Whole Exome Sequence of Pakistani Acute Lymphocytic Leukemia Patient from Pakhtuns Ancestry Reveal the Novel Genetic Variant Characterization in the GLDC Gene
Article Information
Shahid Ullah1, Alex Tonks2, Maryam A Halawi3, Alruwaili A M4, Alkuwaykibi M S5, Azhar A Halawi6, Amir Hayat1, Hedib Alkoumi H Alrawili7, Maryam Alanazi8, Abdul Wadood1, Asifullah Khan1, Muhammad Arif Lodhi1,*
1Department of Biochemistry Abdul Wali Khan University Mardan-23200, KP, Pakistan
2Department of Hematology, Division of Cancer & Genetics, School of Medicine Cardiff University UK
3Clinical pharmacy, college of Pharmacy, Jazan University, Jazan Saudi Arabia; Department of Hematology, Division of Cancer & Genetics, School of Medicine Cardiff University UK
4Medical Laboratory Technology Department, College of Applied Medical Sciences, Northern Border University, Arar 91431, Saudi Arabia
5General Medicine and Surgery, College of Medicine, Northern Border University, Arar 91431, Saudi Arabia
6Family Medicine Specialist, Ahad Almasreha Hospital, Jazan, Saudi Arabia
7Wolfson College, School of Medicine, Oxford University, England UK
8Department of Clinical Laboratory Sciences, College of Applied Medical Science, Shaqra University, Shaqra 15572, Saudi Arabia
*Corresponding author: Muhammad Arif Lodhi, Department of Biochemistry Abdul Wali Khan University Mardan-23200, KP, Pakistan.
Received: 30 August 2023; Accepted: 06 September 2023; Published: 29 September 2023
Citation: Shahid Ullah, Alex Tonks, Maryam A Halawi, Alruwaili A M, Alkuwaykibi M S, Azhar A Halawi, Amir Hayat, Hedib Alkoumi H Alrawili, Maryam Alanazi, Abdul wadood, Asifullah Khan, Muhammad Arif Lodhi. Whole Exome Sequence of Pakistani Acute Lymphocytic Leukemia Patient from Pakhtun Ancestry Reveal the Novel Genetic Variant Characterization in the GLDC Gene. Journal of Biotechnology and Biomedicine. 6 (2023): 409-420.
View / Download Pdf Share at FacebookAbstract
Background: Acute Lymphoblastic Leukemia (ALL) is the most common malignant disease in children and often involves numerical chromosomal abnormalities, fusion genes, or minor localized deletions that are significant in the development of leukemia. Glycine Decarboxylase (GLDC) gene overexpression and mutation is associated with oncogenic activity in various cancers. However, the pathophysiological roles and structural consequences of GLDC in acute lymphocytic leukemia have not been investigated.
Objective: We aimed to identify novel variant in acute lymphocytic leukemia through whole exome sequencing. Methods: This study employs whole exome sequencing to examine seven pediatric patients with Acute Lymphoblastic Leukemia (ALL) in Pakistan. The patients under investigation are of Pakistani origin. The deleterious effect was predicted by SIFT, PolyPhen2, CADD, FATHMM, HOPE, and Mutation Assessors. Structure stability assessment was performed using the I-Mutant-3.0server. The atomic structure of the Single Nucleotide Polymorphism (SNP) was analyzed utilizing the Molecular Dynamics (MD) with WEBGRO server.
Results: The present study identified a novel pathogenic heterozygous variant NM_000170.2:p.Ser551Cys/c.1651A>T in GLDC gene of early stage diagnose ALL patient the variant was not present in the dbSNP & 1000Genome Project databases. Structural instability, disrupted function, and altered 3D structure were observed in the mutant GLDC protein model compared to the wild-type structure.
Conclusion: The novel SNP was found in a highly conserved region of the GLDC protein and is predicted to be a high-risk candidate for leukemia. This variant greatly affects the stability of the protein.
Keywords
SNP; Glycine Decarboxylase; Acute lymphocytic leukemia; Gene–gene interaction
SNP articles SNP Research articles SNP review articles SNP PubMed articles SNP PubMed Central articles SNP 2023 articles SNP 2024 articles SNP Scopus articles SNP impact factor journals SNP Scopus journals SNP PubMed journals SNP medical journals SNP free journals SNP best journals SNP top journals SNP free medical journals SNP famous journals SNP Google Scholar indexed journals Glycine Decarboxylase articles Glycine Decarboxylase Research articles Glycine Decarboxylase review articles Glycine Decarboxylase PubMed articles Glycine Decarboxylase PubMed Central articles Glycine Decarboxylase 2023 articles Glycine Decarboxylase 2024 articles Glycine Decarboxylase Scopus articles Glycine Decarboxylase impact factor journals Glycine Decarboxylase Scopus journals Glycine Decarboxylase PubMed journals Glycine Decarboxylase medical journals Glycine Decarboxylase free journals Glycine Decarboxylase best journals Glycine Decarboxylase top journals Glycine Decarboxylase free medical journals Glycine Decarboxylase famous journals Glycine Decarboxylase Google Scholar indexed journals Acute lymphocytic leukemia articles Acute lymphocytic leukemia Research articles Acute lymphocytic leukemia review articles Acute lymphocytic leukemia PubMed articles Acute lymphocytic leukemia PubMed Central articles Acute lymphocytic leukemia 2023 articles Acute lymphocytic leukemia 2024 articles Acute lymphocytic leukemia Scopus articles Acute lymphocytic leukemia impact factor journals Acute lymphocytic leukemia Scopus journals Acute lymphocytic leukemia PubMed journals Acute lymphocytic leukemia medical journals Acute lymphocytic leukemia free journals Acute lymphocytic leukemia best journals Acute lymphocytic leukemia top journals Acute lymphocytic leukemia free medical journals Acute lymphocytic leukemia famous journals Acute lymphocytic leukemia Google Scholar indexed journals Gene?gene interaction articles Gene?gene interaction Research articles Gene?gene interaction review articles Gene?gene interaction PubMed articles Gene?gene interaction PubMed Central articles Gene?gene interaction 2023 articles Gene?gene interaction 2024 articles Gene?gene interaction Scopus articles Gene?gene interaction impact factor journals Gene?gene interaction Scopus journals Gene?gene interaction PubMed journals Gene?gene interaction medical journals Gene?gene interaction free journals Gene?gene interaction best journals Gene?gene interaction top journals Gene?gene interaction free medical journals Gene?gene interaction famous journals Gene?gene interaction Google Scholar indexed journals childhood cancer articles childhood cancer Research articles childhood cancer review articles childhood cancer PubMed articles childhood cancer PubMed Central articles childhood cancer 2023 articles childhood cancer 2024 articles childhood cancer Scopus articles childhood cancer impact factor journals childhood cancer Scopus journals childhood cancer PubMed journals childhood cancer medical journals childhood cancer free journals childhood cancer best journals childhood cancer top journals childhood cancer free medical journals childhood