ISSN: 0973-7510
E-ISSN: 2581-690X
This study was designed to explore the incidence of blaOXA-1 amongst Klebsiella pneumoniae isolates with resistant to carbapenem. Between December 2014 and April 2015, one hundred samples were taken from two hospitals: Babylon Teaching Hospital for Maternity and Pediatric / Babylon Province (clinical, umbilical infections, n= 40; environmental, n=20) and Karbala Hospital for Pediatric / Karbala Province (40 stool samples). All patients were hospitalized or attended these hospitals, all under 1 year of age. Seventeenth (17%) isolates were identified as Klebsiella pneumoniae. The antibiotic resistance profile of isolates was tested using disk diffusion method. High-level of resistance was recorded with ampicillin (94.1%) and piperacillin (88.2%) antibiotics. Resistance to carbapenem was reported in two K.pneumoniae isolates, these were investigated for the existence of OXA-1β-lactamase using Polymerase Chain Reaction (PCR) technique. Two (100%) isolates gave positive result. Transference of this gene was studied by conjugation experiment. The blaOXA-1 gene conjugated successfully in 1 (50%) isolate only.
Klebsiella pneumoniae, Carbapenem resistance, OXA-1 β-lactamase, PCR, Conjugation
Antimicrobial resistance is a major public health problem worldwide. Infections caused by multi-drug resistance organisms due to long hospital stay, antibiotics treatment and poor hygiene are in continuous increase and linked with high rates of mortality and morbidity 1,2. The possible resistance mechanism in Klebsiella spp is the production of extended spectrum beta-lactamases (ESBLs). These enzymes are capable of hydrolyzing penicillin, cephalosporin (3rd and 4th generation), monobactams, but have no effect on cephamycins or carbapenem3,4.
The predominant mechanisms for resistance to inhibitor penicillin combinations are : class C chromosomal β-lactamase production, overproduction of TEM-1 and TEM-2 type β-lactamases and OXA-1 β-lactamase production5,6,7.
OXA-1 β-lactamase has the ability to hydrolyzes amino, ureidopenicillins (piperacillin), cloxacillin, oxacillin and methicillin in significant mean while it hydrolyzes cephalosporins (narrow–spectrum) weakly. Moreover, it hydrolyzes broad-spectrum cephalosporins, mediated diminished susceptibility to antibiotics like cefepime and cefpirome8,9. OXA-1 β- lactamase distributed widely among Enterobacteriaceae family and a major reason for resistance to amoxicillin /clavulanic acid combination mainly in Escherichia coli and Salmonella enterica10,11,12.
The present work was attempted to evaluate the frequency of Klebsiella pneumoniae among clinical and environmental specimens, characterize resistant isolates, detect blaOXA-1 gene using Polymerase Chain Reaction (PCR) technique in isolates showed resistance to carbapenem and test its transmissibility by conjugation experiment.
Sample collection
In a five months period (December,2014 to April, 2015), 100 different specimens were recovered from two hospitals namely :Babylon Teaching Hospital for Maternity and Pediatric / Babylon Province (clinical: umbilical infections, n=40; environmental: n=20) and Karbala Hospital for Pediatric / Karbala Province (40 stool samples). Collected samples were cultured on different prepared media. Suspected K.pneumoniae isolates were identified based on their colonial, morphological characteristics and microbiological procedures as mentioned previously13,14,15.
Antimicrobial susceptibility testing
To determine the resistance profiles of K.pneumoniae isolates, the antimicrobial susceptibility to thirteen antimicrobial agents were analyzed by Kirby –Bauer disk diffusion method on plates with Mueller- Hinton agar medium (Oxiod, England)16. The selected agents included: ampicillin (AMP), piperacillin (PRL), amoxicillin- clavulanic acid (AMC), cefotaxime (CTX), ceftazidime (CAZ), ceftriaxone (CRO), cefepime (FEP), cefoxitin (FOX), gentamicin (CN), imipenem (IMP), meropenem (MEM), levofloxacin (LE5) and norfloxacin (NOR). The results of susceptibility were interpreted according to the Clinical and Laboratory Standards Institute (CLSI) guidelines17. The Escherichia coli ATCC 25922 (University of Kufa, College of Medicine,) was used as the control strain in antimicrobial susceptibility testing.
