Differences in overall survival and mutation prevalence between right- and left-sided colorectal adenocarcinoma
Introduction
Previous retrospective studies, including both retrospective cohort studies and post-hoc analyses of clinical trials, have demonstrated inferior outcomes for patients with right-sided colon cancer (RCC) compared to patients with left-sided colorectal cancer (LCC) (1-4). Among the most expansive of these reports is a systematic review and metanalysis including 66 prior studies and a total of over 1.4 million patients, which demonstrated a lower risk of death among patients with left-sided primaries [hazard ratio (HR) 0.82, 95% confidence interval (CI): 0.79–0.84] compared to those with right-sided primaries at a median follow-up of 65 months (5). Differences in treatment response based on disease sidedness have also been reported (6).
Intriguingly, differences in prognosis remain even among patients with metastatic disease (6). These findings suggest that anatomy or stage at diagnosis alone cannot explain all of the observed differences in survival between left- and right-sided disease. Moreover, these differences have led to the suggestion that disease sidedness should be included among other prognostic factors when making decisions regarding treatment intensity for patients with CRC (4,5,7).
These findings are perhaps not surprising, as genetic differences between RCC and LCC have long been described (8). More recently, genetic differences have been described on a larger scale, including descriptions of disproportionate prevalence of KRAS and BRAF mutations among RCC, as well as increased rates of microsatellite instability (7). LCC, on the other hand, has been associated with higher prevalence of p53 and NRAS mutations, as well as chromosomal instability. However, despite such reports, the genetic and molecular drivers underlying differences in prognosis and treatment response remain incompletely understood.
Methods
In this setting, we conducted a retrospective review of overall survival and mutation prevalence in LCC vs. RCC at a single academic medical center. Relevant cases were identified from the tumor mutation database maintained by the Center for Personalized Diagnostics (CPD) at the University of Pennsylvania Health System. This database contains results from next-generation sequencing (NGS) panels ordered in routine clinical practice by practitioners within the system’s oncology practice. Relevant samples were identified by querying the CPD’s database for gastrointestinal malignancies between 2013 and 2016.
We reviewed clinic records to confirm the nature of the patient’s primary malignancy, with colorectal adenocarcinoma being our only malignancy of interest. More extensive chart review was then conducted for those patients identified as having a CRC to determine primary location (left- vs. right-sided disease), date of pathologic diagnosis, stage at diagnosis, date of death, and date of last contact with our medical system. We defined RCC as arising from the cecum to splenic flexure and LCC as arising from the descending colon to rectum (5).
Survival was calculated from date of pathologic diagnosis to death or last follow-up via the Kaplan-Meier method. Censorship was assumed not to affect the probability of survival. Survival curves were generated via the Real Statistics Resource Pack (v. 4.14, Real Statistics, Trento, Italy) for Excel 2016 (Microsoft, Redmond, WA, USA), with statistical significance determined via log-rank test. Comparisons between mutation prevalence were made via two-sided Z test for proportions, and 95% CIs were calculated around point estimates of mutation prevalence. Finally, we compared the mutation prevalence for individual genes in our sample with those found in the Catalogue of Somatic Mutations in Cancer (COSMIC) database as a means of external validation.
NGS was performed at the CPD, and reported results included 38 genes (ABL1, AKT1, APC, ATM, BRAF, CDH1, CSF1R, CTNNB1, EGFR, ERBB2, ERBB4, FBXW7, FGFR1, FGFR2, FGFR3, FLT3, GNA11, GNAQ, GNAS, HRAS, IDH1, IDH2, JAK3, KDR, KIT, KRAS, MET, NRAS, PDGFRA, PIK3CA, PTEN, RB1, RET, SMAD4, SMO, STK11, TP53, and ZRSR2). Prior to sequencing analysis, formalin fixed-paraffin embedded (FFPE) sections were examined for adequacy by a surgical pathologist. To be considered adequate for mutational analysis, samples were required to have a tumor volume of greater than 10%. Genomic DNA was extracted from FFPE tissue according to manufacturer’s instructions (Qiagen, Inc., Hilden, Germany). Targeted analysis for mutations was performed via enrichment of specific loci using the Illumina TruSeq Amplicon (Illumina Inc., San Diego, CA, USA) or a custom Agilent Haloplex assay (Agilent Technologies, Santa Clara, CA, USA). Sequencing of enriched libraries was performed on the Illumina MiSeq and HiSeq platforms using multiplexed, paired-end reads. Analysis and interpretation utilized a customized bioinformatics process, and variant classifications were made using the hg19 genome build (9).