cancer famous journals childhood cancer Google Scholar indexed journals lymphoid progenitor cells articles lymphoid progenitor cells Research articles lymphoid progenitor cells review articles lymphoid progenitor cells PubMed articles lymphoid progenitor cells PubMed Central articles lymphoid progenitor cells 2023 articles lymphoid progenitor cells 2024 articles lymphoid progenitor cells Scopus articles lymphoid progenitor cells impact factor journals lymphoid progenitor cells Scopus journals lymphoid progenitor cells PubMed journals lymphoid progenitor cells medical journals lymphoid progenitor cells free journals lymphoid progenitor cells best journals lymphoid progenitor cells top journals lymphoid progenitor cells free medical journals lymphoid progenitor cells famous journals lymphoid progenitor cells Google Scholar indexed journals glycine-serine articles glycine-serine Research articles glycine-serine review articles glycine-serine PubMed articles glycine-serine PubMed Central articles glycine-serine 2023 articles glycine-serine 2024 articles glycine-serine Scopus articles glycine-serine impact factor journals glycine-serine Scopus journals glycine-serine PubMed journals glycine-serine medical journals glycine-serine free journals glycine-serine best journals glycine-serine top journals glycine-serine free medical journals glycine-serine famous journals glycine-serine Google Scholar indexed journals carcinogenic articles carcinogenic Research articles carcinogenic review articles carcinogenic PubMed articles carcinogenic PubMed Central articles carcinogenic 2023 articles carcinogenic 2024 articles carcinogenic Scopus articles carcinogenic impact factor journals carcinogenic Scopus journals carcinogenic PubMed journals carcinogenic medical journals carcinogenic free journals carcinogenic best journals carcinogenic top journals carcinogenic free medical journals carcinogenic famous journals carcinogenic Google Scholar indexed journals untransformed articles untransformed Research articles untransformed review articles untransformed PubMed articles untransformed PubMed Central articles untransformed 2023 articles untransformed 2024 articles untransformed Scopus articles untransformed impact factor journals untransformed Scopus journals untransformed PubMed journals untransformed medical journals untransformed free journals untransformed best journals untransformed top journals untransformed free medical journals untransformed famous journals untransformed Google Scholar indexed journals pediatric patients articles pediatric patients Research articles pediatric patients review articles pediatric patients PubMed articles pediatric patients PubMed Central articles pediatric patients 2023 articles pediatric patients 2024 articles pediatric patients Scopus articles pediatric patients impact factor journals pediatric patients Scopus journals pediatric patients PubMed journals pediatric patients medical journals pediatric patients free journals pediatric patients best journals pediatric patients top journals pediatric patients free medical journals pediatric patients famous journals pediatric patients Google Scholar indexed journals
Article Details
Introduction
Acute lymphoblastic leukaemia (ALL) is the most prevalent form of childhood cancer and the leading factor in pediatrics having a high mortality rate [1]. It is characterized by the uncontrolled proliferation of lymphoid progenitor cells, which are immature white blood cells that would normally develop into lymphocytes. In ALL, these cells do not mature properly and accumulate in the bone marrow, blood, and other tissues, leading to a range of symptoms and complications [2]. B-ALL of the B-cell precursor is a lymphoid progenitor cell cancer that exhibits significant biology and clinical variability [3]. Adults are more prone than children to develop high-risk B-ALL disease, and despite aggressive chemotherapy and/or allogeneic stem cell transplantation therapies, adult long-term disease-free survival rates are only 40%. This stands in stark contrast to pediatric ALL, where advanced treatment protocols have led to cure rates that are close to 80% [4]. Nevertheless, despite this success, some children with ALL have a dismal prognosis; 15% of them die from ALL relapses [5]. Unfortunately, Pakistan has one of the highest infant mortality rates in the world, at 71% [6]. The aggressive proliferation and invasion of tumor cells are assisted by aberrant glycine metabolism, an emerging hallmark of cancer [7]. The GLDC is the enzyme that catabolizes glycine to supply one-carbon metabolism in mitochondria, GLDC is the rate-limiting enzyme in the glycine cleavage system. According to a recent study, it aids in the proliferation and pyrimidine production of tumor-initiating cells, which is critical in the development of tumors [8]. The GLDC oxidoreductase plays a crucial role in the metabolism of amino acids [9]. GLDC overexpression encourages cell proliferation and their conversion to cancer cells through increased glycine-serine metabolism and nucleotide synthesis. Many cancer patients are affected by the carcinogenic effects of abnormal GLDC overexpression, and high GLDC expression levels are linked to greater mortality and worse survival rates in hepatocellular carcinoma (HCC) [10,11] glioma and non-small-cell lung carcinoma (NSCLC) and several other cancer patients (12- 13). Cell viability is unaffected by GLDC expression knockdown in untransformed cells, GLDC is a target that may have a broad therapeutic index, indicating the therapeutic significance. The demographics and prognosis of children with ALL in Pakistan are poorly understood. The present study presents a Novel variant of the GLDC gene found in a Pakistani Pakhtun ALL patient that has a great impact on the protein structure stability and function.
Materials and Methods
The Study ethical Approval
Approval NO. Dir A&R AWKUM 2020/5118 was received for the current study from Abdul Wali Khan University's Institute Review Board (IRB), located in Mardan, KPK, Pakistan.
ALL diagnosis and sample collection
Seven blood samples were collected from pediatric patients with acute lymphocytic leukemia (ALL) within the age range of 2 to 20 years. The samples were obtained from patients who received treatment at the Oncology and Hematology Clinic in Khyber Pakhtunkhwa, Pakistan. Complete blood counts were conducted using the Sysmex XT 4000i Hematology Analyzer (Sysmex Corporation, Japan).
DNA Extraction
The QUBIT DNA BR Kit was utilized to extract DNA from whole blood samples obtained from six pediatric patients diagnosed with acute lymphocytic leukemia and one healthy control, following the manufacturer's instructions. To assess the quality and integrity of the extracted DNA, agarose gel electrophoresis was performed using a 1% (w/v) gel.
