
The effects of LL-37 on virulence factors related to the quorum sensing system of Pseudomonas aeruginosa
Introduction
Pseudomonas aeruginosa (P. aeruginosa) is a complex Gram-negative conditional pathogen that is responsible for both hospital-acquired and community-acquired infections (1-4). Quorum sensing (QS) is a cell density-based intercellular communication system. Pseudomonas has a QS system (5-7) and exhibits significant resistance to antibiotics and the innate immune system. The QS system of Pseudomonas is one of its most important signal transduction systems and is the key to its pathogenicity (8,9). QS has a cascade regulation mechanism. The Las system is located at the top of the regulatory network. It is initially activated after receiving a stimulus signal and can positively regulate the Rhl system (10,11). The synthesis of the Pseudomonas quinolone signal (PQS) (12) is positively regulated by the Las system. PQS regulates the expression of the rhlI-associated genes that depend on the Rhl system, thus linking the Las system and the Rhl system. In this way, the QS system can regulate the expression of virulence factors and trigger biofilm formation (13,14).
The virulence of P. aeruginosa is closely related to its cellular structure (pili, lipopolysaccharides, and flagella) and secreted factors (exotoxins, elastases, pyocyanin, and extracellular polysaccharides). The QS system involves the cell-density dependent accumulation of signal molecules that enable bacteria to modulate the expression of virulence-associated genes (15,16) including elastase, alkaline protease, exotoxin A, rhamnolipid, pyocyanin, and exogenous lectins. A host of virulence factors are regulated by 2 acyl-homoserine lactone (AHL) QS systems: the LasI-LasR (las system) and the RhlI-RhlR (rhl system) (17,18). Virulence factors can alter the balance between host defenses and bacterial virulence, leading to damaged blood vessels, exotoxin spread, systemic inflammatory response syndrome, and ultimately host injury or death (19). This means that the virulence factors in the QS system are key to the pathogenicity of P. aeruginosa.
Previous study found that exotoxin A was a type II secreted extracellular enzyme encoded by the toxA gene. This enzyme alone or synergistically with other hydrolases causes cell death, severe tissue damage, and necrosis in humans (20). Elastase, which is encoded by the lasA gene (staphylolysin), lasB gene (pseudolysin), and secreted extracellularly (21,22), plays an important role in the infection process of P. aeruginosa. Pyocyanin is a redox-active virulence factor that is produced by P. aeruginosa and can easily penetrate biological membranes. Pyocyanin has been shown to play a major role in animal models of acute and chronic infection caused by P. aeruginosa (23,24).
The unreasonable use of antibiotics makes bacteria resistant to these treatments, which can lead to repeated or persistent infections. P. aeruginosa resistance is an issue of public concern. As the prevalence of Pseudomonas bacterial resistance increases, it is critical to find new methods for treating infections. Interfering with the expression of virulence genes regulated by the QS system can effectively regulate the pathogenicity and drug resistance of P. aeruginosa, and may represent a new and promising direction for potential drug therapies.
Since antimicrobial peptides (AMPs), also known as host defense peptides, were discovered in the 1980s, they have been viewed as a promising alternative to conventional antibiotics. AMPs show promise against multi-resistant pathogens due to their combined antibacterial, immunomodulatory, and anti-biofilm effects (25,26). The AMP LL-37 is the only member of the Cathelicidin family that is produced by epithelial cells, macrophages, neutrophils, and lymphocytes in the human body (27,28). LL-37 has been shown to exhibit a broad-spectrum of antimicrobial and immunomodulatory activities, and promotes damage repair and other restorative functions. It is also involved in important physiological functions including cell proliferation, invasion, cell cycle regulation, and apoptosis. LL-37 is highly expressed at barrier sites, serving as an important first-line defense against pathogenic microorganisms (29). Like other AMPs, the primary mechanism of action is membrane disruption. Dorschner et al. (30) reported that LL-37 had an inhibitory effect on Escherichia coli, P. aeruginosa, Enterococcus, and Staphylococcus aureus (S. aureus) in vitro. LL-37 has also been found to inhibit the activity of Staphylococcus epidermidis (31), Pseudomonas (32), and Acinetobacter baumannii (33). The ability to form biofilms is a critical component of chronic P. aeruginosa infection (34,35). LL-37 can inhibit the formation of P. aeruginosa biofilms and destroy already formed biofilms, suggesting that it may be a potential therapy against chronic P. aeruginosa infection (36). Nagant et al. showed that not only LL-37 but also some of its fragments blocked the formation of a biofilm. LL-37 also affected PAO1 within an established biofilm (37). Overhage et al. (38) demonstrated using microarrays that LL-37 can inhibit biofilm formation by reducing cell adhesion, stimulating the twitching motility of bacteria, and influencing the QS system to downregulate genes essential to biofilm development. However, how LL-37 affects the QS system of P. aeruginosa to exert its antibacterial effects has not been well reported. Although several studies have provided promising evidence of the effectiveness of LL-37 in the treatment of infections, further studies are still needed. The goal of this work is to understand the influence of LL-37 on the virulence factors related to the QS of P. aeruginosa. We present the following article in accordance with the MDAR reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-617/rc).
