
A dynamic model for individualized prognosis prediction in patients with avian influenza A H7N9
Introduction
In March 2013, the first case of human avian influenza A (H7N9) was reported by the World Health Organization (1). Ongoing epidemics meant that by March 28, 2018, this infectious disease had resulted in 1,625 confirmed cases and 623 deaths worldwide (2).
In the early stages, H7N9 infection clinically manifests as fever and cough with sputum production. The infection progresses rapidly in patients, leading to severe respiratory illness or multisystem organ failure (3). The overall fatality rate is high, with approximately 40% of diagnoses resulting in death (2). Timely diagnosis and effective treatment are crucial to decreasing the case fatality rate.
Previous studies have found that biomarkers, such as the oxygenation index (OI), neutrophil (NEUT), C-reactive protein (CRP), white blood cell count (WBC), cytokines, plasma angiotensin II, and human leukocyte antigen-DR (HLA-DR) levels of CD14+ cells, play an essential role in H7N9 progression and are independent predictors of survival outcome (4-10). However, measurements in these studies were collected at specific time points, either at baseline, at the end of follow-up, or when values peaked during follow-up. Taking measurements in this way fails to consider the dynamic process of laboratory examinations during hospitalization and may have led to biased estimations.
Dynamic risk prediction uses longitudinal data, which increases the accuracy of the predictions (7). With a dynamic risk prediction model, it is possible to predict the conditional survival probability of individuals, as the model considers changes in risk over time, allowing for the selection of appropriate treatments.
The present study analyzed the clinical data of 96 patients with H7N9 infection from Jiangsu, with the aims of developing a dynamic risk prediction model based on the data and identifying predictors associated with H7N9 progression. A random forest incorporating demographic data, baseline characteristics, and laboratory measurements was used to screen for candidate features. A joint model was then constructed to characterize the time-varying effects of those features and to predict the time-to-event duration in patients with H7N9.
The following article is presented in accordance with the STROBE (Strengthening the Reporting of OBservational studies in Epidemiology) reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-21-4126/rc).
Methods
Study population and design
For this study, patients with H7N9 infection were recruited from hospitals in Suzhou, Wuxi, Huai’an, and Taizhou in Jiangsu, China, between January 2016 and May 2017. The epidemiological investigation of patients was performed based on a demographic and epidemiological survey on human infections with the H7N9 virus. Demographic and clinical data for each laboratory-confirmed patient were collected by epidemiological field investigation teams from the Jiangsu Provincial Center for Disease Control and Prevention (Jiangsu CDC). Patients’ demographic data included the age, sex, body mass index (BMI), and history of chronic diseases. Clinical data were obtained from the healthcare facilities and included status at illness onset (including symptoms and initial lung infection on admission and at the time of onset), H7N9 diagnosis and treatment, laboratory testing during hospitalization (such as routine blood tests, blood biochemistry tests, and blood gas analysis), and clinical outcomes (death or recovery). All the data in this study were acquired from questionnaires and official case investigation reports. The accuracy of the data was further verified before their inclusion in an electronic database.
Confirmation and outcome
For patients with severe bilateral pneumonia, leukopenia, and lymphocytopenia whose upper respiratory tract specimens (pharyngeal swabs) or deep respiratory tract specimens (sputum or bronchoalveolar lavage fluid) tested positive for the nucleic acid of the H7N9 virus, specimens were collected for quantitative PCR. If the H7N9 virus strain could be isolated from the specimens, H7N9 infection was confirmed. Death during hospitalization was the primary endpoint for this study. Recovery and discharge from hospital were considered as the censored outcome. The follow-up time was defined as the interval from hospital admission to either death or discharge.
Ethical statement
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The ethics review committee of the Jiangsu Provincial Center for Disease Control and Prevention confirmed that this work was routinely performed by infectious disease surveillance personnel and did not require ethical review. All data in this study were de-identified, and no patient informed consent was required due to the study’s retrospective nature.