Whole Exome Sequencing
Raw sequencing data were filtered to produce clean reads, which were then mapped to the human reference genome to generate a preliminary comparison file in BAM format using Burrows-Wheeler Aligner (BWA) [14,15]. The Genome Analysis Toolkit (GATK) was used to ensure accurate variant calling, following the recommended Best Practices for variant examination. GATK was also used to mark duplicate reads and recalibrate the base quality score. To ensure accurate sequencing data, a comprehensive quality control (QC) system was implemented into the workflow [16]. Single nucleotide polymorphisms (SNPs) and insertions/deletions (InDels) were detected using the Haplotype Caller of GATK, followed by a filtering process to obtain variant calls with high confidence. The SnpEff program was used for variant annotation http://snpeff.sourceforge.net/SnpEff manual HTML. To detect structural variation (SV) in the genome, we used Break Dancer, while CNVnator was used to detect copy number variation (CNV). The results of SV and CNV were annotated using Ensemble-VEP [17]. Additional analyses were conducted to identify cancer susceptibility genes, driver gene clones, drug-targeted annotations, high-frequency mutations, homology, hyper-mutated samples, loss of heterozygosity, molecular classification, mutation signatures, and neoantigen prediction.
Variant effect predictor
To assess the potential impact of variants, including single nucleotide polymorphisms (SNPs) and structural variants (SVs), we used the Variant Effect Predictor (VEP) to predict the disease-relatedness of GLDC non-synonymous SNPs [16]. Additionally, we used the SIFT algorithm (https://sift.bii.a-star.edu.sg/) to predict the functional effect of amino acid substitutions. The SIFT score cut-off of 0.05 was used to determine whether a substitution was tolerable or deleterious (18).
We also employed PolyPhen (http://genetics.bwh.harvard.edu/pph2/) to evaluate the impact of amino acid substitutions on protein structure and function. The PolyPhen score ranges from 0 to 1, with a threshold of 0.5 for benign and 0.5 for deleterious variants. A high PolyPhen score indicates a stronger negative impact on protein structure for missense SNPs.
SNPs&GO
The SNPs&GO server, available at https://snps.biofold.org/snps-and-go/snps-and-go.html, is a bioinformatics tool that predicts the potential impact of single nucleotide polymorphisms (SNPs) on protein function based on various data sources, including gene ontology annotation, protein function, sequence, 3D structures, and protein sequence profile databases [19]. To utilize this tool, we input the UniProt accession ID (P23378 for GLDC) along with the specific mutation positions and amino acid changes of interest. The output from SNPs&GO includes a prediction of whether the SNP is likely to be disease-causing or neutral, along with a reliability index (RI) score ranging from 0 (non-reliable) to 10 (reliable), which can be used to assess the confidence of the prediction. It is important to note that SNP prediction tools, like SNPs&GO, can be useful for identifying potentially disease-causing variants. However, caution should be exercised when interpreting the results as these tools are not always accurate. Additional experimental validation is usually required to confirm the impact of a specific SNP on protein function and disease risk.
I-Mutant
The I-Mutant v2.0 server (https://folding.biofold.org/i-mutant/i-mutant2.0.html) is a bioinformatics tool used to assess the potential impact of amino acid substitutions on protein structure and stability. It employs an SVM-based prediction algorithm to estimate the effect of mutations on the thermodynamic stability of proteins [20, 21]. I-Mutant uses the ProTherm-derived repository, a thermodynamic database for proteins and mutations, containing experimental data on the thermodynamic stability of proteins under various conditions such as changes in temperature, pH, and solvent composition. By comparing the wild-type and mutant protein sequences and their associated thermodynamic data from the ProTherm-derived repository, I-Mutant can predict whether a mutation is likely to destabilize or stabilize the protein structure.
SNAP2
The SNAP2 service, available at https://rostlab.org/services/snap2web/, is a specialized tool that utilizes neural networks to distinguish between potentially disease-causing and neutral nsSNP variants by examining various sequence and variant-related features [22]. In this study, the FASTA sequence of the GLDC protein served as input for the SNAP2 service. The potential pathogenicity of the predicted amino acid changes was determined by establishing a threshold value of >1.
Prediction and Visualization of 3D structure
To generate 3D structural models of the native GLDC protein, we utilized the AlphaFold protein structure database. Specifically, we obtained the GLDC protein accession ID P23378 from UniProt and used it to model the 1020 amino acid structure. Additionally, we generated mutated 3D structure models using Chimera 1.16 and subsequently refined them using the Galaxy Refine 2 server (https://galaxy.seoklab.org/cgi-bin/submit.cgi?type=REFINE) for further structural improvement [23]. To validate the resulting native and mutated GLDC models, we performed a Ramachandran plot analysis using the SAVESv6.0 server. Finally, we visualized the 3D models using the Discovery Studio Visualizer tool.
Protein-Protein Interaction
The bioinformatics tool, STRING dB (https://string-db.org/), was utilized to identify and analyze protein-protein interactions (PPIs). By employing its default settings, the server provided a PPI network for the GLDC gene, displaying the different proteins that interact with GLDC and the strength of their interaction. The prediction score for each interaction was derived from various genomics and proteomics resources, including biological databases and scientific literature [24].
Analysis of MD Simulation
Molecular dynamics (MD) simulations provide a detailed understanding of the structure-to-function interactions of macromolecular proteins in biologically relevant environments. Our investigation aimed to comprehend the dynamic characteristics of the macromolecules, including their conformational ensembles and fluctuations due to residue substitutions. To achieve this, we utilized the WEBGRO UAMS server (https://simlab.uams.edu/index.php) to conduct MD simulation analyses on both the native and mutant GLDC models. The complex trajectories were recorded during a 20-ns MD simulation of the native and mutant structural models. We employed the built-in features of WEBGRO UAMS, such as gmx rmsd, gmx hbond, gmx gyrate, and gmx rmsf functions, to calculate parameters such as root-mean-square deviation (RMSD), hydrogen bond, radius of gyration (Rg), and root-mean-square fluctuation (RMSF), respectively.
GLDC Gene–Gene Interaction
The interaction of the gene of interest was investigated using Gene MANIA (https://genemania.org/) [25, 26]. This tool employs co-expression, protein domain similarity, co-localization, and pathway analysis to identify potential gene interactions. A gene list containing the official symbols for each gene was input to identify core genes associated with the gene of interest.
HOPE
To construct a model of the desired protein, HOPE utilizes a homologous structure. In this study, the Yasara & WHAT IF Twinset was utilized for this purpose. Structural information was obtained from various resources, including the UniProt database, the Reprof software, and the WHAT IF Web services. The UniProt entry for the relevant GLDC protein is P23378, which can be accessed through the https://www3.cmbi.umcn.nl/hope/ website [27].