Methods
Bacterial strains, media, reagents, and instruments
The wild-type P. aeruginosa strain PAO1 was donated by Zou Lin from Children’s Hospital of Chongqing Medical University. The double mutant PA-ΔlasI/rhII (39) strain was constructed via gene homologous recombination and verified with polymerase chain reaction (PCR) sequencing, southern blot, and high performance liquid chromatography mass spectrometry (HPLC MS). PAO1 and PA-ΔlasI/rhII were grown at 37 ℃ in Luria-Bertani broth (LB). Total RNA was extracted using RNAiso Plus reagent (Takara, Dalian, Liaoning, China). Reverse transcription was performed with the PrimeScript RT Reagent Kit (Takara, Dalian, Liaoning, China; code No. RR047A). Azithromycin (AZM) (Guangdong Drug Inspection Institute, Guangzhou, China), and real-time PCR kit (Thermo Fisher Scientific, Waltham, MA, USA) were used. LL-37 (LLGDFFRKSKEKIGKEFKRIVQRIKDFLRNLVPRTES; lot: JT69395) was purchased from Nanjing Jeptide Biotechnology Co. A Multiskan FC microplate reader (Thermo Fisher Scientific, Waltham, MA, USA) and Rotor-Gene Q real-time quantitative PCR (Thermo Fisher Scientific, Waltham, MA, USA) were used.
Determination of MIC
The MICs of LL-37 for the PAO1 and PA-ΔlasI/rhlI strains were measured using the micro broth dilution method in accordance with the Clinical and Laboratory Standards Institute (CLSI) M07-A9 guidelines: (I) MIC was measured during the logarithmic growth phase; (II) 100 μL of bacterial solution at a concentration of 1×105 CFU/mL was added to 96-well microtiter plates; (III) two-fold serial dilutions of LL-37 ranging from 1,024 to 0.25 μg/mL were tested; (IV) the final culture concentrations of LL-37 were 1,024, 512, 256, 128, 64, 32, and 16 μg/mL; (V) sterile physiological saline was used as a blank control and a bacterial solution well with no AMP was used as a negative control; (VI) the plates were incubated in a 35 ℃ carbon dioxide incubator for 18–20 h. Following the incubation period, the plates were visually evaluated for the presence of bacterial growth. The lowest concentration of LL-37 that inhibited bacterial growth was defined as the MIC value. Measurement of the MIC values of the QS inhibitor AZM was then performed in the same manner so that AZM could be used as a positive control.
P. aeruginosa growth curve
Single colonies of PAO1 and PA-ΔlasI/rhlI were inoculated in 3 mL of LB medium and cultured overnight at 37 ℃ with continuous shaking (200 r/min). Various concentrations of LL-37 were combined with a range of P. aeruginosa cultures: (I) 20 μL of bacterial culture was added into 3 mL of LB medium and cultured with continuous shaking at 37 ℃; (II) the culture was then treated with LL-37 at concentrations of 250, 125, 62.5, and 31.25 μg/mL. OD600nm values (performed in triplicate) were measured 2, 4, 6, 8, 10, 12, 14, and 16 h after treatment to draw growth curves.