Molecular detection of blaOXA-1 gene
Deoxyribonucleic acid (DNA) of carbapenem resistant K.pneumoniae was extracted based on the method mentioned with some modifications18. Conventional Polymerase Chain Reaction technique was applied to amplification blaOXA-1 gene using specific primers (Bioneer, Korea) OXA-1/F (F: ATA TCT CTA CTG TTG CAT CTC C) and OXA-1/R (R: AAA CCC TTC AAA CCATCC) (619 bp). All amplifications were implemented in a total volume of 25 µl consisted of 12.5 µl Go Taq Green Master Mix 2X (Promega, USA), 5 µl of extracted DNA, 2.5 µl forward and reverse primer (10 pmol/ µl) each and 2.5 µl nuclease-free water. The DNA template was denatured at 94°C for 5 min, followed by 30 cycles of denaturation ( 94°C for 50 sec), annealing ( 55°C for 50 sec), extension ( 72°C for 1 min) and the final extension (72°C at 10 min)19 . The PCR reaction product was separated by gel electrophoresis (1.5% agarose gel stained with ethidium bromide solution, 0.5 mg/ml) at 70 volts for 2-3 hrs, PCR product was examined using UV-Transilluminator, and photographed with Gel documentation system. The size of DNA band was estimated using DNA Ladder,100 bp (Bioneer, Korea).
Conjugation experiment
To test the transmissibility of blaOXA-1 gene, two carbapenem-resistant K.pneumoniae harboring OXA-1 gene (donors) and rifampicin resistant Escherichia coli MM294 (University of Kufa, College of Medicine) (recipient),were selected. Conjugation experiment was attempted by liquid mating assay20,21. All the transconjugants were screened for the existence of this gene using PCR assay with same primers applied in the procedure. The Minimum inhibitory concentrations (MICs) for ampicillin , cefotaxime, ceftazidime, imipenem and meropenem were detected using HiComb Minimum Inhibitory Concentration (HiComb MIC) (Himedia, India) and Minimum Inhibitory Concentration Evaluator (M.I.C.E) (Oxoid, England) tests in accordance with the guidelines of Clinical and Laboratory Standards Institute17.
During study period (From December,2014 to April, 2015), 17(17%) strains were belonged to Klebsiella pneumoniae ,12 (70.6%) were recovered from stool samples and 5(29.4%) from umbilical infections, (Table 1). Recently, K.pneumoniae from stool samples was documented among children attending different hospitals in Dar es Salaam, Tanzanid22 . Another report identified 12(2%) prevalence rate for Klebsiella spp. isolated from newborns with omphalitis in Pakistan23.
Table (1):
Prevalence of17 K.pneumoniae isolates obtained from various samples.
| Hospital’s name | Source of samples | Types and samples No. | K.pneumoniae isolates No.(%) |
|---|---|---|---|
| Babylon Teaching Hospital for Maternity and Pediatric | Clinical | Umbilical infections (n=40) | 5 (29.4%) |
| Environmental | floor (n=10) | 0 (0 %) | |
| beds (n= 6) | 0 (0 %) | ||
| walls (n=4) | 0 (0 %) | ||
| Karbala Hospital for Pediatric | clinical | Stool (n=40) | 12 (70.6%) |
| Total | 100 | 17 (100) | |
However, K.pneumoniae from environmental samples was not detected in this study. The reason may be related to low number of tested samples. One study in Hillah city identified the species in various clinical and environmental samples24. Also, Abbas25 proved the detection of K.pneumoniae from burn unit environment of Al-Hillah teaching hospital.
In this study, all K.pneumoniae isolates presented higher resistance against penicillin antibiotics (ampicillin, piperacillin) with (94.1%) and (88.2%) resistance rates, respectively, (table -2). One work documented high level of resistance (98.6%) for ampicillin by K.pneumoniae among septicemic patients in India9. The higher frequency of resistance can be attributed to excessive consumption of these drugs in clinical settings.