Statistical analysis
Normally distributed continuous variables were expressed as the mean and standard deviation (mean ± SD), while abnormally distributed continuous variables were shown as the median and interquartile range (median, IQR). Comparisons between continuous variables were made using Wilcoxon rank-sum and t-tests. Categorical variables were described by frequency (%) and compared between survivors and non-survivors when appropriate using a Chi-square or Fisher’s exact test. The K-nearest neighboring method (KNN) was used to impute the missing laboratory values during hospitalization using the R package DMwR2 (available at https://mirrors.tuna.tsinghua.edu.cn/CRAN/) (11).
Feature selection
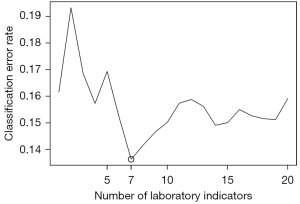
For each patient, abnormal percentages and the proportion of abnormal times to total measurement times during hospitalization were calculated for each of the biomarkers. The significance of baseline characteristics and the abnormal percentages of biomarkers during hospitalization were calculated using a random forest, which is an ensemble and decision-driven learning method (12). The Gini importance of each predictor was obtained from the random forest, and then the sliding windows sequential forward feature selection (SWSFS) algorithm was used to determine the number of significant biomarkers (7). With the demographic and baseline characteristics frozen in the model, the SWSFS algorithm added the biomarkers one by one to the random forest model in the order of their Gini importance. The error rates from 300 random forests, which included identical predictors, were averaged to plot the average classification error rate. The average classification error rate measured the performance of each model consisting of a different number of biomarkers and identified a subset of indicators with the lowest classification error rates for further analysis.
Time-dependent coefficients for biomarkers
This study aimed to examine whether the effects of the biomarkers varied during the course of hospitalization. Therefore, the time-varying effects of the biomarkers were estimated to validate the establishment of
a constant, in the Cox model equation:
Joint model construction
Joint modeling is used to investigate how a marker (such as a biomarker) that is repeatedly measured in time is associated with the time to an event of intewrest (13). The joint model in this study consisted of two sub-models: a linear mixed model that obtained the trajectory of predictors and a Cox regression model with a penalized spline risk function that estimated the hazard ratio. Parameters were obtained from the respective posterior distributions under the multivariate joint model using the JMbayes package in R (available at https://mirrors.tuna.tsinghua.edu.cn/CRAN/web/packages/JMbayes/index.html) (14). Also, a multivariate joint model was used to estimate the survival probability distribution for each patient (15). Covariates, adjusted in the joint model, included baseline age, sex, BMI, previous cardiovascular disease history, and the presence or absence of complications.
Assessment of accuracy
The performance of the joint model at different time points during hospitalization was determined based on the time-dependent area under the receiver-operator characteristic curve (AUC) (13). A time-dependent AUC aligns individuals to a common start time and compares them at a fixed follow-up window . The time-dependent AUC at with the fixed window of is defined as the probability of concordance by which the model assigns a higher survival probability to the participant who did not have the event of interest within the fixed follow-up window (16,17). Further, a 3-fold internal cross-validation procedure was performed 10 times for the AUC estimations to ensure the stability of the predictions. In this study, the calculation of the time-dependent AUCs was performed on days 6 to 15 of hospitalization, with a prediction window of 3 days.
All statistical analyses were performed using R software (version 4.0.2) (available at https://cran.r-project.org/mirrors.html). A P value of less than 0.05 was considered to be statistically significant.