The detection of exotoxin A
Exotoxin A activity was measured with the NAD (Coenzyme I) method: (I) 1mL of the bacterial solution was centrifuged at 10,000 r/min at 4 ℃ for 1 h; (II) 10 μL of the supernatant and 2 μL of NAD solution was added to each Eppendorf tube and mixed via pipetting; (III) the mixture was incubated at 25 ℃ for 30 min; (IV) in order to prepare for the corresponding solution in each measuring tube of the color development system, 0.2 mL of nitrotetrazolium blue (NTB), 0.8 mL of phenazine dimethyl ester (PMS), 0.1 mL of lactate dehydrogenase (LDH), and 1.8 mL of sodium lactate were added in advance; (V) the mixture was then placed in a water bath at 37 ℃ for 10 min; (VI) supernatant-treated NAD was then added to the corresponding test tubes of the color development system in addition to 0.2 mL of 10 mmol/L phosphate buffered saline; (VII) after mixing each of the above measuring tube solutions with the previously prepared solution, the samples were placed in a water bath at 37 ℃ for 5 min; (VIII) finally, 0.1 mL of hydrochloric acid (2 mol/L) was added to each tube to stop the reaction. The experiment was set with negative and positive controls. The negative control tube did not contain NAD, while the positive control tube contained NAD but did not include a toxin. When the OD490nm colorimetric measurement was performed, the negative control tube was used as the zero, and the color inhibition rate was calculated according to the formula RI = 100 − (OD490nm value of toxin-treated tube/OD490nm value of positive reference tube) ×100%. Three parallel experiments were performed for statistical analysis.
Elastase detection
Elastase activity was detected with the elastin-Congo red (ECR) method (40,41): (I) 1 mL of bacterial solution was centrifuged at 12,000 r/min at 4 ℃ for 1 h; (II) 100 μL of the supernatant was added into a new Eppendorf tube; (III) 900 μL of ECR and ECR buffer was added and mixed by pipetting to achieve an ECR concentration of 20 g/L; (IV) the reaction was placed in a 37 ℃ incubator for 16 h; (V) the reaction was stopped by adding 100 μL of ethylenediaminetetraacetic acid (EDTA), then centrifuging at 10,000 r/min at 4 ℃ for 20 min. OD495nm absorbance was measured with a microplate reader. LB medium was used as a negative control. All experiments were performed in triplicate.
Pyocyanin detection
Pyocyanin detection was performed using the chloroform extraction method (42): (I) 5 mL of bacterial solution was centrifuged at 5,000 r/min at 4 ℃ for 1 h; (II) 3 mL of chloroform was added to 3 mL of the supernatant for extraction; (III) the blue layer was re-extracted into 1 mL of 0.2 mol/L hydrochloric acid for the extract, yielding a red layer on the top. Absorbance was measured at 520 nm with a microplate reader, and the concentration of pyocyanin in the absence and presence of LL-37 was recorded.
Quantitative PCR (qPCR)
The RNA of P. aeruginosa strains was extracted for qPCR analysis according to the RNA reagent instructions (Promega, Madison, WI, USA): (I) 1 μg of the extracted RNA was reverse transcribed into cDNA with a reverse transcription reagent kit (TaKaRa, Dalian, Liaoning, China); (II) following the instructions of the SYBR Green qPCR Master Mix (Thermo Fisher Scientific, Waltham, MA, USA), relative real-time PCR was performed with the ABI ViiATM 7Dx system (Applied Biosystems, Foster, CA, USA) to detect cDNA; (III) the housekeeping gene ribosomal protein rpoD was used as an internal control for normalization. Relative changes in gene expression levels were calculated with the 2−ΔΔCt method. Primer sequences are shown in Table 1.
Table 1
| Gene | Forward (5'-3') | Reverse (5'-3') |
|---|---|---|
| lasA | CTGCTGGCTTTCAAGGTTTC | CCAGCAAGACGAAGAGGAAC |
| lasI | CGTGCTCAAGTGTTCAAGGA | AAAACCTGGGCTTCAGGAGT |
| rhlA | AGCTGGGACGAATACACCAC | GACTCCAGGTCGAGGAAATG |
| rhlB | GAGCGACGAACTGACCTACC | CGTACTTCTCGTGAGCGATG |
| toxA | TGCTGCACTACTCCATGGTC | ACACCTTGATGTTCGAAGGC |
| rpoD | CTGATCCAGGAAGGCAACAT | TGAGCTTGTTGATCGTCTCG |
Statistical analysis
All treatments were performed in triplicate to confirm reproducibility. SPSS 16.0 (IBM, Armonk, NY, USA) statistical software was used for analysis. Data are expressed as mean ± standard deviation. Comparisons between two groups were performed using grouped t-tests. Comparisons between three groups were performed using one-way ANOVA. Differences were considered statistically significant at P<0.05.