Gene ontology Blast2GO
The Blast2GO (B2G) tool is a valuable resource for mining sequence data using Gene Ontology (GO) even in cases where GO annotation is not readily available. B2G utilizes statistically based similarity searches, visualization tools, and directed acyclic graphs to facilitate GO annotation. This tool is particularly useful for functional genomics research in non-model species. The B2G desktop application is interactive and user-friendly, allowing for monitoring and comprehension of the entire annotation and analysis process [28]. The annotation process involves a 3-step procedure consisting of blast, mapping, and annotation. GO annotations are obtained by merging and converting InterPro data obtained from InterProScan at EBI. Following annotation, both combined and individual graphs can be generated [29]. The pie chart included in this study was created using Origin software.
Results
Clinical characteristics
Seven samples were selected for Whole Exome Sequencing (WES), including two samples from patients diagnosed at the initial stage, two from remission patients, two from relapse patients, and one from a healthy child. The patients selected had blast cells above 20%, high leukocyte count, and low Hb, with some exceptions for remission samples. Clinical symptoms included weakness, lethargy, exhaustion, dyspnea, fever, weight loss, or bleeding. Hepatosplenomegaly or adenopathy could also be caused by blasts infiltrating lymph nodes or organs. The demographics and prognosis of children with ALL in Pakistan of Pakhtun ancestry are poorly understood. Using WES, we identified an average of 106,442 SNPs, of which 98.27% were represented in dbSNP, and 92.71% of these variations were annotated in the 1000 Genomes Project database. We discovered 1,812 novel SNPs, with a Transversion to Transition ratio (Ti/Tv) of 2.32. Table 1 summarizes the statistics of the identified SNPs.
Table 1: Summary and statistics of SNPs identified.
Prediction of Damaging Effects of GLDC novel Varient.
To evaluate the potential pathogenicity of variants identified in a patient with acute lymphoblastic leukemia (ALL), we employed several computational tools, including PolyPhen2, SIFT, Condel, Mutation Assessor, FATHMM, CADD, and MetalR. These tools were utilized to predict the impact of amino acid substitutions on protein structure and function. The deleterious effect of the identified novel variant on GLDC protein was assessed and summarized in Table 2. The result of SNAP2 analysis is presented in Figure 1A, while PolyPhen2 predicted the nsSNP to be "probably damaging," as depicted in Figure 1B.
|
Tools |
Score |
Prediction |
Threshold |
|
Poly phene 2 |
0.935(D) |
Damaging |
0 to 1 |
|
SIFT |
0.001(D) |
Damaging |
≥ 0.05 |
|
Mutation accessor |
2.615(M) |
Mutation |
0 to 1 |
|
Meta SVM |
1.0997(D) |
Damaging |
|
|
FATHMM |
-4.74(D) |
Damaging |
0.5 |
|
MetaLR |
0.9517(D) |
Damaging |
o.8 |
|
VEST3 |
0.899 |
Pathogenic |
0 benign1pathogenic |
|
CADD |
6.372671 |
Damaging |
0 to 99 |
Table 2: PolyPhen2, SIFT, FATHMM, CADD, VEST3, Metal LR, and Mutation accessor results for the selected GLDC Varient.
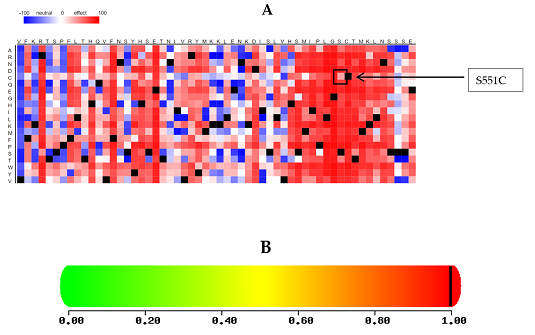
Figure 1: (A) represents the heat map of SNAP2 (B) shows polyphen predicted mutation with a score of 0.998. A score of 0.998 suggests that the mutation is predicted to be damaging.
Gene ontology of GLDC gene
GO (Gene Ontology) is a community-driven bioinformatics resource that uses ontologies to describe biological knowledge and provides information about the functions of genes and gene products. The mutant residue identified in our study is in a region that is essential for the protein's function and is adjacent to another domain that is also critical for that activity. It is possible that the mutation may disrupt the interaction between these domains, thereby affecting the protein's ability to perform its function properly. The GO resource consists of three distinct categories, namely cellular components, biological processes, and molecular function [30]. In our study, we utilized the Blast 2GO software to investigate the biological process, molecular function, and cellular component of the GLDC gene (as shown in Figure 2A). Our findings suggest that GLDC contributes significantly to metabolic processes. At the molecular level, GLDC is involved in oxidoreductase activity, binding activity, and catalytic activity. Regarding cellular components, GLDC activity was observed in mitochondrial and other membrane-bound organelles, cytoplasm, complexes, and membranes.
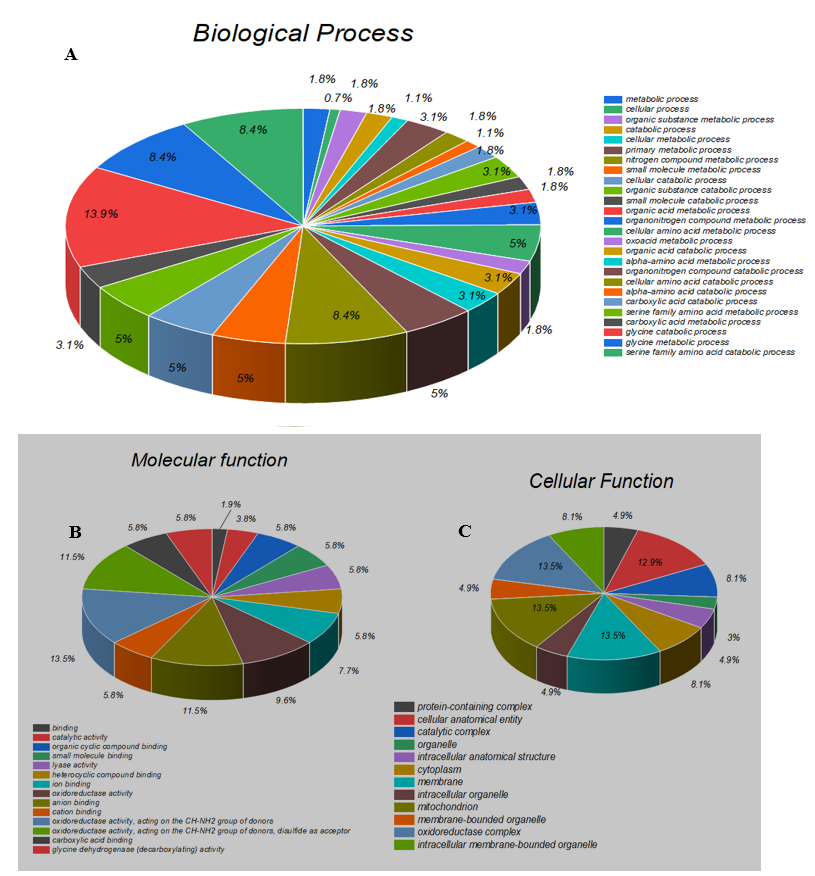
Figure 2: A shows the biological processes of GLDC the Sequences were analyzed by Blast2GO and Gene Ontology (GO). B Represent the molecular function While Figure C represents the cellular function of GLDC.