Results
Baseline characteristics of the study population
The study population included 96 patients who were admitted to hospitals in Jiangsu between January 2016 and May 2017, and were laboratory confirmed as being infected with the H7N9 virus. The median follow-up time was 22 days. Of the 96 patients, 54 (56.25%) patients died of H7N9 during hospitalization, while the other 42 patients (43.75%) recovered and were discharged. The median age of the participants was 57.0 (IQR, 45.75–65.25) years. There were more males (74.0%) than females (26.0%) in the research population (Table 1). Comorbidities, including cardiovascular disease (33/96, 34.38%), metabolic diseases (14/94, 14.89%), and diabetes (12/96, 12.50%) were present in 44.21% of the cohort (Table 1). The most common symptom upon admission was cough (88.54%), followed by weakness (41.67%), muscle ache (18.75%), and pharyngalgia (15.62%) (Table 1). The baseline laboratory measurements and comparisons between survivors and non-survivors are presented in Table 2. All patients received antibiotics and antiviral treatments (Table S1). The use of corticosteroids and invasive mechanical ventilation showed no significant difference between survivors and non-survivors. Of the associated complications, respiratory failure displayed the highest prevalence (65.96%), followed by acute respiratory distress syndrome (48.94%), hepatic insufficiency (38.30%), toxic shock (29.03%), renal insufficiency (25.53%), and heart failure (12.09%). With the exception of hepatic insufficiency, the rates of complications were significantly higher in non-survivors than in survivors (Table S1).
Table 1
| Baseline characteristics | No. (%) | P value | ||
|---|---|---|---|---|
| Total (N=96) | Survivor (N=42) | Non-survivor (N=54) | ||
| Age, median (IQR) | 57.0 (45.75–65.25) | 53.5 (41.25–60.0) | 60.0 (47.25–68.0) | |
| Mean ± SD | 55.49±15.07 | 52.14±14.45 | 58.09±15.16 | 0.053 |
| Range | 21–91 | 21–89 | 25–91 | |
| ≤39 years | 16 (16.7%) | 9 (21.4%) | 7 (12.9%) | |
| 40–49 years | 16 (16.7%) | 8 (19.1%) | 8 (14.8%) | |
| 50–59 years | 24 (25.0%) | 13 (30.9%) | 11 (20.4%) | |
| 60–69 years | 25 (26.0%) | 8 (19.1%) | 17 (31.5%) | |
| ≥70 years | 15 (15.6%) | 4 (9.5%) | 11 (20.4%) | |
| Sex | 0.095 | |||
| Female | 25 (26.0%) | 15 (35.7%) | 10 (18.5%) | |
| Male | 71 (74.0%) | 27 (64.3%) | 44 (81.5%) | |
| BMI (mean ± SD) | 24.24±4.28 | 23.73±3.93 | 24.63±4.53 | 0.297 |
| Smoking status | 0.999 | |||
| Yes | 13 (13.6%) | 5 (11.9%) | 8 (14.8%) | |
| No | 56 (58.3%) | 20 (47.6%) | 36 (66.7%) | |
| Missing | 27 (28.1%) | 17 (40.5%) | 10 (18.5%) | |
| Drinking status | 0.461 | |||
| Yes | 8 (8.3%) | 4 (9.6%) | 4 (7.4%) | |
| No | 53 (55.2%) | 19 (45.2%) | 34 (63.0%) | |
| Missing | 35 (36.5%) | 19 (45.2%) | 16 (29.6%) | |
| Any comorbidity | 42/95 (44.21%) | 14/41 (34.15%) | 28 (51.85%) | 0.130 |
| Chronic lung disease | 6/94 (6.38%) | 2 (4.76%) | 4/52 (7.69%) | 0.688 |
| Chronic kidney disease | 5/95 (5.26%) | 1/41 (2.44%) | 4 (7.41%) | 0.386 |
| Chronic liver disease | 3 (3.12%) | 1 (2.38%) | 2 (3.70%) | 0.999 |
| Cardiovascular disease | 33 (34.38%) | 9 (21.43%) | 24 (44.44%) | 0.032 |
| Metabolic diseases | 14/94 (14.89%) | 5/41 (12.20%) | 9/53 (16.98%) | 0.723 |
| Diabetes | 12 (12.50%) | 5 (11.90%) | 7 (12.96%) | 0.999 |
| Signs and symptoms | ||||
| Cough | 85 (88.54%) | 39 (92.86%) | 46 (85.19%) | 0.338 |
| Pharyngalgia | 15 (15.62%) | 7 (16.67%) | 8 (14.81%) | 0.999 |
| Weak | 40 (41.67%) | 19 (45.24%) | 21 (38.89%) | 0.676 |
| Muscle ache | 18 (18.75%) | 8 (19.05%) | 10 (18.52%) | 0.999 |
Data are presented as median (IQR), mean ± SD, n (%), or n/N (%), where N is the total number of patients with available data. The P value for differences between survivors and non-survivors was tested using a t-test (continuous) or a Chi-square test (categorical). IQR, interquartile range; SD, standard deviation; BMI, body mass index.