Results
Minimum inhibitory concentration (MIC) of LL-37 against P. aeruginosa
A sub-minimum inhibitory concentration (sub-MIC) was used to test the effect of LL-37 on the virulence factors regulated by the QS system. The wells exposed to LL-37 concentrations of 1,024, 512, and 256 μg/mL were transparent, while turbidity appeared in wells treated with a concentration of 128 μg/mL, as shown in Table 2. The MIC of LL-37 against both PAO1 and PA-ΔlasI/rhII was therefore determined to be 256 μg/mL. As a positive control, the MIC of azithromycin (AZM) was 128 μg/mL, and the concentration of AZM used against all of the strains in this study was 2 μg/mL.
Table 2
| Group | 1 | 2 | 3 | 4 | 5 | 6 | 7 | Negative control | Blank |
|---|---|---|---|---|---|---|---|---|---|
| LL-37 concentration (μg/mL) | 1,024 | 512 | 256 | 128 | 64 | 32 | 16 | 0 | 0 |
| PAO1 | − | − | − | + | + | + | + | + | − |
| PA-ΔlasI/rhII | − | − | − | + | + | + | + | + | − |
“+”: turbidity and bacterial growth; “−”: clear fluid and bacterial inhibition. PAO1, wild-type P. aeruginosa strain; PA-ΔlasI/rhII, double mutant P. aeruginosa strain; P. aeruginosa, Pseudomonas aeruginosa; Negative control, untreated group; Blank, sterile saline.
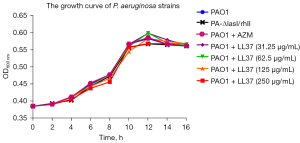
The growth curves of P. aeruginosa strains in the setting of sub-MIC LL-37
Four different concentration levels of LL-37 sub-MIC were selected as the treatment group, and PAO1, PA-ΔlasI/rhII, and PAO1 + AZM without LL-37 were used as controls. Since AZM is a QS inhibitor, it was used as a positive control. A concentration of 2 μg/mL of AZM did not affect the growth of the strains. PA-ΔlasI/rhII had a complete deletion of the coding domain. As shown in Figure 1, there were no significant differences in proliferation between the groups at any timepoint (P>0.05).
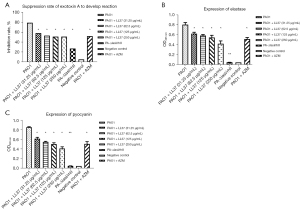
Influence of LL-37 on the expression of P. aeruginosa virulence factors
To confirm the expression of P. aeruginosa virulence factors, we examined exotoxin A, elastase, and pyocyanin expression induced by LL-37. As shown in Figure 2, the expression of exotoxin A, elastase, and pyocyanin was significantly reduced in the PA-ΔlasI/rhII strain compared with the wild-type PAO1 strain (P<0.05). There were no significant differences between the PA-ΔlasI/rhII strain and the negative control group. Notably, all of the different concentrations of LL-37 influenced the expression of exotoxin A, elastase, and pyocyanin, as shown in Figure 2A-2C. After interference with LL-37, the PAO1 strain exposed to a different sub-MIC concentration of LL-37 showed a significant decrease in exotoxin A, elastase, and pyocyanin expression compared with the PAO1 control group (P<0.05). With increasing concentrations of LL-37, reduced activity of exotoxin A, elastase, and pyocyanin were again demonstrated, but differences between the groups were not significant (P>0.05).
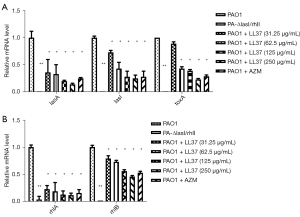
Influence of LL-37 on the expression of PAO1 virulence genes
The expression of the QS-related virulence genes of P. aeruginosa were significantly decreased compared with the PAO1 strain at a sub-MIC concentration of LL-37 (Figure 3A,3B). With increased LL-37 concentrations, the expression levels of lasA, lasI, toxA, rhlA, and rhlB genes significantly decreased (P<0.05) to a degree that was consistent with the expression of virulence factors at sub-MIC concentrations of LL-37. Low QS-related gene expression of the PA-ΔlasI/rhII group further suggested that a double gene deletion strain model was successfully constructed (P<0.01).
