo DS Assunção LP Barbosa AM Costa IR de Curcio JS Moraes D Oliveira LN Santos TG Silva CTX Silva KSF Silva MG
Corresponding Author
Silva KSFBiological Sciences Institute, Federal University of Goiás, UFG, GO, Brazil
Figures & Tables
All figures presented here show polar interactions for the amino acid residues predicted as hot spots within the interface of interaction of the proteins under study (Table 2). A – Arg474; B – Gln476; C – Trp480; D – Arg70; E – Arg175; F – His178; G – His179; H – Arg181. Blue: eNOS; red: p53; yellow: hot spot residues.
All figures presented here show polar interactions for the amino acid residues predicted as hot spots within the interface of interaction of the proteins under study (Table 3). A – Trp322; B – Leu326; C – His178. Blue: eNOS; red: p53; yellow: hot spot residues.
All figures presented here show polar interactions for the amino acid residues predicted as hot spots within the interface of interaction of the proteins under study (Table 3). A – Trp322; B – Asn239; C – Arg280. Blue: eNOS; red: p53; yellow: hot spot residues.

A – Interface of interaction between eNOS and p53 for a balanced coefficient of energy. B – Peptide rationally designed to modulate the eNOS-p53 mode of interaction based on balanced coefficients. C – Interface of interaction for the electrostatic-favored coefficient. D – Peptide rationally designed based on the electrostatic-favored coefficient. E – Interface of interaction for the hydrophobic-favored coefficient. F – Peptide rationally designed based on the hydrophobic-favored coefficient. G – Interface of interaction for the Van der Waals-favored coefficient. H – Peptide rationally designed action based on the Van der Waals-favored coefficient
Table 1: Hotspot residues that significantly contribute to the free-energy of binding through balanced coefficients of energy
|
Residue |
Scorea |
Scoreb |
|
|
A |
Arg70 |
0.36 |
0.04 |
|
Trp244 |
1.38 |
0.29 |
|
|
Gln476 |
0.52 |
0.06 |
|
|
B |
Asp478 |
1.37 |
0.01 |
|
Trp480 |
1.28 |
0.29 |
|
|
His178 |
1.21 |
0.04 |
|
|
a |
Met243 |
1.14 |
0.22 |
|
Arg280 |
0.47 |
0.07 |
Scorea – Score based on conformation
Scoreb – Score based on biochemical properties
Table 2: Hotspot residues that significantly contribute to the free-energy of binding through electrostatic-favored coefficient of energy.
|
Residue |
Scorea |
Scoreb |
|
|
A |
Phe105 |
0.63 |
0.04 |
|
Trp244 |
1.40 |
0.32 |
|
|
A |
Arg474 |
0.99 |
0.14 |
|
Gln476 |
0.55 |
0.02 |
|
|
Trp480 |
1.66 |
0.31 |
|
|
B |
Arg70 |
0.80 |
0.16 |
|
a |
Arg175 |
0.78 |
0.36 |
|
His178 |
1.20 |
0.17 |
|
|
His179 |
1.46 |
0.29 |
|
|
a |
Arg181 |
0.72 |
0.06 |
|
a |
Asn239 |
1.18 |
0.01 |
Scorea – Score based on conformation
Scoreb – Score based on biochemical properties
Table 3: Hotspot residues that significantly contribute to the free-energy of binding through hydrophobic-favored coefficients of energy
|
Residue |
Scorea |
Scoreb |
|
|
Trp322 |
1.46 |
0.25 |
|
|
A |
Leu326 |
0.60 |
0.14 |
|
His178 |
1.64 |
0.26 |
|
|
His179 |
0.52 |
0.18 |
Scorea – Score based on conformation
Scoreb – Score based on biochemical properties
Table 4: Hotspot residues that significantly contribute to the free-energy of binding through Van Der Waals-favored coefficients of energy