Mutations were classified as pathogenic, variants of uncertain significance, or benign by the CPD based on a literature review and query of publicly available databases (including dbSNP, COSMIC, ExAC, and the 1000 Genomes Project). Pathogenic variants were defined as those with known or predicted loss or gain of function of the protein products.
This retrospective study was approved by the Institutional Review Board at the University of Pennsylvania prior to the collection of data.
Results
Among 288 cases identified via the above review, there were 167 left-sided and 103 right-sided cases (Table 1). In addition, there were 18 cases of patients with synchronous bilateral disease or otherwise without clear primary, which were excluded from all subsequent analyses. Selected demographic and disease characteristics are summarized in Table 2. Patients with RCC were disproportionately older [mean 61.0 years (95% CI: 58.5–63.6) vs. 55.4 years (95% CI: 53.5–57.3) for LCC]. There were more males among patients with LCC (53% vs. 45%), though this difference was not statistically significant. The majority of patients (n=150) had metastatic disease at time of diagnosis. Among these patients, 90 had LCC and 60 had RCC. Consequently, a higher proportion of patients with RCC had metastatic disease at the time of diagnosis [58% (95% CI: 49–68%) vs. 53% (95% CI: 46%–61%) for LCC], though this disparity was also not statistically significant.
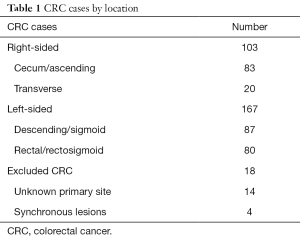
Full table

Full table
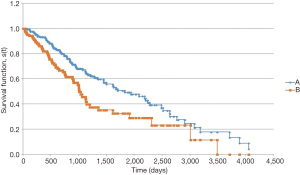
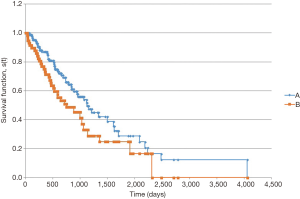
In the survival analysis, patients with LCC had a longer overall survival (Figure 1). Median overall survival from date of pathologic diagnosis was 1,823 days for patients with LCC vs. 1,006 days for RCC (P=0.004). Among patients with metastatic disease at diagnosis, the survival advantage for patients with LCC persisted (Figure 2). Median overall survival for these patients was 1,124 days for LCC and 750 days for RCC (P=0.047).
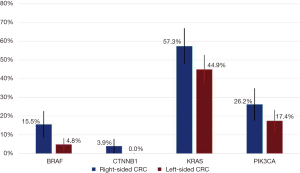
Among the assessed genes, BRAF and CTNNB1 mutations were more prevalent in RCC (Figure 3). BRAF was mutated in 15.5% of RCC samples (95% CI: 8.5–22.5%) compared to 4.8% (95% CI: 1.6–8.0%) (P=0.003). CTNNB1 was mutated in 3.9% of RCC (95% CI: 0.2–7.6%) compared to no instances of CTNNB1 mutations in LCC (P=0.01). Among RCC, there was also a trend toward more KRAS mutations at 57.3% (95% CI: 47.7–66.8%) vs. 44.9% (95% CI: 37.4–52.5%) and more PIK3CA mutations at 26.2% (95% CI: 17.7–34.7%) vs. 17.4% (95% CI: 11.6–23.1%). The prevalence of other mutations was similar between the two groups (data not shown).