Prediction and Validation of 3D structure
The 3D structure of Glycine decarboxylase (GLDC) (PDB ID: P23378) was used as a template to predict the 3D structure of the GLDC protein using Alpha Fold. Chimera 1.16 was employed to generate mutant models through amino acid substitution at position 551 of GLDC. The structural data were further refined using Galaxy refined software, with Saves 0.6 being used for structure validation. A Ramachandran plot analysis indicated that 87% of residues in both the wild-type and mutant protein structures are in regions where torsion angles are permitted. The overall quality factor of the wild-type and mutant structural models was determined as 96.2% based on the Errat score.
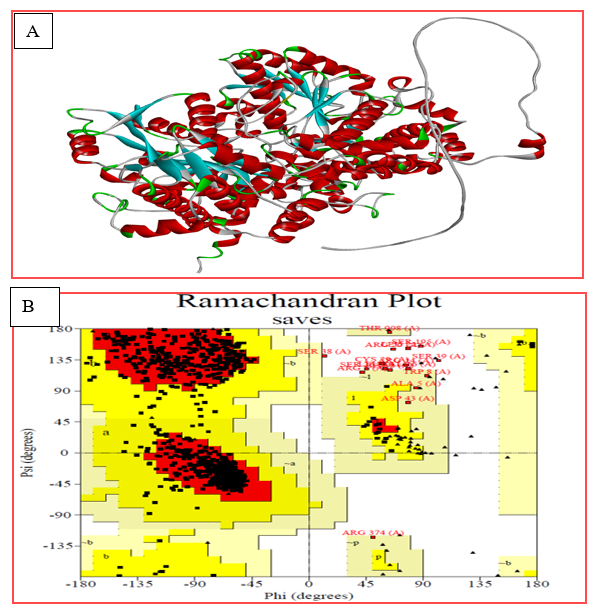
Figure 3: (A) shows a 3D model structure of the GLDC native protein in solid ribbon style, where the blue color represents beta sheets, red represents alpha helices, grey represents coils, and green represents turns. (B) shows a Ramachandran plot of the GLDC protein structure model. The Ramachandran plot is a graphical representation of the energetically allowed regions for the backbone dihedral angles of amino acid residues in protein structures.
Analysis of evolutionary conservation and the stability of protein structure
The stability of the protein structure was analyzed using the I-Mutant server, which predicted a decrease in stability with a reported Reliability Index of 6.0. To investigate the conserved nature of the S551C mutation in highly conserved regions of the protein, the ConSurf web server was employed. As shown in Figure 3, the score for Glycine is 8. The ConSurf web server was used to handle the conserved nature of the prioritized nsSNP. The S551C mutation was predicted to occur in a highly conserved region, indicating its significance for the structure and function of the GLDC protein
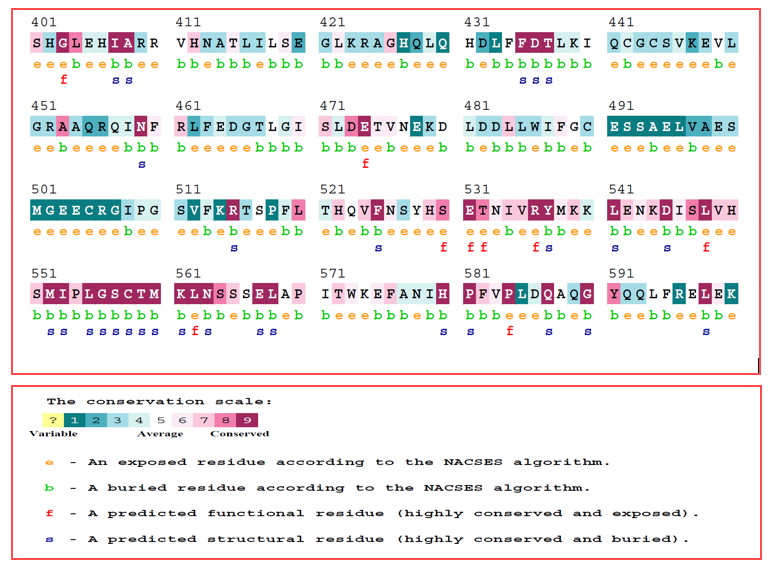
Figure 4: The figure displays the conserved GLDC protein residue sequence, with the color codes ranging from blue to purple. The purple regions indicate high conservation, while the blue regions indicate variability. Serine at position 551, which is a highly conserved residue that is substituted by cysteine in the mutant protein. This information is important for understanding the functional implications of the mutation and how it affects the overall structure and function of the protein.
Characteristics of Wild and Mutant Amino Acids
The GLDC protein gene has undergone a mutation from Serine to Cysteine at position 551, as depicted in Figure 4. The backbone of each amino acid is the same, whereas the side chain is unique and imparts specific characteristics such as size, charge, and degree of hydrophobicity. In this study, we aimed to investigate the effect of the newly introduced mutant residue on the stability and ligand interactions of the protein. Our findings indicate that the mutant residue has distinct properties compared to the wild-type residue, with higher hydrophobicity being one of the key differences.
The change in hydrophobicity due to the mutation may impact the local stability of the protein, which can, in turn, affect the interactions between nearby residues and ligands. For instance, in the wild-type protein, Histidine at position 550 and Glycine at position 889 form a hydrogen bond, but the difference in hydrophobicity between the wild-type and mutant residues can influence hydrogen bond formation.
Further analysis revealed that the mutation causes a loss of hydrogen bonds in the protein's core, leading to improper folding and destabilization of the protein.
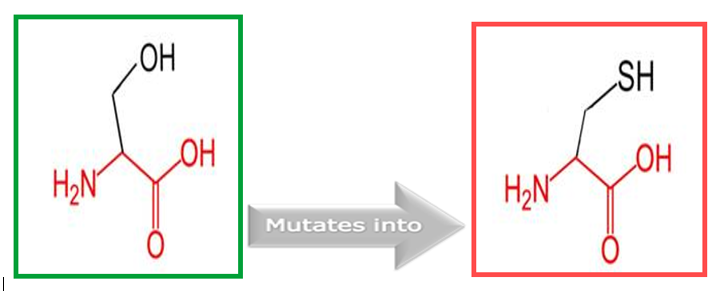
Figure 5: Show the original (left) and mutant (right) amino acids' schematic structures.