|
Residue |
Scorea |
Scoreb |
|
|
Trp322 |
0.47 |
0.10 |
|
|
a |
Asn 239 |
0.67 |
0.06 |
|
Arg280 |
0.52 |
0.04 |
Scorea – Score based on conformation
Scoreb – Score based on biochemical properties
References
- Barter P (2003) Introduction. Lipids inflammation and CVD--understanding the basics to break the atherosclerosis cycle. Int J Clin Pract Suppl 2-4. [Crossref]
- Ross R (1999) Atherosclerosis--an inflammatory disease N Engl J Med 340: 115-126. [Crossref]
- Hansson GK, Hermansson (2011) A The immune system in atherosclerosis. Nat Immunol 12: 204-212. [Crossref]
- Head T, Daunert S, Goldschmidt Clermont PJ The Aging Risk and Atherosclerosis: A Fresh Look at Arterial Homeostasis. Front Genet 8: 216. [Crossref]
- GBD 2015 Disease Injury Incidence Prevalence Collaborators (2016) Global, regional, and national incidence, prevalence, and years lived with disability for 310 diseases and injuries, 1990-2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet 388: 1545-1602. [Crossref]
- Yusuf S, Hawken S, Ounpuu S, Dans T Averzum A et al. (2004) Effect of potentially modifiable risk factors associated with myocardial infarction in 52 countries (the INTERHEART study): case-control study. Lancet 364: 937-952. [Crossref]
- Goldstein JL Brown MS (2015) A century of cholesterol and coronaries: from plaques to genes to statins. Cell 161: 161-172. [Crossref]
- Szabó GV (2013) The role and importance of gene polymorphisms in the development of atherosclerosis. Interv Med Appl Sci 5: 46-51. [Crossref]
- Biros E, Karan M, Golledge J (2008) Genetic Variation and Atherosclerosis. Curr Genomics 9: 29-42. [Crossfer]
- Jayashree Shanker,Vijay V Kakkar (2017) Implications of Genetic Polymorphisms in Inflammation-Induced Atherosclerosis. Open Cardiovasc Med J 4: 30-37. [Crossref]
- Brown TM, Bittner V (2008) Biomarkers of atherosclerosis: clinical applications. curr cardiol Rep 10: 497-504. [Crossref]
- Soeki T, Sata M (2016) Inflammatory Biomarkers and Atherosclerosis. Int Heart J 57: 134-139. [Crossref]
- W van Lammeren G, L Moll F, Borst GJD, de Kleijn DP, PM de Vries JP et al. (2011) Atherosclerotic Plaque Biomarkers: Beyond the Horizon of the Vulnerable Plaque. Curr Cardiol Rev 7: 22-27. [Crossref]
- Yeo CQX, Alexander I, Lin Z, Lim S, Aning OA et al. (2016) p53 Maintains Genomic Stability by Preventing Interference between Transcription and Replication. Cell Rep 15: 132-146. [Crossref]
- 15.Ashley B Williams, Bjorn Schumacher (2016) p53 in the DNA-Damage-Repair Process. Cold Spring Harb Perspect Med 6: a026070. [Crossref]
- Shaw PH (1996) The role of p53 in cell cycle regulation. Pathol Res Pract 192: 669-675. [Crossref]
- Chen J (2016) The Cell-Cycle Arrest and Apoptotic Functions of p53 in Tumor Initiation and Progression. Cold Spring Harb Perspect Med 6: a026104. [Crossref]
- Rajan Gogna, Esha Madan, Mahmood Khan, Uttam Pati, Periannan Kuppusamy (2013) p53’s choice of myocardial death or survival: Oxygen protects infarct myocardium by recruiting p53 on NOS3 promoter through regulation of p53-Lys118 acetylation. EMBO Mol Med 5: 1662-1683. [Crossref]
- Kimata M, Matoba S, Iwai-Kanai E, Nakamura H, Hoshino A et al. (2010) p53 and TIGAR regulate cardiac myocyte energy homeostasis under hypoxic stress. Am J of Physiolo Heart Circ Physiol 299: H1908-H1916. [Crossref]