The prevalence of CTNNB1 mutations in our cohort (1.5%, 95% CI: 0.0–2.9%) was lower than that seen among large intestine adenocarcinomas in the COSMIC database (5.0%, 95% CI: 4.5–5.6%). The overall prevalence of BRAF mutations in our sample (8.9%, 95% CI: 5.5–12.3%) was consistent with the prevalence of BRAF mutations in COSMIC (10.6%, 95% CI: 10.4–10.8%). All observed BRAF mutations in this cohort were previously described pathogenic mutations. The majority (4 of 8 among LCC and 11 of 16 among RCC) harbored V600E mutations. Multiple mutations were noted at the 466, 469, and 594 codons, with G466V, N581S, D594N, D594G observed in LCC cases and L597R, G466A, G469A, G469V, D594N observed in RCC samples.
Discussion
The results obtained in this study for overall survival in LCC vs. RCC were consistent with those seen in prior studies, namely, that RCC carried a worse prognosis than did LCC. Also, as seen in prior studies, stage at diagnosis does not seem to explain this finding in isolation, as our cohort demonstrated a statistically significant difference in overall survival even among those patients who had metastatic disease at the time of diagnosis.
The differing mutation prevalence noted here may implicate these genetic pathways in the mechanisms underlying the discrepant outcomes and treatment responses between RCC and LCC described in this study. V600E BRAF mutations, the most common mutation seen in this gene, are well established as conferring a worse prognosis in CRC, with poorer results reported in BRAF-mutant CRC in multiple studies. These findings have been reported in both metastatic (10-14) and earlier-stage disease (15,16), though the impact has been particularly striking in patients with metastatic disease. In addition, RCC has been previously reported to have higher rates of BRAF mutations compared to LCC (7).
Of note, 5 of the 103 RCC samples in this cohort and 4 of the 167 LCC samples harbored non-V600E BRAF mutations. The clinical implications of non-V600E mutations are less clear than are those of V600E mutations. Jones et al. recently reported the largest description of clinicopathologic characteristics of non-V600E-mutant CRC (17). In their analysis of 9,643 CRC specimens with available NGS data, non-V600E mutations were associated with a favorable overall prognosis compared to both V600E BRAF-mutant cases as well as compared to patients with BRAF wild-type disease (median overall survival 60.7 vs. 11.4 vs. 43.0 months, respectively). Moreover, non-V600E-mutant disease was less likely to be found in RCC compared to V600E-mutant disease (36% right-sided among all non-V600E-mutant samples in their study vs. 81% right-sided for V600E-mutant samples).
The implications of CTNNB1 mutations are even less clear. CTNNB1 mutations are relatively rare in CRC. In our cohort, 1.5% of samples harbored a CTNNB1 mutation, which is lower than the 5.0% rate reported in COSMIC, but similar to rates described in other sources in the literature (18). The clinical relevance of these mutations is uncertain, particularly as CTNNB1 mutations are rarely found in isolation. Malapelle et al. found that mutations in CTNNB1 in CRC are associated with constitutive RAF/MEK/ERK pathway signaling, typically via association with mutations in other cancer-related genes (18). However, studies that have assessed correlations between CTNNB1 mutational status and clinical outcomes have not reported consistent trends (19).
The present study, like most of the previously reported results regarding survival and CRC sidedness, is a retrospective cohort study and therefore subject to the biases associated with retrospective studies. Moving forward, the implementation of broader NGS approaches in a prospective fashion may help to identify other gene variants related to the survival differences noted in this and other studies. Most prior prospective studies have been limited to RAS testing. In addition, the impact of broader measures, such as tumor mutational burden requires assessment in a prospective fashion.
The present study was conducted at a single referral center and consequently may not be generalizable to other centers. In addition, while our study demonstrated worse outcomes for patients with right-sided disease as demonstrated in their worse overall survival from time of diagnosis compared to patients with LCC, the patients with RCC were significantly older than patients with LCC, as noted above. Consequently, the worse outcomes of the patients with RCC may have been partially driven by factors other than disease biology. In addition, the impact of other possible prognostic markers including immunohistochemistry for CDX2 and microsatellite instability markers was not assessed in this study, in part due to limited availability of this data.
The relative timing of NGS data collection with respect to prior therapy was also not assessed in this study, though most NGS assays were performed on patients’ initial pathology specimens. Previous studies have demonstrated an impact of systemic therapy on clonal evolution of CRC, which may have implications for treatment outcomes and survival (20). For instance, hypothesis-generating data from the REVERCE study suggest the sequencing of cetuximab and regorafenib in RAS wild-type CRC may impact survival due to earlier generation of RAS mutations when EGFR inhibitors are utilized (21).