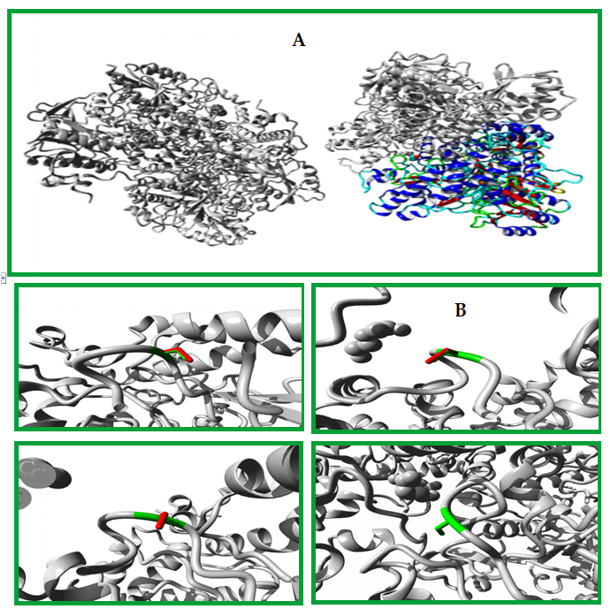
Figure 6: Shows an overview of the protein structure in ribbon presentation. The different elements of the protein are color-coded, with the α-helix shown in blue, β-strand in red, turn in green, 3/10 helix in yellow, and random coil in cyan. Other molecules in the complex are colored grey.A closer look at the mutation is provided in (B), which is shown from a slightly different angle. The side chains of the mutant residue and its wild-type counterpart are highlighted, with the mutant residue colored green and the wild-type residue colored red. The protein itself is colored grey in this figure. Overall, this figure provides a clear visualization of the protein structure and the location of the mutation, allowing the reader to better understand the impact of the mutation on the protein structure and function.
Wild and mutant systems dynamics stability and residual flexibility
We performed molecular dynamics simulations for 20ns on native and mutant GLDC model structures to investigate their dynamics and stability. The results are presented in Figure 7. Figure 7A shows the root-mean-square deviation (RMSD) values for Ca atoms in the mutant and native structures over time. The mutant structure exhibited higher RMSD values than the native structure, indicating that the mutant structure was more flexible and underwent more structural changes during the simulation. In Figure 7B, we present the root mean square fluctuation (RMSF) values for the carbon alpha throughout the simulation. The RMSF values for the mutant structure were generally higher than those of the native structure, suggesting that the mutant structure exhibited greater flexibility and local structural changes. Figure 7C shows the overall H-bond counts for both structures during the simulation. The native structure formed more H-bonds than the mutant structure, indicating that the mutant structure had a weaker ability to form and maintain H-bonds. Finally, Figure 7D displays the radius of gyration (Rg) of the protein backbone over time. The mutant structure exhibited a higher Rg value than the native structure, indicating that the mutant structure had a more extended conformation. Taken together, our findings suggest that the mutant GLDC structure had greater flexibility, weaker ability to form and maintain H-bonds, and a more extended conformation compared to the native GLDC structure. These results have important implications for understanding the effects of nsSNP-based substitutions on protein structure and dynamics and may inform the development of new therapeutic approaches.
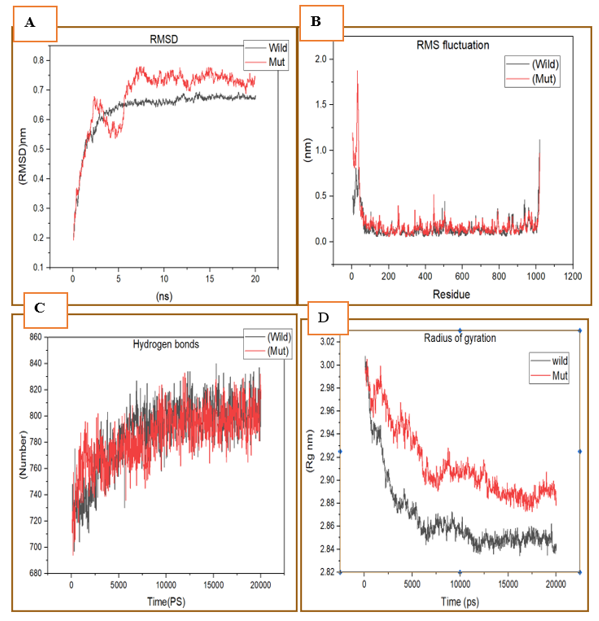
Figure 7: Native and mutant GLDC model structures at 20 ns were analyzed using molecular dynamics. (A) show RMSD values for Ca atoms in mutant and natural structures. The Y-axis represents RMSD (nm), while the X-axis represents time (ns). (B) RMSF values for the carbon alpha throughout the whole simulation. The X-axis is residue, while the Y-axis is RMSF (nm). (C) H-bond counts overall for both native and mutant structures during the simulation. (D) Rg of the protein backbone during the simulation's full run. The X-axis represents time, while the Y-axis is Rg (nm) (ns). (The native GLDC is represented by (black colour), while the mutant by (red colour).
Non-bonding Interactions
Non-bonding interactions play a significant role in stabilizing the secondary structure of proteins. To investigate the effect of non-bonding interactions on the structure of the GLDC protein, we analyzed the interactions between native and mutant amino acid residues and their close interacting residues in their respective 3D predicted models using Discovery Studio Visualizer. Our structural modeling revealed that nsSNP-based substitutions result in changes to the original structure and interactions with nearby amino acid residues, potentially impacting the function of the GLDC protein. Specifically, the novel variant disrupts the normal interaction of wild GLDC, which may affect its functioning. To further understand the effects of these mutations, we performed MD simulations on both wild-type and mutant GLDC models. Our analysis showed that the mutant models exhibited higher levels of fluctuations compared to the wild-type models. We then analyzed the non-bonding interactions of the mutant models in the context of the native GLDC residues at corresponding positions. Interestingly, while both wild and mutant models formed similar H-bond interactions with Gly (A) 889 and Met (A) 552, there were additional two alkyl bonds present in the mutant models with Phe 890 and Ile 546, which were absent in the wild-type GLDC. These findings are depicted in Figure 8.
Taken together, our results suggest that the nsSNP-based substitutions alter the non-bonding interactions within the GLDC protein, potentially impacting its function.