Table (2):
Resistance profiles of the 17 Klebsiella pneumoniae isolates.
| Antimicrobial class | Antimicrobial tested | Resistant isolates No.(%) |
|---|---|---|
| Penicillins | ampicillin | 16(94.1%) |
| piperacillin | 15(88.2%) | |
| β –lactams ∕β- lactamase inhibitor combinations | amoxicillin-clavulanic acid | 14 (82.3%) |
| Cephems | cefotaxime | 13 (76.5%) |
| ceftazidime | 12 (70.6%) | |
| cefriaxone | 1 2(70.6%) | |
| cefepime | 14(82.4%) | |
| cefoxitin | 13(76.5%) | |
| Aminoglycosides | gentamicin | 10(58.8%) |
| Penems | imipenem | 2 (11.8%) |
| meropenem | 2(11.8%) | |
| Quinolones | levofloxacin | 4 (23.5%) |
| norfloxacin | 4 (23.5%) |
Additionally, the lower frequency was observed with imipenem (11.8%) and meropenem (11.8%), (Table 2).One report carried out by Hashemi et al.26 documented (20%) as a rate of resistance against imipenem and meropenem for K.pneumoniae isolated from two hospitals in Tehran, Iran. Other research characterized (28.57%) resistance rate for meropenem antibiotic by clinical isolates of K.pneumoniae in Southeastern Nigeria27.
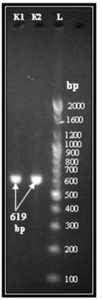
Fig. 1. Conventional PCR for amplification blaOXA-1 gene in K.pneumoniae isolates with resistant to carbapenem. Lane (L), 100- bp DNA Ladder. Lane (1,2) are OXA-1 positive isolates for blaOXA-1 gene (619 bp)
All carbapenem- resistant K.pneumoniae were positive for OXA-1 gene using PCR technique (Fig.1). According to Flores et al.28 blaOXA-1 gene was demonstrated in 42(60%) K.pneumoniae isolated from rectal swabs of patients settings intensive care unit, Brazil. The occurrence of K.pneumoniae harboring blaOXA-1 gene (34.4%) was previously reported in Malaysia29.
Conjugation has been regarded as a very efficient method for horizontal transfer of resistance genes in bacteria with higher frequency in nature than under laboratory conditions30,31. In current research, conjugative transfer of blaOXA-1 gene was successful for only 1 (50%) isolate of K.pneumoniae (K2) which was selected as a donor for conjugation (Table-3, Fig.2).Successful transfer of OXA-1 gene by conjugation was previously reported in a spanish isolates of K.pneumoniae32. Also, Rakotonirino et al.33 documented the transfer of this gene in 6 isolates of K.pneumoniae obtained from four hospitals and medical centers in Antananarivo, Madagascar. The widespread of OXA-1 gene may be related to localization of this gene on variable region of integrin (class I) that also contain other resistance determinants like aac(6) Ib, CTX-M ESBL type and carbapenemases32.
Table (3):
Characteristics of carbapenem-resistant K.pneumoniae clinical isolates and their conjugates.
| Isolate | OXA-1 β-lactamase confirmed by PCR | MIC (μg /ml) | ||||
|---|---|---|---|---|---|---|
| AMP (>32) |
CTX (>64) |
CAZ (>32) |
IMP (≥16) |
MEM (≥16) |
||
| K. pneumoniae K1 (clinical isolate) | + | > 256 | >240 | >256 | >32 | >32 |
| E.coli transconjugant (TCK1) |
– | – | – | – | – | – |
| K. pneumoniaeK2 (clinical isolate) | + | > 256 | >240 | >256 | >32 | >32 |
| E.coli transconjugant (TCK2) |
+ | > 256 | >240 | >256 | 2 | 2 |
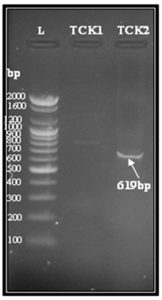
Fig. 2. Agarose gel electrophoresis for blaOXA-1 gene (619 bp) of transconjugant E.coli isolates TCK1 and TCK2. Lane (L), DNA molecular weight marker (100- bp Ladder). Lane (TCK2) showing positive result with blaOXA-1 gene. Lane (TCK1) showing negative result
Antibiotics susceptibilities of the transconjugant (TCK2) revealed higher resistance to ampicillin, cefotaxime and ceftazidime as its donor isolates with MIC values (>256, >240, >256) µg/ml, respectively,(table-3).Such higher resistance among transconjugant may be explained that OXA-1 gene was transferred and expressed .
The MIC of imipenem and meropenem for transconjugat (TCK2) was relatively lower (2 µg /ml) than the donor isolate (table -3).This finding suggest the presence of other resistant mechanism like chromosomal -mediated resistance genes in the donor that can not transferred by conjugation .