This study could benefit from additional statistical power, and the preceding analysis could be repeated as additional data is generated at our institution. Alternatively, a similar analysis could be conducted using larger pooled databases, similar to the methods utilized by Jones et al. (17). The overall prevalence of BRAF mutations in our sample (8.9%, 95% CI: 5.5–12.3%) was consistent with the prevalence of BRAF mutations among large intestine adenocarcinomas in the COSMIC database, as well as rates described elsewhere in the literature (22), lending some support to the external validity of these data. Unfortunately, the reference database we used (COSMIC) does not readily contain location data (left- vs. right-sided) for most of its documented cases of colorectal cancer, making leveraging this database for a similar analysis problematic.
Overall, the present study re-demonstrates the discrepant prognosis of left- vs. right-sided CRC reported in prior studies. Also seen are signals of differing prevalence of mutations in multiple genes between LCC and RCC, most notably BRAF and CTNNB1. These genetic pathways may have relevance to prognosis in other CRC cohorts. For instance, the association between BRAF mutational status and primary sidedness described by Jones et al. may have some explanatory power for the discrepancies in prognosis between LCC and RCC seen in prior studies (17). However, other reports that have stratified their results by BRAF mutational status have found persistent differences in outcomes based on CRC sidedness (6). Further work is therefore needed to more clearly elucidate genetic differences between RCC and LCC and mechanistic relationship between these mutations and differences in prognosis. Further research is also needed regarding non-genetic biologic differences between LCC and RCC that may have prognostic significance. For instance, tumor burden at the time of diagnosis, which was not captured in this study, may have some explanatory power beyond stage alone. A better understanding of such factors is needed to guide both discussions of prognosis as well as treatment decisions.
Acknowledgements
The authors wish to acknowledge the assistance of Dr. Jennifer Morrissette in obtaining data from the Center for Personalized Diagnostics.
Footnote
Conflicts of Interest: Disclosures for A Loaiza-Bonilla include: Speakers Bureau: Celgene, Bristol Myers-Squibb, Guardant, Caris Life Science, Eisai; Advisory Board: Bayer, Astra Zeneca; Consulting: Massive Bio; Research: Ipsen. CE Jensen and JY Villanueva have no conflicts of interest to declare.
Ethical Statement: The study was approved by the Institutional Review Board at the University Pennsylvania prior to the collection of data (approval number 827031). Data was collected retrospectively, and the study was deemed to represent minimal risk to patients, so informed consent was not obtained; however, patients’ data has been secured according to requirements of IRB protocol. Findings did not affect any treatment decisions.
References
- Tejpar S, Stintzing S, Ciardiello F, et al. Prognostic and predictive relevance of primary tumor location in patients with RAS wild-type metastatic colorectal cancer: Retrospective analyses of the CRYSTAL and FIRE-3 trials. JAMA Oncol 2016;3:194-201. [Crossref] [PubMed]
- Schrag D, Weng S, Brooks G, et al. The relationship between primary tumor sidedness and prognosis in colorectal cancer. J Clin Oncol 2016;34:abstr 3505.
- Loupakis F, Yang D, Yau L, et al. Primary tumor location as a prognostic factor in metastatic colorectal cancer. J Natl Cancer Inst 2015;107:1-9. [Crossref] [PubMed]
- Yahagi M, Okabayashi K, Hasegawa H, et al. The worse prognosis of right-sided compared with left-sided colon cancers: a systematic review and meta-analysis. J Gastrointest Surg 2016;20:648-55. [Crossref] [PubMed]
- Petrelli F, Tomasello G, Borgonovo K, et al. Prognostic survival associated with left-sided vs right-sided colon cancer: a systematic review and meta-analysis. JAMA Oncol 2016;3:211-9. [Crossref] [PubMed]
- Venook AP, Ou FS, Lenz HJ, et al. Primary (1°) tumor location as an independent prognostic marker from molecular features for overall survival (OS) in patients (pts) with metastatic colorectal cancer (mCRC): analysis of CALGB / SWOG 80405 (Alliance). J Clin Oncol 2017;35:abstr 3503.