Discussion
This study sought to understand whether the antibacterial effects of LL-37 are through the regulation of virulence factors associated with the QS system. Human cathelicidin was found to be more potent than conventional antibiotics at killing extra- and intracellular S. aureus (43). LL-37 was previously found to inhibit the inflammatory pathway by reducing the release of IL-6 and TNF-α and inhibiting the phosphorylation of Akt and JNK (44). Furthermore, Uhlmann et al. showed that LL-37 affected the virulence factors of Streptococcus pyogenes (45). It was therefore of interest to determine whether the potency of LL-37 translated to the treatment of P. aeruginosa, which commonly develops multidrug resistant strains and would therefore benefit from novel adjunct treatments.
To develop an accurate model to evaluate the impact of LL-37 on Pseudomonas function, it was necessary to confirm that sub-MIC concentrations of this vector did not affect the growth characteristics of the PA-ΔlasI/rhII and wild-type PAO1 strains. The present work showed that LL-37 at concentrations below 256 μg/mL did not affect the growth curve of P. aeruginosa in its early stages (0–16 h in culture) of proliferation. It is therefore possible to attribute changes in virulence factor expression to the effects of LL-37 alone, not altered proliferation.
Patients with P. aeruginosa have increased expression of virulence factors. Inhibiting their activity is one of the main targets for P. aeruginosa treatment. The present work suggests that LL-37 has an inhibitory effect on P. aeruginosa exotoxin A, elastase, and pyocyanin. P. aeruginosa possesses the ability to form biofilms to facilitate the colonization of medical devices, thereby contributing to the spread of drug-resistant strains in the hospital environment (46). Pyocyanin is one of the primary virulence factors responsible for biofilm formation (47). In addition to biofilm development, QS has been linked to the regulation of other physiological processes, including virulence factor production, metabolic adjustment, and host-microbe interactions (48). The expression of pyocyanin was significantly inhibited by LL-37. Mishra and Wang (49) in their study of P. aeruginosa emphasized the importance of antibiofilm peptides for early infections.
The results of the present work suggest that LL-37 plays a vital role in the antibiofilm process. It is also possible that the antibiofilm effects of LL-37 shown against P. aeruginosa may be similarly effective against other bacteria that produce biofilms (50). LL-37 significantly reduced Pseudomonas-associated QS virulence factors, and as the concentration of LL-37 increased, its absorbance gradually decreased and its toxicity was reduced. This may suggest that the anti-infective effect of LL-37 can only be achieved by inhibiting the production of P. aeruginosa virulence factors. The effects of LL-37 on other virulence factors may be of importance as well, and will be evaluated in future work.
Using PAO1 as a control group, the mRNA transcription levels of the virulence factor genes lasA, lasI, toxA, rhlA, and rhlB, which are involved in QS systems, were significantly reduced by LL-37. The transcription level of these factors also decreased with increased concentrations of LL-37, suggesting that there is a dose-dependent antibacterial effect. This change is consistent with the altered phenotype of virulence factors caused by LL-37. As these virulence factors are Las (lasA, lasI, toxA) and Rhl (rhlA, rhlB) system-related genes (51), it is possible that if both the Las and rhl cell-to-cell signaling systems are congested, P. aeruginosa may not be able to re-establish the production of cell-to-cell signaling-dependent virulence factors. This approach may efficiently reduce virulence factor production and the high death rates associated with P. aeruginosa.
In agreement with published data (52), the present work demonstrates that LL-37 can downregulate the expression of P. aeruginosa QS system virulence factors in vitro, and that these inhibitory effects are dose-dependent. Our study emphasizes the importance of LL-37 in the pathogenesis of P. aeruginosa. LL-37 may therefore be of clinical potential in the future.
There are several limitations to this study. For example, it only focused on a single bacterial species and tested a relatively small number of virulence factors. Future work should include S. aureus, Klebsiella pneumoniae, and other biofilm-producing bacteria. However, the experimental data presented in this work provides valuable insights into the function of LL-37, especially on P. aeruginosa.
Conclusions
In conclusion, the AMP LL-37 is a potential inhibitor of the QS system and the pathogenicity of P. aeruginosa. The newly uncovered therapeutic potential of LL-37 against this challenging pathogen merits further investigation (53,54). In-depth study of the QS system will not only help to enrich our understanding of the mechanism of P. aeruginosa resistance, but also how to optimize the antimicrobial properties of LL-37, which may improve our ability to treat bacterial resistance. Future work will seek to further evaluate the effects of sub-antimicrobial concentrations of LL-37 on other regulatory pathways under the control of the lasRI/rhlRI system, clarify the regulatory pathway that contributes to this drug’s efficacy, and identify additional new therapeutic targets for treating clinical bacterial infections.