- Matsusaka H, Ide T, Matsushima S, lkeuchi M, Kubota T et al. (2016) Targeted deletion of p53 prevents cardiac rupture after myocardial infarction in mice. Cardiovasc Res 70: 457-465. [Crossref]
- Goodman JE, Hofseth LJ, Hussain SP, Harris CC (2004) Nitric oxide and p53 in cancer-prone chronic inflammation and oxyradical overload disease. Environ Mol Mutagen 44: 3-9. [Crossref]
- Wang X, Michael D, de Murcia G, Oren M (2002) p53 Activation by nitric oxide involves down-regulation of Mdm2. J Biol Chem 277: 15697-15702. [Crossref]
- Hermida N, Balligand JL (2014) Low-density lipoprotein-cholesterol-induced endothelial dysfunction and oxidative stress: the role of statins. Antioxid Redox Signal 20: 1216-1237. [Crossref]
- Konukoglu D, Uzun H (2017) Endothelial Dysfunction and Hypertension. Adv Exp Med Biol 956: 511-540. [Crossref]
- Avogaro Avogaro, Mattia Albiero, Lisa Menegazzo L, Saula de Kreutzenberg, Gian Paolo Fadini (2011) Endothelial Dysfunction in Diabetes. Diabetes Care 34: S285–S290. [Crossref]
- Golbidi S, Edvinsson L, Laher I (2018) Smoking and Endothelial Dysfunction. Curr Vasc Pharmacol. [Crossref]
- 27.Schneider A, Neas L, Herbst MC, Case M, Williams RW et al. (2008) Endothelial dysfunction: associations with exposure to ambient fine particles in diabetic individuals. Environ Health Perspect 116: 1666-1674. [Crossref]
- Ghisi GL, Durieux A, Pinho R, Benetti M (2010) Physical exercise and endothelial dysfunction. Arq Bras Cardiol 95: e130-e137. [Crossref]
- Ma WQ, Han XQ, Wang X, Wang Y, Zhu Y et al. (2016) Associations between XRCC1 Gene Polymorphisms and Coronary Artery Disease: A Meta-Analysis. PLoS One 11: e0166961. [Crossref]
- Au A, Griffiths LR, Irene L, Kooi CW, Wei LK (2017) The impact of APOA5, APOB, APOC3 and ABCA1 gene polymorphisms on ischemic stroke: Evidence from a meta-analysis. Atherosclerosis 265: 60-70. [Crossref]
- Barbosa AM, Silva KSF, Lagares MH, Rodrigues DA, da Costa et al. (2017) Atherosclerosis: analysis of the eNOS (T786C) gene polymorphism. Genet Mol Res 16. [Crossref]
- Maiorana A, O’Driscoll G, Taylor R, Green D (2003) Exercise and the nitric oxide vasodilator system. Sports Med 33: 1013-1035. [Crossref]
- Godo S, Shimokawa H (2017) Divergent roles of endothelial nitric oxide synthases system in maintaining cardiovascular homeostasis. Free Radic Biol Med 109: 4-10. [Crossref]
- Tsutoshi Shimamura, Yue Zhu, Shimin Zhang, Maeng Bong Jin, Naoki Ishizaki et al. (1999) Protective Role of Nitric Oxide in Ischemia and Reperfusion Injury of the Liver. J Am Coll Surg 188: 43-52. [Crossref]
- Napoli C, Paolisso G, Casamassimi A, AI Omran M, Barbieri M et al.(2013) Effects of nitric oxide on cell proliferation: novel insights. J Am Coll Cardiol 62: 89-95. [Crossref]
- Hemish J, Nakaya N, Mittal V, Enikolopov G (2003) Nitric oxide activates diverse signaling pathways to regulate gene expression. J Biol Chem 278: 42321-42329. [Crossref]
- Tripathi P (2007) Nitric oxide and immune response. Indian J Biochem Biophys 44: 310-319. [Crossref]
- Neves JA, Oliveira R de CM et al. (2016) Biomarkers of endothelial function in cardiovascular diseases: hypertension. Jornal Vascular Brasileiro 15: 224-233.