- Shen H, Yang J, Huang Q, et al. Different treatment strategies and molecular features between right-sided and left-sided colon cancers. World J Gastroenterol 2015;21:6470-8. [Crossref] [PubMed]
- Birkenkamp-Demtroder K, Olesen SH, Sorensen FB, et al. Differential gene expression in colon cancer of the caecum versus the sigmoid and rectosigmoid. Gut 2005;54:374-84. [Crossref] [PubMed]
- Daber R, Sukhadia S, Morrissette JJ. Understanding the limitations of next generation sequencing informatics, an approach to clinical pipeline validation using artificial data sets. Cancer Genet 2013;206:441-8. [Crossref] [PubMed]
- Loupakis F, Cremolini C, Masi G, et al. Initial therapy with FOLFOXIRI and bevacizumab for metastatic colorectal cancer. N Engl J Med 2014;371:1609-18. [Crossref] [PubMed]
- Di Nicolantonio F, Martini M, Molinari F, et al. Wild-type BRAF is required for response to panitumumab or cetuximab in metastatic colorectal cancer. J Clin Oncol 2008;26:5705-12. [Crossref] [PubMed]
- Richman SD, Seymour MT, Chambers P, et al. KRAS and BRAF mutations in advanced colorectal cancer are associated with poor prognosis but do not preclude benefit from oxaliplatin or irinotecan: results from the MRC FOCUS trial. J Clin Oncol 2009;27:5931-7. [Crossref] [PubMed]
- Van Cutsem E, Kohne CH, Lang I, et al. Cetuximab plus irinotecan, fluorouracil, and leucovorin as first-line treatment for metastatic colorectal cancer: updated analysis of overall survival according to tumor KRAS and BRAF mutation status. J Clin Oncol 2011;29:2011-9. [Crossref] [PubMed]
- Tran B, Kopetz S, Tie J, et al. Impact of BRAF mutation and microsatellite instability on the pattern of metastatic spread and prognosis in metastatic colorectal cancer. Cancer 2011;117:4623-32. [Crossref] [PubMed]
- Roth AD, Tejpar S, Delorenzi M, et al. Prognostic role of KRAS and BRAF in stage II and III resected colon cancer: results of the translational study on the PETACC-3, EORTC 40993, SAKK 60-00 trial. J Clin Oncol 2010;28:466-74. [Crossref] [PubMed]
- Sinicrope FA, Shi Q, Smyrk TC, et al. Molecular markers identify subtypes of stage III colon cancer associated with patient outcomes. Gastroenterology 2015;148:88-99. [Crossref] [PubMed]
- Jones JC, Renfro LA, Al-Shamsi HO, et al. Non-V600 BRAF mutations define a clinically distinct molecular subtype of metastatic colorectal cancer. J Clin Oncol 2017;35:2624-30. [Crossref] [PubMed]
- Malapelle U, Pisapia P, Sgariglia R, et al. Less frequently mutated genes in colorectal cancer: evidences from next-generation sequencing of 653 routine cases. J Clin Pathol 2016;69:767-71. [Crossref] [PubMed]
- Morikawa T, Kuchiba A, Yamauchi M, et al. Association of CTNNB1 (beta-catenin) alterations, body mass index, and physical activity with survival in patients with colorectal cancer. JAMA 2011;305:1685-94. [Crossref] [PubMed]
- Braxton DR, Zhang R, Morrissette JD, et al. Clinicopathogenomic analysis of mismatch repair proficient colorectal adenocarcinoma uncovers novel prognostic subgroups with differing patterns of genetic evolution. Int J Cancer 2016;139:1546-56. [Crossref] [PubMed]
- Shitara K, Yamanaka T, Denda T, et al. Reverce: randomized phase II study of regorafenib followed by cetuximab versus the reverse sequence for metastatic colorectal cancer patients previously treated with fluoropyrimidine, oxaliplatin, and irinotecan. J Clin Oncol 2018;36:abstr 557.
- Tie J, Gibbs P, Lipton L, et al. Optimizing targeted therapeutic development: analysis of a colorectal cancer patient population with the BRAF(V600E) mutation. Int J Cancer 2011;128:2075-84. [Crossref] [PubMed]