Acknowledgments
Funding: This research was funded by grants from the Guangdong Hospital of Traditional Chinese Medicine (No. YN2016QJ13) and the Traditional Chinese Medicine Bureau of Guangdong Province (No. 20221197 and No. 20192027).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-617/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-22-617/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-617/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Mathee K, Narasimhan G, Valdes C, et al. Dynamics of Pseudomonas aeruginosa genome evolution. Proc Natl Acad Sci U S A 2008;105:3100-5. [Crossref] [PubMed]
- Silby MW, Winstanley C, Godfrey SA, et al. Pseudomonas genomes: diverse and adaptable. FEMS Microbiol Rev 2011;35:652-80. [Crossref] [PubMed]
- Gellatly SL, Hancock RE. Pseudomonas aeruginosa: new insights into pathogenesis and host defenses. Pathog Dis 2013;67:159-73. [Crossref] [PubMed]
- Balasubramanian D, Schneper L, Kumari H, et al. A dynamic and intricate regulatory network determines Pseudomonas aeruginosa virulence. Nucleic Acids Res 2013;41:1-20. [Crossref] [PubMed]
- Fuqua C, Parsek MR, Greenberg EP. Regulation of gene expression by cell-to-cell communication: acyl-homoserine lactone quorum sensing. Annu Rev Genet 2001;35:439-68. [Crossref] [PubMed]
- Smith RS, Harris SG, Phipps R, et al. The Pseudomonas aeruginosa quorum-sensing molecule N-(3-oxododecanoyl)homoserine lactone contributes to virulence and induces inflammation in vivo. J Bacteriol 2002;184:1132-9. [Crossref] [PubMed]
- Schuster M, Lostroh CP, Ogi T, et al. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J Bacteriol 2003;185:2066-79. [Crossref] [PubMed]
- Glessner A, Smith RS, Iglewski BH, et al. Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of twitching motility. J Bacteriol 1999;181:1623-9. [Crossref] [PubMed]
- Müller MM, Hörmann B, Syldatk C, et al. Pseudomonas aeruginosa PAO1 as a model for rhamnolipid production in bioreactor systems. Appl Microbiol Biotechnol 2010;87:167-74. [Crossref] [PubMed]
- Lee J, Zhang L. The hierarchy quorum sensing network in Pseudomonas aeruginosa. Protein Cell 2015;6:26-41. [Crossref] [PubMed]
- Lu Y, Zeng J, Wu B, et al. Quorum sensing N-acyl homoserine lactones-SdiA suppresses Escherichia coli-Pseudomonas aeruginosa conjugation through inhibiting traI expression. Front Cell Infect Microbiol 2017;7:7. [Crossref] [PubMed]
- Déziel E, Lépine F, Milot S, et al. Analysis of Pseudomonas aeruginosa 4-hydroxy-2-alkylquinolines (HAQs) reveals a role for 4-hydroxy-2-heptylquinoline in cell-to-cell communication. Proc Natl Acad Sci U S A 2004;101:1339-44. [Crossref] [PubMed]
- Das MC, Sandhu P, Gupta P, et al. Attenuation of Pseudomonas aeruginosa biofilm formation by Vitexin: A combinatorial study with azithromycin and gentamicin. Sci Rep 2016;6:23347. [Crossref] [PubMed]
- Chen R, Déziel E, Groleau MC, et al. Social cheating in a Pseudomonas aeruginosa quorum-sensing variant. Proc Natl Acad Sci U S A 2019;116:7021-6. [Crossref] [PubMed]
- Smith RS, Iglewski BH P.. aeruginosa quorum-sensing systems and virulence. Curr Opin Microbiol 2003;6:56-60. [Crossref] [PubMed]
- Schuster M, Greenberg EP. A network of networks: quorum-sensing gene regulation in Pseudomonas aeruginosa. Int J Med Microbiol 2006;296:73-81. [Crossref] [PubMed]
- McKnight SL, Iglewski BH, Pesci EC. The Pseudomonas quinolone signal regulates rhl quorum sensing in Pseudomonas aeruginosa. J Bacteriol 2000;182:2702-8. [Crossref] [PubMed]