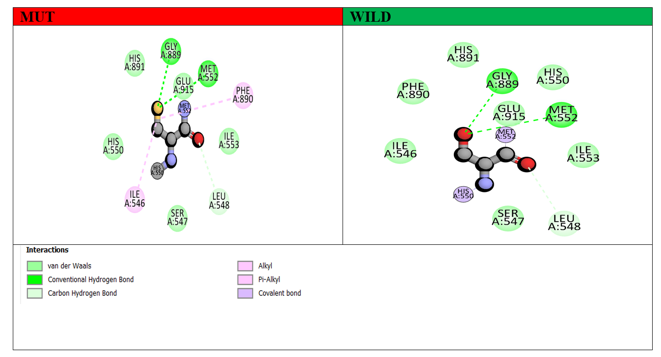
Figure 8: Demonstrates the interaction of wild-type (green) and mutant (red) GLDC proteins with certain residues at position (551). The different colors in this 2D image represent various molecular interactions. The hydrogen bond is represented by the green dot line, the carbon-hydrogen bond is represented by the cyan dot line, and the Pi-alkyl interaction is represented by the pink dot line.
Protein-protein interaction
To investigate potential interactions of GLDC genes with ALL-associated SNPs, we utilized two community-based bioinformatics tools, STRING and GeneMANIA, which predict protein-protein interactions and gene-gene interactions, respectively. The vast majority of molecular mechanisms and biological processes rely on protein interactions. The results from both tools were documented after inputting gene symbols. String predicted interactions of GLDC with 10 other genes including AMT, GCSH, SHMT1, SHMT2, DLD, SARDH, AGXT, AGXT2, PIPOX, and GNMT. The cutoff value was set at 0.1, with the best score being 0.9 and the lowest score being 0. GeneMANIA builds a composite gene-gene functional interaction network, and Fig. 9 displays the predicted interaction network of GLDC. The predictions made by GeneMANIA included co-localization (3.63%), co-expression (8.01%), physical interactions (77.64%), predicted (5.37%), shared protein domains (0.60), pathways (1.88), and genetic relationships (2.87). The additional genes that were identified as candidates for a potential role in the pathogenesis of GLDC include GCSH, AMT, TRAPPC5, HSPDI, GRPEL1, AKR1B10, TLR10, EFL1, SNU13, PSMC1, TNFAIP3, DLD, PCK2, APOA1, SHMT1, POLG2, ETNPPL, OAT, and SPATA13, as shown in Figure 9.
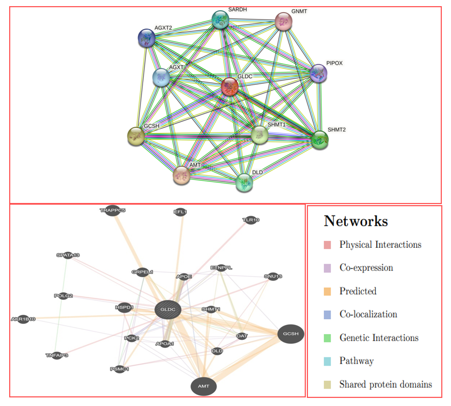
Figure 9: A. protein-protein interaction model of GLDC genes using STRING.
Gene MANIA created a network of gene-gene interactions for all possible types of interactions.
Discussion
SNPs play a significant role in the pathophysiology of diseases and can also contribute to altered pharmacological responses when located in the target's active site [31] et al. In this study, we conducted whole-exome sequencing (WES) of six patients with acute lymphoblastic leukemia (ALL) and one healthy child with clinical features, and identified a novel pathogenic heterozygous variant, NM_000170.2: p. Ser551Cys/c.1651A>T, in the GLDC gene that segregated with disease phenotypes. Bioinformatics techniques projected that the missense mutation is pathogenic, and the I-Mutant v2.0 servers indicated that GLDC (Ser551Cys) decreases the stability of GLDC protein. The structural instability generated by this mutation may result in functional abnormality of GLDC. ConSurf was used to generate the conservation of GLDC and predicted that the functional and structural consequences of nsSNPs could have negative impacts on human health by being localized at buried residues of score 8, which employs solvent accessibility together with evolutionary conservation data [32]. We evaluated the root mean square deviation (RMSD) for the wild-type and mutant protein backbones relative to the native structure during molecular dynamics (MD) simulation to learn more about the structure and function of the GLDC protein. The RMSD values of (Ser551Cys) were unstable compared to the wild-type protein, suggesting that the mutant deviates greatly from the wild type and is significantly destabilized. Furthermore, GLDC (Ser551Cys) showed abnormal fluctuation, less compactness, high structural alteration, and volatility compared to wild-type GLDC. Structural modeling showed that nsSNP-based alterations alter the original structure and interactions with surrounding amino acid residues, which may affect the GLDC protein's ability to perform its intended function. The novel variant disrupts the normal interaction of wild GLDC and may affect the normal functioning of GLDC. The Gene Ontology (GO) tool, which uses ontologies to express biological knowledge and provides details on the roles of GLDC genes and gene products, was used to infer useful details about the functions of gene products [33]. GO is divided into three main categories: biological process, cellular component (CC), and molecular function (MF). The cellular component is where gene products are active (BP; pathways or larger processes that multiple gene products are involved in) [34]. In the pathophysiology of multi-gene hereditary illnesses, gene-gene interaction is a significant phenomenon. Numerous genes interact with GLDC and have both known and unknown pathogenesis patterns. To anticipate several gene-gene interaction mechanisms, we employed Gene MANIA and STRING databases. We discovered that 10 genes were located in the core region based on STRING predictions Figure 9, while 20 genes were identified to be interactive in pathways, and Gene MANIA predicted type-specific interactions. GLDC is predicted to be a novel drug target.
Conclusions
In this study, we discovered a new heterozygous missense Varient, Ser 551 Cys, in the GLDC gene that is linked to the disease phenotype of ALL. Molecular dynamics simulations demonstrated that the mutated form of the protein exhibited significant structural and functional changes. The location of this mutation in a highly conserved region of the protein suggests its vital role in maintaining protein function. Our findings indicate that this mutation may be the possible high-risk candidate for leukaemia and severely impacts the protein's stability, which may contribute to the development of the disease. In short, the deleterious novel SNP significantly affects the normal folding and structural stability of proteins. The combination of bioinformatics techniques, such as WES, I-Mutant v2.0, ConSurf, and MD simulations, along with community-based tools like GO, GeneMANIA, and STRING, provides a powerful approach to identify and understand the potential pathogenic mechanisms and therapeutic targets for genetic disorders such as ALL. These results provide valuable insights into the molecular mechanisms underlying the disease and may have implications for its diagnosis, treatment, and prevention.