- Campedelli FL, E Silva KSF, Rodrigues DA, Martin JVM, Costa IR et al. (2017) Polymorphism of the gene eNOS G894T (Glu298Asp) in symptomatic patients with aterosclerosis. Genet Mol Res 16. [Crossref]
- Bhanoori M (2011) Endothelial nitric oxide synthase (eNOS) variants in cardiovascular disease: pharmacogenomic implications. Indian J Med Res 133: 464-466. [Crossref]
- Yang J, Yan R, Roy A, Xu D, Poisson J, et al. (2015) The I-TASSER Suite: protein structure and function prediction. Nat Methods 12: 7-8. [Crossref]
- Sitao Wu, Yang Zhang (2007) LOMETS: a local meta-threading-server for protein structure prediction. Nucleic Acids Res 35: 3375-3382. [Crossref]
- Swendsen null, Wang JS (1986) Replica Monte Carlo simulation of spin glasses. Phys Rev Lett 57: 2607-2609. [Crossref]
- Zhang Y, Skolnick J (2004) SPICKER: a clustering approach to identify near-native protein folds. J Comput Chem 25: 865-871. [Crossref]
- Yang J, Roy A, Zhang Y (2013) Protein-ligand binding site recognition using complementary binding-specific substructure comparison and sequence profile alignment. Bioinformatics 29: 2588-2595. [Crossref]
- Ghoorah AW, Devignes MD, Smail Tabbone M, Ritche DW (2016) Classification and Exploration of 3D Protein Domain Interactions Using Kbdock. Methods Mol Biol 1415: 91-105. [Crossref]
- Finn RD, Attwood TK, Babbitt PC, Bork P, Bridge AJ et al. (2017) InterPro in 2017-beyond protein family and domain annotations. Nucleic Acids Res 45: D190-D199. [Crossref]
- Kozakov D, Hall DR, Xia B, Porter KA, Padhorony D et al. (2017) The ClusPro web server for protein-protein docking. Nat Protoc 12: 255-278. [Crossref]
- Zhu X, Mitchell JC (2011) KFC2: a knowledge-based hot spot prediction method based on interface solvation, atomic density, and plasticity features. Proteins 79: 2671-2683. [Crossref]
- Wan X, Liu P, Jin X, Xin X, Li P et al. (2018) Electrospun PCL/keratin/AuNPs mats with the catalytic generation of nitric oxide for potential of vascular tissue engineering. J Biomed Mater Res A 106: 3239-3247. [Crossref]
- Wu D, Hu Q, Zhu D (2018) An Update on Hydrogen Sulfide and Nitric Oxide Interactions in the Cardiovascular System. Oxid Med Cell Longev 2018: 4579140. [Crossref]
- Liao Q, Huang YM, Fan W, Li C, Yang H (2016) Endothelial nitric oxide synthase deficiency influences normal cell cycle progression and apoptosis in trabecular meshwork cells. Int J Ophthalmol 9: 799-803. [Crossref]
- Kaminski A, Pohl CB, Sponholz C, Ma N, Stamm C et al. (2004) Up-regulation of endothelial nitric oxide synthase inhibits pulmonary leukocyte migration following lung ischemia-reperfusion in mice. Am J Pathol 164: 2241-2249. [Crossref]
- Santizo RA, Xu H-L, Galea E, Muyskens S, Baughman VL et al. (2002) Combined endothelial nitric oxide synthase upregulation and caveolin-1 downregulation decrease leukocyte adhesion in pial venules of ovariectomized female rats. Stroke 33: 613-616. [Crossref]
- Kader KN, Akella R, Ziats NP, Lakey LA, Harasaki H et al. (2000) eNOS-overexpressing endothelial cells inhibit platelet aggregation and smooth muscle cell proliferation in vitro. Tissue Eng 6: 241-251. [Crossref]
- Lim K-H, Ancrile BB, Kashatus DF, Counter CM (2008) Tumour maintenance is mediated by eNOS. Nature 452: 646-649. [Crossref]
- Yakovlev VA (2018) Nitric Oxide: Genomic Instability And Synthetic Lethality. Redox Biol 5: 414. [Crossref]
- Mobaraki RN, Karimi M, Alikarami F, Farhari E, Amini et al. RITA induces apoptosis in p53-null K562 leukemia cells by inhibiting STAT5, Akt, and NF-κB signaling pathways. Anticancer Drugs 29: 847-853. [Crossref]
- Santos TR, Silva KSF e, Silva RCPC (2018) Infertility caused by an association between Arg72Pro polymorphism of the p53 gene and Glu298Asp of the eNOS gene in patients with endometriosis. Genetics and Molecular Research 17.