Table 2
| Biomarker | Survivors (N=42) | Non-survivors (N=54) | P value |
|---|---|---|---|
| ALT, U/L | 45.00 (32.22, 68.25) | 38.00 (28.05, 57.50) | 0.323 |
| AST, U/L | 77.00 (55.00, 110.00) | 85.00 (57.00, 127.00) | 0.580 |
| BE, mmol/L | −0.44 (−2.41, 1.22) | −2.45 (−5.08, 0.85) | 0.076 |
| BUN, μmol/L | 5.60 (3.64, 6.92) | 6.60 (5.03, 10.50) | 0.009 |
| PCO2, mmHg | 32.00 (28.25, 34.82) | 32.05 (28.58, 37.48) | 0.277 |
| CPK, U/dL | 197.50 (91.00, 492.70) | 420.20 (179.00, 702.00) | 0.069 |
| CRP, mg/L | 66.80 (31.65, 106.59) | 94.40 (36.40, 141.00) | 0.043 |
| FiO2 | 0.49 (0.35, 0.60) | 0.75 (0.53, 1.00) | <0.001 |
| Lac, mmol/L | 1.50 (1.00, 1.89) | 1.85 (1.20, 2.70) | 0.027 |
| LDH, U/L | 766.5 (520.0, 1,072.0) | 726.0 (528.0, 1,230.0) | 0.831 |
| PCT, ng/mL | 0.27 (0.16, 0.80) | 1.10 (0.34, 3.04) | 0.002 |
| PH | 7.46 (7.43, 7.49) | 7.44 (7.40, 7.47) | 0.014 |
| PaO2, mmHg | 65.75 (55.08, 77.25) | 53.95 (44.55, 62.20) | 0.003 |
| SCr, μmol/L | 66.00 (53.20, 89.30) | 84.30 (61.30, 113.43) | 0.060 |
| SaO2, % | 94.10 (90.07, 97.10) | 87.10 (79.65, 95.00) | 0.001 |
| MONO, ×109 per L | 0.13 (0.06, 0.28) | 0.19 (0.10, 0.34) | 0.325 |
| RR, per min | 24.00 (20.00, 28.00) | 25.00 (20.00, 30.00) | 0.512 |
| LYMPH, ×109 per L | 0.52 (0.34, 0.71) | 0.41 (0.23, 0.59) | 0.017 |
| WBC, ×109 per L | 3.76 (2.91, 6.38) | 4.86 (2.71, 7.66) | 0.306 |
| Hr, per min | 90.00 (82.00, 98.00) | 96 (82.50, 111.25) | 0.131 |
| OI | 167.56 (119.40, 206.96) | 70.86 (52.80, 99.84) | <0.001 |
| NEUT, ×109 per L | 3.00 (2.25, 5.50) | 4.18 (2.23, 7.11) | 0.091 |
Data are presented as median (IQR), and P values were calculated using a Wilcoxon rank sum test. ALT, alanine aminotransferase; AST, aspartate aminotransferase; BE, base excess; BUN, blood urea nitrogen; PCO2, partial pressure of carbon dioxide; CPK, creatine phosphokinase; CRP, C-reactive protein; FiO2, fraction of inspired oxygen; Lac, blood lactic acid; LDH, lactate dehydrogenase; PCT, procalcitonin; PH, potential of hydrogen; PaO2, arterial oxygen partial pressure; SCr, serum creatinine; SaO2, oxygen saturation; MONO, monocyte count; RR, respiratory rate; LYMPH, lymphocyte count; WBC, white blood cell count; Hr, heart rate; OI, oxygenation index; NEUT, neutrophils; IQR, interquartile range.