The current study documents the presence of K.pneumoniae carrying ESBL of OXA-1 gene. The ability of this gene for transference pose a significant challenge for therapeutic options currently in use. Therefore, effective prevention and control strategies must be applied to prevent occurrence and dissemination of these strains.
ACKNOWLEDGMENTS
The author is grateful to all the medical staff in Hospitals for their assistance and for providing facilities to complete this research.
FUNDING
None.
ETHICS STATEMENT
Not applicable.
AVAILABILITY OF DATA
All datasets generated or analyzed during this study are included in the manuscript.
- Garner JS, Jarvis WR, Emori TG, Horan TC, Hughes JM. CDC Definitions for Nosocomial Infections. In: R. N. Olmsted, Ed. APIC Infection Control and Applied Epidemiology: Principles and Practice, Mosby, St Louis. 1996:A1-A20.
- Diriba K, Awulachew E, Tekele L, Ashuro Z. Fecal carriage rate of extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae among apparently health food handlers in Dilla University Student Cafeteria. Infect. Drug Resist. 2020; 13:3791-3800.
Crossref - Bush K, Jacoby GA. Updated functional classification of b-lactamases. Antimicrob. Agents Chemother. 2010; 54(3): 969-976.
Crossref - Lakshmi R, Nuserin KS, Ann GS, Sreelakshmi KS. Role of beta lactamases in antibiotic resistance : a review. Int. Res. J.Pharm. 2014;5(2):37-40.
Crossref - Bergstrom S, Normark S. Beta-Lactam resistance in clinical isolates of Escherichia coli caused by elevated production of the AmpC-mediated chromosomal beta-lactamase. Antimicrob.Agents Chemother. 1979; 16: 427-433.
Crossref - Shannon K, Williams H, King A, Philipps I. Hyperproduction of TEM-1 beta-lactamase in clinical isolates of Escherichia coli serotype O15. FEMS Microbiol. Lett. 1990; 67: 319-323.
Crossref - Zhou XY, Bordon F, Sirot D, Kitzis MD, Gutmann L. Emergence of clinical isolates of Escherichia coli producing TEM-1derivatives or an OXA-1 beta-lactamase conferring resistance to beta-lactamase inhibitors. Antimicrob. Agents Chemother. 1994; 38: 1085-1089.
Crossref - Naas T, Nordmann P. OXA-type beta lactmases. Curr. Pharm. Des. 1999; 5 (11): 865-879.
- Sugumar M, Kumar KM, Manoharan A, Anbarasu A, Ramaiah S. Detection of OXA-1 beta-lactamase gene of Klebsiella pneumoniae from blood stream infections (BSI) by conventional PCR and in-silico analysis to understand the mechanism of OXA mediated resistance. PLoS ONE. 2014; 9(3): e91800.
Crossref - Machado E, Coque TM, Cantón R, Baquero F, Sousa JC, Peixe L. Dissemination in Portugal of CTX-M-15-, OXA-1-, and TEM-1-producing Enterobacteriaceae strains containing the aac(6′)-Ib-cr gene, which encodes an aminoglycoside and fluoroquinolone-modifying enzyme. Antimicrob. Agents. Chemother. 2006; 50:3220-3221.
Crossref - Oteo J, Cuevas O, López-Rodríguez I, et al. Emergence of CTX-M-15-producing Klebsiella pneumoniae of multilocus sequence types 1, 11, 14, 17, 20, 35 and 36 as pathogens and colonizers in newborns and adults. J. Antimicrob. Chemother. 2009; 64:524-528.
Crossref - Jamali S, Tavakoly T, Mojtahedi A, Shenagari M. The Phylogenetic relatedness of bla NDM-1 harboring Extended-Spectrum b-Lactamase producing uropathogenic Escherichia coli and Klebsiella pneumoniae in the North of Iran. Infect. Drug Resist. 2020; 13: 651-657.
Crossref - Holt JG, Krieg NR, Sneath HA, Stanley JT, Williams ST. Bergeys manual of determinative bacteriology. 9th Ed. Baltimore, Wiliams and Wilkins, USA. 1994.
- Collee JG, Fraser AG, Marmiom BP, Simmon A. Mackie and McCarteny Practical Medical Microbiology. 4th Ed. Churchill Livingstone Inc., USA. 1996.
