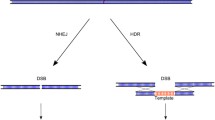
Abstract
One problem in the study of regulatory mechanisms in living systems is the difficulty in analysis of regulatory elements in their natural context. One promising way to solve this problem is direct in situ modification of regulatory element sequences. The new technology of gene modification based on the bacterial CRISPR/Cas system allows one to quickly and accurately modify any genomic fragment, in particular, in cells of an adult organism. This review considers principles of the CRISPR/Cas technology and its application to the study of regulatory elements, as well as the potential of using this technology in studies of regulatory systems in pancreas tumors.
Similar content being viewed by others
References
Birney, E., The making of ENCODE: Lessons for bigdata projects, Nature, 2012, vol. 489, no. 7414, pp. 49–51.
Chung, I.–M., Ketharnathan, S., Kim, S.–H., et al., Making sense of the tangle: Insights into chromatin folding and gene regulation, Genes, 2016, vol. 7, no. 10, p. 71.
Visscher, P.M., Brown, M.A., McCarthy, M.I., and Yang, J., Five years of GWAS discovery, Am. J. Hum. Genet., 2012, vol. 90, no. 1, pp. 7–24.
Seruggia, D., Fernandez, A., Cantero, M., et al., Functional validation of mouse tyrosinase non–coding regulatory DNA elements by CRISPR–Cas9–mediated mutagenesis, Nucleic Acids Res., 2015, vol. 43, no. 10, pp. 4855–4867.
Andrey, G., Montavon, T., Mascrez, B., et al., A switch between topological domains underlies HoxD genes collinearity in mouse limbs, Science, 2013, vol. 340, no. 6137, p. 1234167.
Reid, L.H., Shesely, E.G., Kim, H.S., and Smithies, O., Cotransformation and gene targeting in mouse embryonic stem cells, Mol. Cell. Biol., 1991, vol. 11, no. 5, pp. 2769–2777.
Sagai, T., Hosoya, M., Mizushina, Y., et al., Elimination of a long–range cis–regulatory module causes complete loss of limb–specific Shh expression and truncation of the mouse limb, Development, 2005, vol. 132, no. 4, pp. 797–803.
Ishii, A., Kurosawa, A., Saito, S., and Adachi, N., Analysis of the role of homology arms in gene–targeting vectors in human cells, PLoS One, 2014, vol. 9, no. 9, p. e108236.
Hsu, P.D., Lander, E.S., and Zhang, F., Development and applications of CRISPR–Cas9 for genome engineering, Cell, 2014, vol. 157, no. 6, pp. 1262–1278.
Barrangou, R., Fremaux, C., Deveau, H., et al., CRISPR provides acquired resistance against viruses in prokaryotes, Science, 2007, vol. 315, no. 5819, pp. 1709–1712.
Marraffini, L.A. and Sontheimer, E.J., CRISPR interference: RNA–directed adaptive immunity in bacteria and archaea, Nat. Rev. Genet., 2010, vol. 11, no. 3, pp. 181–190.
Jansen, R., Embden, J.D., Gaastra, W., and Schouls, L.M., Identification of genes that are associated with DNA repeats in prokaryotes, Mol. Microbiol., 2002, vol. 43, no. 6, pp. 1565–1575.
Lander, E.S., The Heroes of CRISPR, Cell, 2016, vol. 164, nos. 1–2, pp. 18–28.
Pennisi, E., The CRISPR craze, Science, 2013, vol. 341, no. 6148, pp. 833–836.
Jinek, M., Chylinski, K., Fonfara, I., et al., A programmable dual–RNA–guided DNA endonuclease in adaptive bacterial immunity, Science, 2012, vol. 337, no. 6096, pp. 816–821.
Jiang, F., Taylor, D.W., Chen, J.S., et al., Structures of a CRISPR–Cas9 R–loop complex primed for DNA cleavage, Science, 2016, vol. 351, no. 6275, pp. 867–871.
Kleinstiver, B.P., Prew, M.S., Tsai, S.Q., et al., Engineered CRISPR–Cas9 nucleases with altered PAM specificities, Nature, 2015, vol. 523, no. 7561, pp. 481–485.
Ran, F.A., Cong, L., Yan, W.X., et al., In vivo genome editing using Staphylococcus aureus Cas9, Nature, 2015, vol. 520, no. 7546, pp. 186–191.