Feature selection
In this study, 22 biomarkers were measured repeatedly during hospitalization. Among the biomarkers, lactic acid and the OI had high missing rates (26% and 43%, respectively), and were therefore excluded from further analyses. A total of 59 patients with complete clinical covariate data and abnormal biomarker percentages were included in the random forest model for feature selection. Details of the abnormal biomarker percentages are given in online available: https://cdn.amegroups.cn/static/public/atm-21-4126-01.pdf. The SWSFS algorithm identified 7 top biomarkers that exhibited minimal classification error rates: CRP, blood urea nitrogen (BUN), procalcitonin (PCT), base excess (BE), lymphocyte count (LYMPH), WBC, and creatine phosphokinase (CPK) (Figure 1).
Time-varying effects
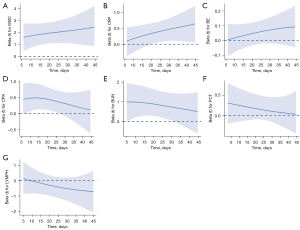
The time-varying effects of the 7 selected biomarkers were described in Bayesian P-splines after adjusting for age, sex, BMI, previous cardiovascular disease history, and presence of complications. WBC, CRP, CPK, and BUN were significantly associated with the risk of death during hospitalization (Figure 2): WBC and CRP were associated with an increased risk of death, whereas CPK and BUN were associated with a decrease in the risk of death over time.
Joint model estimates and assessment
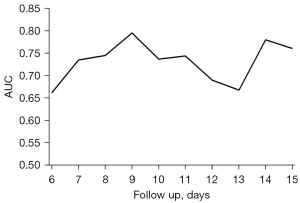
The multivariate joint model was constructed using all 7 selected biomarkers combined with 5 baseline covariates: age, sex, BMI, previous cardiovascular disease history, and presence of complications. With a 3-day prediction window, the accuracy of the joint model at day 6 to 15 of hospitalization was shown by the time-dependent AUCs with 3-fold internal cross-validation (Figure 3). A stable trend was observed in the AUCs, which ranged from 0.66 to 0.80, remaining stable at about 0.75, and reaching 0.80 when the longitudinal data were collected in the first 9 days of hospitalization and used to predict the time to event for the patients in the following 3 days.
Individualized predictions
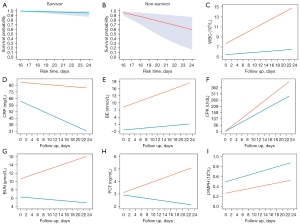
Survival probabilities for each participant were predicted using the posterior means for the fixed and random effects from the linear mixed model. Specifically, the longitudinal measurements collected during the first two-thirds of a patient’s hospitalization were considered as prior information and were used to predict the survival probability in the final third of the patient’s hospitalization. The dynamic survival predictions of two patients (one survivor and one non-survivor) in the final third of hospitalization are shown in Figure 4A,4B. The predicted longitudinal measurements during hospitalization were characterized using smooth curves (Figure 4C-4I). Certain biomarkers showed clear differences, especially in the later period. For instance, the expression levels of PCT and BUN decreased in the surviving patient but increased in the patient who died.