- Lagares MH, Silva KSF, Barbosa AM, Rodrigues DA, Costa IR, et al. (2017) Analysis of p53 gene polymorphism (codon 72) in symptomatic patients with atherosclerosis. Genet Mol Res 16. [Crossref]
- Silva KS, Moura KK (2016) Genetic polymorphisms in patients with endometriosis: an analytical study in Goiânia (Central West of Brazil). Genet Mol Res 15. [Crossref]
- Staal FJ, van der Burg M, Wessels LF, Barendregt BH, Baert MR et al. (2003) DNA microarrays for comparison of gene expression profiles between diagnosis and relapse in precursor-B acute lymphoblastic leukemia: choice of technique and purification influence the identification of potential diagnostic markers. Leukemia 17: 1324-1332. [Crossref]
- Silva K (2018) Hot spots and single nucleotide polymorphisms on the interaction interface of RAD51 and p53 complex. J Tre Bio Res 1: 1-5.
- Tannous I, Santos T, de Curcio J (2018) Involvement of Protein-Protein Interactions of eNOS and Genetic Polymorphisms in Coronary Artery Disease. Int J Clin Cardiol Res 2: 067-071.
- de Curcio JS, Lima RM, Oliveira, LN (2018) Structure-based design of TFF3-PAR2 inhibitor peptides as a promising new therapeutic approach for endometriosis patients. M J Gyne 3: 06.
- Achary MS, Reddy AB, Chakrabarti S, Panicker SG, Mandal Ak et al. (2006) Disease-causing mutations in proteins: structural analysis of the CYP1b1 mutations causing primary congenital glaucoma in humans. Biophys J 91 4329-4339. [Crossref]
- Norel R, Sheinerman F, Petrey D, Honig B (2001) Electrostatic contributions to protein–protein interactions: fast energetic filters for docking and their physical basis. Protein Sci 10: 2147-2161. [Crossref]
- Zhang Z, Shawn Witham, Emil Alexov (2011) On the role of electrostatics on protein-protein interactions. Phys Biol 8: 035001. [Crossref]
- Li L, Jia Z, Peng Y, Godar S, Getov I et al. (2017) Forces and Disease: Electrostatic force differences caused by mutations in kinesin motor domains can distinguish between disease-causing and non-disease-causing mutations. Sci Rep 7: 8237. [Crossref]
- Li Y, Huang Y, Swaminathan CP, Smith-Gill SJ, Mariuzza RA (2005) Magnitude of the Hydrophobic Effect at Central versus Peripheral Sites in Protein-Protein Interfaces. Structure 13: 297-307. [Crossref]
- Dong JY, Qin LQ, Zhang Z, Zhao Z,Wang J et al. (2011) Effect of oral L-arginine supplementation on blood pressure: a meta-analysis of randomized, double-blind, placebo-controlled trials. Am Heart J 162: 959-965. [Crossref]
- Liao SM, Du QS, Meng JZ, Pang ZW, Huang RB (2013) The multiple roles of histidine in protein interactions. Chem Cent J 7: 44. [Crossref]
- Cauët E, Rooman M, Wintjens R, Liévin J, Biot C (2005) Histidine-Aromatic Interactions in Proteins and Protein-Ligand Complexes: Quantum Chemical Study of X-ray and Model Structures. J Chem Theory Comput 1: 472-483. [Crossref]
- Feng J, Hu Y, Yi H, Han S, Hu J et al. (2013) Two Conserved Arginine Residues from the SK3 Potassium Channel outer vestibule control selectivity of recognition by scorpion toxins. J Biol Chem 288: 12544-12553. [Crossref]
- Tesei G, Vazdar M, Jensen MR (2017) Self-association of a highly charged arginine-rich cell-penetrating peptide. PNAS 114: 11428-11433.