Mali, P., Yang, L., Esvelt, K.M., et al., RNA–guided human genome engineering via Cas9, Science, 2013, vol. 339, no. 6121, pp. 823–826.
Kim, H. and Kim, J.S., A guide to genome engineering with programmable nucleases, Nat. Rev. Genet., 2014, vol. 15, no. 5, pp. 321–334.
Sanchez–Rivera, F.J. and Jacks, T., Applications of the CRISPR–Cas9 system in cancer biology, Nat. Rev. Cancer, 2015, vol. 15, no. 7, pp. 387–395.
Xiao, A., Wang, Z., Hu, Y., et al., Chromosomal deletions and inversions mediated by TALENs and CRISPR/Cas in zebrafish, Nucleic Acids Res., 2013, vol. 41, no. 14, p. e141.
Choi, P.S. and Meyerson, M., Targeted genomic rearrangements using CRISPR/Cas technology, Nat. Commun., 2014, vol. 5, p. 3728.
Wakabayashi, A., Ulirsch, J.C., Ludwig, L.S., et al., Insight into GATA1 transcriptional activity through interrogation of cis elements disrupted in human erythroid disorders, Proc. Natl. Acad. Sci. U. S. A., 2016, vol. 113, no. 16, pp. 4434–4439.
Laursen, K.B., Kashyap, V., Scandura, J., and Gudas, L.J., An alternative retinoic acid–responsive Stra6 promoter regulated in response to retinol deficiency, J. Biol. Chem., 2015, vol. 290, no. 7, pp. 4356–4266.
Li, Y., Rivera, C.M., Ishii, H., et al., CRISPR reveals a distal super–enhancer required for Sox2 expression in mouse embryonic stem cells, PLoS One, 2014, vol. 9, no. 12, p. e114485.
Didych, D.A., Tyulkina, D.V., Pleshkan, V.V., Alekseenko, I.V., and Sverdlov, E.D., Are super–enhancers regulators of regulatory genes of development and cancer?, Mol. Biol. (Moscow), 2015, vol. 49, no. 6, pp. 818–824.
Hsu, P.Y., Hsu, H.K., Hsiao, T.H., et al., Spatiotemporal control of estrogen–responsive transcription in ERalpha–positive breast cancer cells, Oncogene, 2016, vol. 35, no. 18, pp. 2379–2389.
Meyer, M.B., Benkusky, N.A., and Pike, J.W., Selective distal enhancer control of the Mmp13 gene identified through clustered regularly interspaced short palindromic repeat (CRISPR) genomic deletions, J. Biol. Chem., 2015, vol. 290, no. 17, pp. 11093–11107.
Groschel, S., Sanders, M.A., Hoogenboezem, R., et al., A single oncogenic enhancer rearrangement causes concomitant EVI1 and GATA2 deregulation in leukemia, Cell, 2014, vol. 157, no. 2, pp. 369–381.
Flavahan, W.A., Drier, Y., Liau, B.B., et al., Insulator dysfunction and oncogene activation in IDH mutant gliomas, Nature, 2016, vol. 529, no. 7584, pp. 110–114.
Sanborn, A.L., Rao, S.S., Huang, S.C., et al., Chromatin extrusion explains key features of loop and domain formation in wild–type and engineered genomes, Proc. Natl. Acad. Sci. U. S. A., 2015, vol. 112, no. 47, pp. E6456–E6465.
Guo, Y., Xu, Q., Canzio, D., et al., CRISPR inversion of CTCF sites alters genome topology and enhancer/promoter function, Cell, 2015, vol. 162, no. 4, pp. 900–910.
Canver, M.C., Smith, E.C., Sher, F., et al., BCL11A enhancer dissection by Cas9–mediated in situ saturating mutagenesis, Nature, 2015, vol. 527, no. 7577, pp. 192–197.
Diao, Y., Li, B., Meng, Z., et al., A new class of temporarily phenotypic enhancers identified by CRISPR/Cas9–mediated genetic screening, Genome Res., 2016, vol. 26, no. 3, pp. 397–405.
Korkmaz, G., Lopes, R., Ugalde, A.P., et al., Functional genetic screens for enhancer elements in the human genome using CRISPR–Cas9, Nat. Biotechnol., 2016, vol. 34, no. 2, pp. 192–198.
Zuckermann, M., Kawauchi, D., and Gronych, J., Applications of the CRISPR/Cas9 system in murine cancer modeling, Briefings Funct. Genomics, 2017, vol. 16, no. 1, pp. 25–33.