Discussion
H7N9 is a severe disease that can lead to comorbidities, such as severe hypoxemia, tachypnea, and respiratory failure (3). Although H7N9 infection is curable, its mortality rate is still high (7). Therefore, an accurate risk prediction model for prognosis is needed to decrease the case fatality rate. Some studies have focused on the development of prediction models for survival probabilities using clinical data (4,6). However, no studies have collected repeated laboratory test measurements during hospitalization for their predictions, the accuracy of which has, consequently, not been optimum.
In this study, the random forest machine learning technique was used to identify biomarkers that may predict survival progression for patients with H7N9. Bayesian P-splines were used to show the time-varying effects of these potential predictors. Further, a dynamic risk prediction model was developed to provide simple and precise personalized predictions of H7N9 survival, which may assist in individualized medical supervision and treatment decisions.
The multivariate joint model of our study had certain advantages. This dynamic model contained the baseline characteristics of the study participants and repeated measurements of their biomarkers, which increased the reliability of the predictions. The performance of our model was clinically satisfactory, and the AUC reached up to 0.8 on day 9 of hospitalization, with a 3-day prediction window. The model was cost-efficient and showed high accuracy in predicting the prognosis of patients with H7N9 infection. Additionally, the multivariate joint model demonstrated better individualized identification than general forecasting models (7,18,19), because it considered the impact of time-dependent effects and repeated measurements. To facilitate the application of our dynamic prediction model, we developed an online tool, which can be accessed online (available at http://218.2.247.110:19040/H7N9/). With the relevant longitudinal data of patients, the tool was able to output survival probabilities within a specific time frame.
Our study identified 7 significant predictors out of the 20 observed biomarkers. Six of those 7 predictors were associated with the endpoint event, whereas 1 showed a protective effect on H7N9. A high WBC suggested inflammation. Studies have found that the WBC average and range during hospitalization increase in patients who die but decrease among survivors, suggesting that WBC indices could be used to differentiate survivor and non-survivor groups of patients with H7N9 (7). Increases in 2 other inflammatory indicators, CRP and PCT, were also an indicator of H7N9 mortality (20-22). Similarly, WBC was associated with the risk of death (Figure 2A). These results emphasise the need to monitor these indicators during, and particularly in the later period of, hospitalization. Once an increase in the expression levels of these indicators is observed, a suitable treatment strategy should be implemented immediately. Previous studies reported that H7N9 infection may have resulted in transient cardiac injury and led to an increase in CPK expression, which significantly decreased after H7N9 viral tests were returned negative (23,24). Comparable to our research (Figure 2D), a recent study on 130 patients infected with H7N9 found that CPK was an important evaluation index for the severity of pneumonia in these patients and that increased CPK levels were related to a worse prognosis (25). Interestingly, we found that another significant factor, BUN, served as an important biomarker in predicting prognosis. Increased BUN levels are the hallmark of kidney damage and are associated with a more fulminant disease or more debilitated state (26,27). BUN might be used to differentiate survivor and non-survivor groups in patients with H7N9 in the early stage of hospitalization (Figure 2E). Since greater expressions of BUN predict a worse outcome, clinicians should be cautious when the BUN levels of a patient are high at admission. Further, previous findings have proposed LYMPH as a reference index for H7N9 infection diagnosis, with low LYMPH yielding a poor outcome (28), which is consistent with the findings of our study (Figure 2G).
Our study had several strengths. First, random forest has the capacity to avoid overfitting and resisting noise interference to a certain extent. Also, random forest models can detect non-linear relationships between predictors and the outcome, which improves predictive competency. Second, dynamic laboratory changes in laboratory test results during hospitalization strongly impact the progression of the disease; measuring the biomarkers at a single time point provides only a limited reflection of their influence. Hence, our model measured longitudinal biomarkers multiple times, which effectively improved the prediction accuracy. Third, we adopted multivariate joint models to characterize the time-to-event process, and real-time prediction feedback of risk was available on an individual level. Nevertheless, several limitations need to be noted. First, missing information from the laboratory tests and consequent data imputation may have created bias. Second, unmeasured confounders and possible bias may have affected the prediction accuracy of the model. Third, the results of this study were not validated using external data. Therefore, findings may be generalized to the other studies with caution due to our limited sample size.