- Rhys NH, Soper AK, Dougan L (2012) The hydrogen-bonding ability of the amino acid glutamine revealed by neutron diffraction experiments. J Phys Chem B 116: 13308-13319. [Crossref]
- Law MJ, Linde ME, Chambers EJ, Oubrigr C, Katsamba, et al. (2006) The role of positively charged amino acids and electrostatic interactions in the complex of U1A protein and U1 hairpin II RNA. Nucleic Acids Res 34: 275-285. [Crossref]
- de Jesus AJ, Allen TW (2013) The role of tryptophan side chains in membrane protein anchoring and hydrophobic mismatch. Biochimica et Biophysica Acta 1828: 864-876. [Crossref]
- Samanta U, Chakrabarti P (2001) Assessing the role of tryptophan residues in the binding site. Protein Eng Des Sel 14: 7-15. [Crossref]
- Zheng P, Cao Y, Bu T, Straus SK, Li H (2011) Single molecule force spectroscopy reveals that electrostatic interactions affect the mechanical stability of proteins. Biophys J 100: 1534-1541. [Crossref]
- Zhu J, Luther PW, Leng Q, Mixson AJ (2006) Synthetic histidine-rich peptides inhibit Candida species and other fungi in vitro: role of endocytosis and treatment implications. Antimicrob Agents Chemother 50: 2797-2805. [Crossref]
- Dill KA (1990) Dominant forces in protein folding. Biochemistry 29: 7133-7155.
- Chaturvedi D, Mahalakshmi R (2014) Juxtamembrane tryptophans have distinct roles in defining the OmpX barrel-micelle boundary and facilitating protein-micelle association. FEBS Lett 588: 4464-4471. [Crossref]
- Gupta A, Zadafiya P, Mahalakshmi R (2014) Differential contribution of tryptophans to the folding and stability of the attachment invasion locus transmembrane β-barrel from Yersinia pestis. Sci Rep 4: 6508.
- Pace CN, Fu H, Fryar KL, Landua J, Trevino SR et al. (2011) Contribution of hydrophobic interactions to protein stability. J Mol Biol 408: 514-528. [Crossref]
- Yan C, Wu F, Jernigan RL, Dobbs D, Honavar V (2008) Characterization of protein-protein interfaces. Protein J 27: 59-70. [Crossref]
- C M Roth, B L Neal, A M Lenhoff (1996) Van der Waals interactions involving proteins. Biophys J 70: 977-987. [Crossref]
- Roy S, Das TK (2015) Study of Interaction Between Tryptophan, Tyrosine, and Phenylalanine Separately with Silver Nanoparticles by Fluorescence Quenching Method. J Appl Spectrosc 82: 598-606.
- Word JM, Lovell SC, Richardson JS, Richardson DC (1999) Asparagine and glutamine: using hydrogen atom contacts in the choice of side-chain amide orientation. J Mol Biol 285: 1735-1747. [Crossref]
- Yokota A, Tsumoto K, Shiroishi M, Nakanishi T, Kondo H et al. (2010) Contribution of asparagine residues to the stabilization of a proteinaceous antigen-antibody complex, HyHEL-10-hen egg white lysozyme. J Biol Chem 285: 7686-7696. [Crossref]
- Yusufaly TI, Li Y, Singh G, Olson WK (2014) Arginine-phosphate salt bridges between histones and DNA: intermolecular actuators that control nucleosome architecture. J Chem Phys 141: 165102. [Crossref]
- Zhang Z, Liu L, Gomez-Casal R, Wang X, Hayashi R et al. (2016) Targeting cancer stem cells with p53 modulators. Oncotarget 7: 45079-45093. [Crossref]
- Karina Krotova, Hanbo Hu, Shen Ling Xia, Leonid Belayev, Jawaharlal M Patel et al (2006) Peptides modified by myristoylation activate eNOS in endothelial cells through Akt phosphorylation. Br J Pharmacol. 148: 732-740. [Crossref]