Author Contributions
Conceptualization, Shahid Ullah and Asif Ullah khan.; methodology, Shahid Ullah.; software, Maryam Halawi, validation, Shahid Ullah. Abdulsalam M Alruwaili ; formal analysis, Shahid Ullah.; investigation, Shahid Ullah; resources, .; data curation, Alex Tonks.; writing—original draft preparation, Shahid Ullah.; writing—review and editing,; Abdul Wadood visualization,; Supervision, Muhammad Arif Lodhi and Asif Ullah Khan.; Project administration, Muhammad Arif Lodhi.; All authors have read and agreed to the published version of the manuscript.”
Funding
This research received no external funding.
Institutional Review Board Statement
Approval NO. Dir A&R AWKUM 2020/5118 was received for the current study from Abdul Wali Khan University's Institute Review Board (IRB), located in Mardan, KPK, Pakistan.
Acknowledgments:
This research was supported by the Higher education commission of Pakistan, School of Medicine, Cardiff University UK, and the Biochemistry Department of Abdul Wali Khan University Mardan Pakistan.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Xu H, Yang W, Perez-Andreu V, et al. Novel susceptibility variants at 10p12.31-12.2 for childhood acute lymphoblastic leukemia in ethnically diverse populations. J Natl Cancer Inst 105 (2013): 733-774
- Bhojwani D, Pei D, Sandlund JT, et al. 26 (2013): 265-270.
- Roberts KG, Gu Z, Payne-Turner D, et al. High frequency and poor outcome of Philadelphia chromosome-like acute lymphoblastic leukemia in adults. J Clin Oncol 35 (2017): 394-401.
- Paulsson K, Jonson T, Øra I, et al. Characterizations of genomic translocation breakpoints and identification of an alternative TCF3/PBX1 fusion transcript in t(1;19) (q23;p13)-positive acute lymphoblastic leukemias. Br. J Haematol 138 (2007): 196–201.
- Leitão LPC, de Carvalho DC, Rodrigues JCG, et al. Identification of genomic variants associated with the risk of acute lymphoblastic leukemia in Native Americans from Brazilian Amazonia. J Pers Med 12 (2022): 856.
- Awan T, Iqbal Z, Aleem A, et al. Five most common prognostically important fusion oncogenes are detected in the majority of Pakistani pediatric acute lymphoblastic leukemia patients and are strongly associated with disease biology and treatment outcome. Asian Pac. J Cancer Prev 13 (2012): 5469-5475.
- Leidy
 [Genetic changes]. NIH Public Access. Bone 23 (2011): 1-7.
[Genetic changes]. NIH Public Access. Bone 23 (2011): 1-7. - Zhang WC, Ng SC, Yang H, et al. Glycine decarboxylase activity drives non-small cell lung cancer tumor-initiating cells and tumorigenesis. Cell 148 (2012): 259-272.
- Jiang TJ, Jiang JJ, Xu JL, et al. Clinical and genetic analyses of a family with atypical nonketotic hyperglycinemia caused by compound heterozygous mutations in the GLDC gene. Chin. J. Contemp. Pediatr 19 (2017): 1087-1091.
- Zhuang H, Li Q, Zhang X, et al. Downregulation of glycine decarboxylase enhanced cofilin-mediated migration in hepatocellular carcinoma cells. Free Radic Biol Med 120 (2018): 1-12.
- Berezowska S, Galván JA, Langer R, et al. Glycine decarboxylase and HIF-1α expression are negative prognostic factors in primary resected early-stage non-small cell lung cancer. Virchows Arch 470 (2017): 323-330.
- Kim SK, Jung WH, Koo JS. Differential expression of enzymes associated with serine/glycine metabolism in different breast cancer subtypes 9 (2014).
- Sun WY, Kim HM, Jung WH, et al. Expression of serine/glycine metabolism-related proteins is different according to the thyroid cancer subtype. J Transl Med 14 (2016): 1-12.
- Chen Y, Chen Y, Shi C, et al. A MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. Gigascience 7 (2018): 1-6.
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler Transform. Bioinformatics 25 (2009): 1754-1760.
- McLaren W, Gil L, Hunt SE, et al. The Ensembl Variant Effect Predictor. Genome Biol 17 (2016): 1-14.
- McPherson A, Chen K, Wu C, et al. An algorithm for high-resolution mapping of genomic structural variation. Nature Methods Nat Methods 6 (2009): 677-681.
- Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res 11 (2001): 863-874.
- Calabrese R, Capriotti E, Fariselli P, et al. Functional annotations improve the predictive score of human disease-related mutations in proteins. Hum Mutat 30 (2009): 1237-1244.
- Capriotti E, Fariselli P, Casadio R. I-Mutant2.0: Predicting stability changes upon mutation from the protein sequence or structure. Nucleic Acids Res 33 (2005): 306-310.
- Choi Y, Chan AP. PROVEAN web server: A tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics 31 (2015): 2745-2747.
- Hecht M, Bromberg Y, Rost B. Better prediction of functional effects for sequence variants from VarI-SIG 2014: Identification and annotation of genetic variants in the context of structure, function, and disease. BMC Genomics 16 (2015): 1-12.
- Heo L, Park H, Seok C. GalaxyRefine: Protein structure refinement driven by side-chain repacking. Nucleic Acids Res 41 (2013): 384-388.
- Franceschini A, Szklarczyk D, Frankild S, et al. STRING v9.1: Protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res 41 (2013): 808-815.
- Warde-Farley D, Donaldson SL, Comes O, et al. The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res 38 (2010): 214-220.
- Gasteiger E, Gattiker A, Hoogland C, et al. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res 31 (2003): 3784-3788.
- Venselaar H, te Beek TAH, Kuipers RKP, et al. Protein structure analysis of mutations causing inheritable diseases. An e-Science approach with life scientist-friendly interfaces. BMC Bioinformatics 11 (2010): 548.
- Conesa A, Götz S. Blast2GO: A comprehensive suite for functional analysis in plant genomics. Int J Plant Genomics 2008 (2008).
- Peng J, Bai K, Shang X, et al. Predicting disease-related genes using integrated biomedical networks. BMC Genomics 18 (2017): 1-11.
- Zhang Y, Skolnick J. TM-align: A protein structure alignment algorithm based on the TM-score. Nucleic Acids Res 33 (2005): 2302-2309.
- Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: Server and survey. Nucleic Acids Res 30 (2002): 3894-3900.
- Blake JA, Christie KR, Dolan ME, et al. Gene Ontology Consortium: Going forward. Nucleic Acids Res 43 (2015): D1049–D1056.
- Cacchiarelli D, Trapnell C, Ziller MJ, et al. Predicting disease-related genes using integrated biomedical networks. BMC Genomics 18 (2017): 1-11.


 Impact Factor: * 5.3
Impact Factor: * 5.3 CiteScore: 2.9
CiteScore: 2.9  Acceptance Rate: 11.01%
Acceptance Rate: 11.01%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 