Abstract
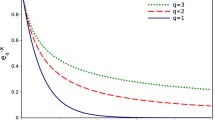
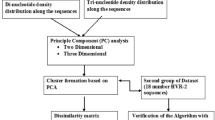
This paper makes two generalizations of the previously presented algorithms of the author based on the principles of information coding in molecular genetics. This is an account of the frequency characteristics of subalphabetic representations of polynucleotides and a generalization of an algorithm for processing arbitrary information presented in a quaternary code. The second generalization indicates the general significance of the proposed algorithms, which the author called molecular genetic or DNA algorithms, emphasizing their difference from the well-known genetic algorithms of the Holland type. An example of displaying the results of the operation of DNA algorithms in the frequency domain with visualization of the cluster structure is given. The example makes it possible to trace a structure that is quite common for DNA, which consists of a main cluster and several satellite clusters. Natural language texts processed by DNA algorithms in the structural and frequency domains are analyzed and compared.



Similar content being viewed by others
REFERENCES
Shmulevich, I. and Dougherty, E.R., Genomic Signal Processing, Princeton: Princeton Univ. Press, 2014.
Weighill, D., et al., Wavelet-based genomic signal processing for centromere identification and hypothesis generation, Front. Genet., 2019, vol. 10, p. 487.
Petoukhov, S.V., Genetic coding and united-hypercomplex systems in the models of algebraic biology, Biosystems, 2017, vol. 158, pp. 31–46.
Petoukhov, S.V. and He, M., Symmetrical Analysis Techniques for Genetic Systems and Bioinformatics: Advanced Patterns and Applications, Hershey: IGI Global, 2010.
Petukhov, S.V., Hypercomplex numbers and the algebraic system of genetic alphabets, Hypercomplex Numbers Geom. Phys., 2011, vol. 8, no. 2, pp. 118–138.
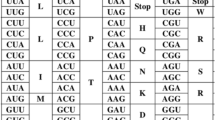
Rumer, Yu.B., Systematization of codons in the genetic code, Rep. Akad. Nauk SSSR, 1968, vol. 183, no. 1, pp. 225–226.
Petoukhov, S.V., Symmetries of the genetic code, Walsh functions and the theory of genetic logical holography, Symmetry Cult. Sci., 2016, vol. 27, pp. 95–98.
Stepanyan, I.V. and Petoukhov, S.V., The matrix method of representation, analysis and classification of long genetic sequences, Information, 2017, vol. 8, no. 1, p. 12.
Stepanyan, I.V., A multiscale model of nucleic acid imaging, Sci. Visualization, 2020, vol. 12, no. 3, pp. 61–78. https://doi.org/10.26583/sv.12.3.06
Stepanyan, I.V., A biomathematical system of methods for describing nucleic acids, Komp’yut. Issled. Model., 2020, vol. 12, no. 2, pp. 417–434.
Zebari, D.A., et al., Multi-level of DNA encryption technique based on DNA arithmetic and biological operations, 2018 International Conference on Advanced Science and Engineering (ICOASE), Duhok: IEEE, 2018, pp. 312–317.
Mendizabal-Ruiz, G., et al., Genomic signal processing for DNA sequence clustering, PeerJ, 2018, vol. 6, p. E4264.
Souza, C.P.E., et al., Epiclomal: Probabilistic clustering of sparse single-cell DNA methylation data, PLoS Comput. Biol., 2020, vol. 16, no. 9, artic. no. e1008270.https://doi.org/10.1371/journal.pcbi.1008270
Truong, H.Q., Ngo, L.T., and Pedrycz, W., Granular fuzzy possibilistic C-means clustering approach to DNA microarray problem, Knowl.-Based Syst., 2017, vol. 133, pp. 53–65.
Scott, T.C., Therani, M., and Wang, X., Data clustering with quantum mechanics, Mathematics, 2017, vol. 5, no. 1, pp. 1–17. https://doi.org/10.3390/math5010005
Huang, D., Wang, C.D., Wu, J.S., Lai, J.H., and Kwoh, C.K., Ultra-scalable spectral clustering and ensemble clustering, IEEE Trans. Knowl. Data Eng., 2019, vol. 32, no. 6, pp. 1212–1226.
Janani, R. and Vijayarani, S., Text document clustering using spectral clustering algorithm with particle swarm optimization, Expert Syst. Appl., 2019, vol. 134, pp. 192–200.
Wang, X., et al., Fast detection and segmentation of partial image blur based on discrete Walsh–Hadamard transform, Signal Process.: Image Commun., 2019, vol. 70, pp. 47–56.
Bonny, T. and Haq, A., Emulation of high-performance correlation-based quantum clustering algorithm for two-dimensional data on FPGA, Quantum Inf. Process., 2020, vol. 19, no. 6, p. 179. https://doi.org/10.1007/s11128-020-02683-9
Hinojosa, C., et al., Single-pixel camera sensing matrix design for hierarchical compressed spectral clustering, 2019 IEEE 29th International Workshop on Machine Learning for Signal Processing (MLSP), Pittsburgh: IEEE, 2019, pp. 1–6.
Petukhov, S.V., The concept of resonances in genetics, Biomashsistemy, 2018, vol. 2, no. 4, pp. 169–221.
Chargaff, E., Lipshitz, R., and Green, C., Composition of the deoxypentose nucleic acids of four genera of sea-urchin, J. Biol. Chem., 1952, vol. 195, no. 1, pp. 155–160.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The authors declare that they have no conflicts of interest.
Additional information
Translated by K. Lazarev
About this article
Cite this article
Stepanyan, I.V. DNA Clustering Algorithms. Autom. Doc. Math. Linguist. 55, 1–7 (2021). https://doi.org/10.3103/S0005105521010039
Received:
Published:
Issue Date:
DOI: https://doi.org/10.3103/S0005105521010039