- MacFaddin JF. Biochemical tests for identification of medical bacteria. 3rd Ed. Lippincott Williams and Wilkins, USA. 2000.
- Bauer AW, Kirby WMM, Sherris JC, Track M. Antibiotic susceptibility testing by standardized single disc method. Am.J. Clin.Pathol. 1966; 45: 493-496.
Crossref - Clinical and Laboratory Standards Institute (CLSI). Performance standards for antimicrobial susceptibility testing. CLSI Supplement M100S. 26th Ed. Wayne,PA. 2016.
- Pospiech T, Neumann J. In genomic DNA isolation Kieser eds. John Innes Center. Norwich NR4 7UH. U.K. 1995.
- Karami N, Hannoun C, Adlerbeth I, Wold AE. Colonization dynamics of ampicillin – resistant Escherichia coli in the infantile colonic microbiota. J. Antimicrob. Chemother. 2008;62:703-708.
Crossref - Sambrook J, Fritshch EF, Maniatis T. Molecular Cloning: A laboratory manual, 2nd Ed. Cold spring Harbor Laboratory Press, Cold Spring Harbor, New York. 1989.
- Sambrook J, Russell DW. Molecular cloning: laboratory manual, 3rd Ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York. 2001.
- Tellevik MG, Blomberg B, Kommedal Ø, Maselle SY, Langeland N, Moyo J. High prevalence of faecal carriage of ESBL-producing Enterobacteriaceae among children in Dar es Salaam, Tanzania. PLOS ONE, 2016; 11(12): e0168024.
Crossref - Mir F, Tikmani SS, Shakoor S, et al. Incidence and etiology of omphalitis in Pakistan: a community-based cohort study. J. Infect. Dev. Ctries. 2011; 5(12):828-833.
Crossref - Al-Charrakh AH, Yousif SY, Al-Janabi HS. Ocurrence and detection of extended spectrum b-lactamases in Klebsiella isolates in Hilla, Iraq. Afri. J. Biotechnology. 2011; 10(4):657-665.
- Abbas FM. Molecular detection of CTX-M extended spectrum beta lactamase among carbapenem- resistant Klebsiella pneumoniae from Al-Hillah Teaching Hospital environment, Babylon Province, Iraq. J. Phys: Conf. Ser. 2019; 1294.
Crossref - Hashemi A, Fallah F, Erfanimanesh S, Hamedani P, Alimehr S, Goudarzi H. Detection of b-lactamases and outer membrane porins among Klebsiella pneumoniae strains isolated in Iran. 2014;2014:726179.
Crossref - Ugwu MC, Shariff M, Nnajide CM, et al. Phenotypic and molecular characterization of b-lactamases among Enterobacterial uropathogens in Southeastern Nigeria. Can. J. Infect. Dis. Med. Microbiol. 2020; 1-9.
Crossref - Flores C, Romao CMCPA, Bianco K, et al. Detection of antimicrobial resistance genes in beta-lactamase- and carbapenemase-producing Klebsiella pneumoniae by patient surveillance cultures at an intensive care unit in Rio de Janeiro, Brazil. J. Bras. Patol. Med. Lab. 2016; 52(5):284-292.
Crossref - Al-Marzooq F, Mohd Yusof MY. Tay ST. Molecular analysis of antibiotic resistance determinants and plasmids in Malaysian isolates of multidrug resistant Klebsiella pneumoniae. PLoS ONE, 2015; 10(7): e0133654.
Crossref - Canton R, Coque TM, Baquero F. Multi-resistant Gram-negative bacilli: from epidemics to endemics. Curr. Opin. Infect. Dis. 2003;16:315-325.
Crossref - Davies J, Davies D. Origins and evolution of antibiotic resistance. Microbiology and Molecular Biology Reviews. 2010; 74: 417-433.
Crossref - Cubero M, Calatayud L, Ayats J, et al. Clonal spread of Klebsiella pneumoniae producing OXA-1 beta lactamase in a Spanish hospital. Int. Microbiol. 2013; 16: 227-233.
Crossref - Rakotonirina H, Garin B, Randrianirina F, Richard V, Talarmin A, Arlet G. Molecular characterization of multidrug -resistant extended spectrum b-lactamase – producing Enterobacteriaceae isolated in Antananarivo, Madagascar. BMC Microbiology. 2013; 13:85.
Crossref
© The Author(s) 2021. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.