Platt, R.J., Chen, S., Zhou, Y., et al., CRISPR–Cas9 knockin mice for genome editing and cancer modeling, Cell, 2014, vol. 159, no. 2, pp. 440–455.
Sanchez–Rivera, F.J., Papagiannakopoulos, T., Romero, R., et al., Rapid modelling of cooperating genetic events in cancer through somatic genome editing, Nature, 2014, vol. 516, no. 7531, pp. 428–431.
Snyder, E.L., Watanabe, H., Magendantz, M., et al., Nkx2–1 represses a latent gastric differentiation program in lung adenocarcinoma, Mol. Cell, 2013, vol. 50, no. 2, pp. 185–199.
Maddalo, D., Manchado, E., Concepcion, C.P., et al., In vivo engineering of oncogenic chromosomal rearrangements with the CRISPR/Cas9 system, Nature, 2014, vol. 516, no. 7531, pp. 423–427.
Blasco, R.B., Karaca, E., Ambrogio, C., et al., Simple and rapid in vivo generation of chromosomal rearrangements using CRISPR/Cas9 technology, Cell Rep., 2014, vol. 9, no. 4, pp. 1219–1227.
Xue, W., Chen, S., Yin, H., et al., CRISPR–mediated direct mutation of cancer genes in the mouse liver, Nature, 2014, vol. 514, no. 7522, pp. 380–384.
Chiou, S.H., Winters, I.P., Wang, J., et al., Pancreatic cancer modeling using retrograde viral vector delivery and in vivo CRIS PR/Cas9–mediated somatic genome editing, Genes Dev., 2015, vol. 29, no. 14, pp. 1576–1585.
Mazur, P.K., Herner, A., Mello, S.S., et al., Combined inhibition of BET family proteins and histone deacetylases as a potential epigenetics–based therapy for pancreatic ductal adenocarcinoma, Nat. Med., 2015, vol. 21, no. 10, pp. 1163–1171.
Zuckermann, M., Hovestadt, V., Knobbe–Thomsen, C.B., et al., Somatic CRISPR/Cas9–mediated tumour suppressor disruption enables versatile brain tumour modelling, Nat. Commun., 2015, vol. 6, p. 7391.
Malina, A., Mills, J.R., Cencic, R., et al., Repurposing CRISPR/Cas9 for in situ functional assays, Genes Dev., 2013, vol. 27, no. 23, pp. 2602–2614.
Khurana, E., Fu, Y., Chakravarty, D., et al., Role of non–coding sequence variants in cancer, Nat. Rev. Genet., 2016, vol. 17, no. 2, pp. 93–108.
Hanahan, D. and Weinberg, R.A., Hallmarks of cancer: The next generation, Cell, 2011, vol. 144, no. 5, pp. 646–674.
Morton, J.P., Jamieson, N.B., Karim, S.A., et al., LKB1 haploinsufficiency cooperates with Kras to promote pancreatic cancer through suppression of p21–dependent growth arrest, Gastroenterology, 2010, vol. 139, no. 2, pp. 586–597.e6.
Vorvis, C., Hatziapostolou, M., Mahurkar–Joshi, S., et al., Transcriptomic and CRISPR/Cas9 technologies reveal FOXA2 as a tumor suppressor gene in pancreatic cancer, Am. J. Physiol.: Gastrointest. Liver Physiol., 2016, vol. 310, no. 11, pp. G1124–G1137.
Diaferia, G.R., Balestrieri, C., Prosperini, E., et al., Dissection of transcriptional and cis–regulatory control of differentiation in human pancreatic cancer, EMBO J., 2016, vol. 35, no. 6, pp. 595–617.
Author information
Authors and Affiliations
Corresponding author
Additional information
Original Russian Text © A.I. Kuzmich, M.V. Zinovyeva, V.K. Potapov, M.B. Kostina, E.D. Sverdlov, 2018, published in Molekulyarnaya Genetika, Mikrobiologiya i Virusologiya, 2018, No. 1, pp. 3–8.
About this article
Cite this article
Kuzmich, A.I., Zinovyeva, M.V., Potapov, V.K. et al. CRISPR/CAS Targeted in vivo Genome Modification for Studying the Functional Role of Genomic Regulatory Elements in Health and Carcinogenesis. Mol. Genet. Microbiol. Virol. 33, 1–7 (2018). https://doi.org/10.3103/S0891416818010081
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.3103/S0891416818010081