- Kostylev M, Kim DY, Smalley NE, et al. Evolution of the Pseudomonas aeruginosa quorum-sensing hierarchy. Proc Natl Acad Sci U S A 2019;116:7027-32. [Crossref] [PubMed]
- Mayer-Hamblett N, Rosenfeld M, Gibson RL, et al. Pseudomonas aeruginosa in vitro phenotypes distinguish cystic fibrosis infection stages and outcomes. Am J Respir Crit Care Med 2014;190:289-97. [PubMed]
- Ahmed AA, Salih FA. Low concentrations of local honey modulate Exotoxin A expression, and quorum sensing related virulence in drug-resistant Pseudomonas aeruginosa recovered from infected burn wounds. Iran J Basic Med Sci 2019;22:568-75. [PubMed]
- Wang H, Chu W, Ye C, et al. Chlorogenic acid attenuates virulence factors and pathogenicity of Pseudomonas aeruginosa by regulating quorum sensing. Appl Microbiol Biotechnol 2019;103:903-15. [Crossref] [PubMed]
- Kuang Z, Hao Y, Walling BE, et al. Pseudomonas aeruginosa elastase provides an escape from phagocytosis by degrading the pulmonary surfactant protein-A. PLoS One 2011;6:e27091. [Crossref] [PubMed]
- Lau GW, Hassett DJ, Ran H, et al. The role of pyocyanin in Pseudomonas aeruginosa infection. Trends Mol Med 2004;10:599-606. [Crossref] [PubMed]
- Fila G, Kawiak A, Grinholc MS. Blue light treatment of Pseudomonas aeruginosa: Strong bactericidal activity, synergism with antibiotics and inactivation of virulence factors. Virulence 2017;8:938-58. [Crossref] [PubMed]
- Tecle T, Tripathi S, Hartshorn KL. Review: Defensins and cathelicidins in lung immunity. Innate Immun 2010;16:151-9. [Crossref] [PubMed]
- Bowdish DM, Davidson DJ, Scott MG, et al. Immunomodulatory activities of small host defense peptides. Antimicrob Agents Chemother 2005;49:1727-32. [Crossref] [PubMed]
- Brown KL, Poon GF, Birkenhead D, et al. Host defense peptide LL-37 selectively reduces proinflammatory macrophage responses. J Immunol 2011;186:5497-505. [Crossref] [PubMed]
- Jacobsen AS, Jenssen H. Human cathelicidin LL-37 prevents bacterial biofilm formation. Future Med Chem 2012;4:1587-99. [Crossref] [PubMed]
- D'Aldebert E, Biyeyeme Bi Mve MJ, Mergey M, et al. Bile salts control the antimicrobial peptide cathelicidin through nuclear receptors in the human biliary epithelium. Gastroenterology 2009;136:1435-43. [Crossref] [PubMed]
- Dorschner RA, Pestonjamasp VK, Tamakuwala S, et al. Cutaneous injury induces the release of cathelicidin anti-microbial peptides active against group A Streptococcus. J Invest Dermatol 2001;117:91-7. [Crossref] [PubMed]
- Hell E, Giske CG, Nelson A, et al. Human cathelicidin peptide LL37 inhibits both attachment capability and biofilm formation of Staphylococcus epidermidis. Lett Appl Microbiol 2010;50:211-5. [Crossref] [PubMed]
- Dosler S, Karaaslan E. Inhibition and destruction of Pseudomonas aeruginosa biofilms by antibiotics and antimicrobial peptides. Peptides 2014;62:32-7. [Crossref] [PubMed]
- Lin MF, Tsai PW, Chen JY, et al. OmpA binding mediates the effect of antimicrobial peptide LL-37 on Acinetobacter baumannii. PLoS One 2015;10:e0141107. [Crossref] [PubMed]
- Singh PK, Schaefer AL, Parsek MR, et al. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 2000;407:762-4. [Crossref] [PubMed]
- Høiby N, Krogh Johansen H, Moser C, et al. Pseudomonas aeruginosa and the in vitro and in vivo biofilm mode of growth. Microbes Infect 2001;3:23-35. [Crossref] [PubMed]
- Pompilio A, Scocchi M, Pomponio S, et al. Antibacterial and anti-biofilm effects of cathelicidin peptides against pathogens isolated from cystic fibrosis patients. Peptides 2011;32:1807-14. [Crossref] [PubMed]
- Nagant C, Pitts B, Nazmi K, et al. Identification of peptides derived from the human antimicrobial peptide LL-37 active against biofilms formed by Pseudomonas aeruginosa using a library of truncated fragments. Antimicrob Agents Chemother 2012;56:5698-708. [Crossref] [PubMed]
- Overhage J, Campisano A, Bains M, et al. Human host defense peptide LL-37 prevents bacterial biofilm formation. Infect Immun 2008;76:4176-82. [Crossref] [PubMed]
- Zeng J, Zhang N, Huang B, et al. Mechanism of azithromycin inhibition of HSL synthesis in Pseudomonas aeruginosa. Sci Rep 2016;6:24299. [Crossref] [PubMed]
- Ohman DE, Cryz SJ, Iglewski BH. Isolation and characterization of Pseudomonas aeruginosa PAO mutant that produces altered elastase. J Bacteriol 1980;142:836-42. [Crossref] [PubMed]
- Rudrappa T, Bais HP. Curcumin, a known phenolic from Curcuma longa, attenuates the virulence of Pseudomonas aeruginosa PAO1 in whole plant and animal pathogenicity models. J Agric Food Chem 2008;56:1955-62. [Crossref] [PubMed]
- Essar DW, Eberly L, Hadero A, et al. Identification and characterization of genes for a second anthranilate synthase in Pseudomonas aeruginosa: interchangeability of the two anthranilate synthases and evolutionary implications. J Bacteriol 1990;172:884-900. [Crossref] [PubMed]
- Noore J, Noore A, Li B. Cationic antimicrobial peptide LL-37 is effective against both extra- and intracellular Staphylococcus aureus. Antimicrob Agents Chemother 2013;57:1283-90. [Crossref] [PubMed]
- Hou M, Zhang N, Yang J, et al. Antimicrobial peptide LL-37 and IDR-1 ameliorate MRSA pneumonia in vivo. Cell Physiol Biochem 2013;32:614-23. [Crossref] [PubMed]
- Uhlmann J, Rohde M, Siemens N, et al. LL-37 triggers formation of Streptococcus pyogenes extracellular vesicle-like structures with immune stimulatory properties. J Innate Immun 2016;8:243-57. [Crossref] [PubMed]
- Mulcahy LR, Isabella VM, Lewis K. Pseudomonas aeruginosa biofilms in disease. Microb Ecol 2014;68:1-12. [Crossref] [PubMed]
- Niemirowicz K, Piktel E, Wilczewska AZ, et al. Core-shell magnetic nanoparticles display synergistic antibacterial effects against Pseudomonas aeruginosa and Staphylococcus aureus when combined with cathelicidin LL-37 or selected ceragenins. Int J Nanomedicine 2016;11:5443-55. [Crossref] [PubMed]
- Moradali MF, Ghods S, Rehm BH. Pseudomonas aeruginosa lifestyle: A paradigm for adaptation, survival, and persistence. Front Cell Infect Microbiol 2017;7:39. [Crossref] [PubMed]
- Mishra B, Wang G. Individual and combined effects of engineered peptides and antibiotics on Pseudomonas aeruginosa biofilms. Pharmaceuticals (Basel) 2017;10:58. [Crossref] [PubMed]
- Neshani A, Zare H, Eidgahi MA, et al. LL-37: Review of antimicrobial profile against sensitive and antibiotic-resistant human bacterial pathogens. Gene Reports 2019;17:100519. [Crossref]
- Das T, Kutty SK, Tavallaie R, et al. Phenazine virulence factor binding to extracellular DNA is important for Pseudomonas aeruginosa biofilm formation. Sci Rep 2015;5:8398. [Crossref] [PubMed]
- Dong L, Pang J, Wang X, et al. Mechanism of pyocyanin abolishment caused by mvaT mvaU double knockout in Pseudomonas aeruginosa PAO1. Virulence 2019;11:57-67. [Crossref] [PubMed]
- Haney EF, Straus SK, Hancock REW. Reassessing the host defense peptide landscape. Front Chem 2019;7:43. [Crossref] [PubMed]
- Chen K, Gong W, Huang J, et al. The potentials of short fragments of human anti-microbial peptide LL-37 as a novel therapeutic modality for diseases. Front Biosci 2021;26:1362-72. [Crossref] [PubMed]
(English Language Editor: C. Betlazar-Maseh)