In conclusion, this study has characterized the time-varying effects of significant biomarkers on the prognostic progression of H7N9 and has provided real-time predictions for individual patients. Our model may serve as a valuable tool for assisting in the treatment decision-making process. Further, early identification of at-risk individuals and early intervention may reduce mortality and the incidence of other complications.
Acknowledgments
Funding: This work was supported by the National Natural Science Foundation of China (grant number 81872709 to YZ) and the Social Development Projects of Jiangsu Province (grant number E2017749 to CB).
Footnote
Reporting Checklist: The authors have completed the STROBE (Strengthening the Reporting of OBservational studies in Epidemiology) reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-21-4126/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-21-4126/dss
Peer Review File: Available at https://atm.amegroups.com/article/view/10.21037/atm-21-4126/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-21-4126/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The ethics review committee of the Jiangsu Provincial Center for Disease Control and Prevention confirmed that this work was routinely performed by infectious disease surveillance personnel and did not require ethical review. All data in this study were de-identified, and no patient informed consent was required due to the study’s retrospective nature.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Centers for Disease Control and Prevention (CDC). Emergence of avian influenza A(H7N9) virus causing severe human illness - China, February-April 2013. MMWR Morb Mortal Wkly Rep 2013;62:366-71. [PubMed]
- Sivanandy P, Zi Xien F, Woon Kit L, et al. A review on current trends in the treatment of human infection with H7N9-avian influenza A. J Infect Public Health 2019;12:153-8. [Crossref] [PubMed]
- EMERGING PATHOGENS. INFLUENZA - H7N9. Dis Mon 2017;63:251-6. [Crossref] [PubMed]
- Cheng Q, Sun Z, Zhao G, et al. Nomogram for the Individualized Prediction of Survival Among Patients with H7N9 Infection. Risk Manag Healthc Policy 2020;13:255-69. [Crossref] [PubMed]
- Zhang Y, Zou P, Gao H, et al. Neutrophil-lymphocyte ratio as an early new marker in AIV-H7N9-infected patients: a retrospective study. Ther Clin Risk Manag 2019;15:911-9. [Crossref] [PubMed]
- Martinez L, Cheng W, Wang X, et al. A Risk Classification Model to Predict Mortality Among Laboratory-Confirmed Avian Influenza A H7N9 Patients: A Population-Based Observational Cohort Study. J Infect Dis 2019;220:1780-9. [Crossref] [PubMed]
- Yang Y, Li X, Birkhead GS, et al. Clinical indices and mortality of hospitalized avian influenza A (H7N9) patients in Guangdong, China. Chin Med J (Engl) 2019;132:302-10. [Crossref] [PubMed]
- Zhou J, Guo X, Fang D, et al. Avian Influenza A (H7N9) viruses isolated from patients with mild and fatal infection differ in pathogenicity and induction of cytokines. Microb Pathog 2017;111:402-9. [Crossref] [PubMed]
- Huang F, Guo J, Zou Z, et al. Angiotensin II plasma levels are linked to disease severity and predict fatal outcomes in H7N9-infected patients. Nat Commun 2014;5:3595. [Crossref] [PubMed]
- Diao H, Cui G, Wei Y, et al. Severe H7N9 infection is associated with decreased antigen-presenting capacity of CD14+ cells. PLoS One 2014;9:e92823. [Crossref] [PubMed]
- Torgo L. Data mining with R: learning with case studies. Boca Raton, FL, USA: Chapman & Hall/CRC, 2011.
- Breiman L. Random Forests. Machine Learning 2001;45:5-32. [Crossref]
- Rizopoulos D. Dynamic predictions and prospective accuracy in joint models for longitudinal and time-to-event data. Biometrics 2011;67:819-29. [Crossref] [PubMed]
- Rizopoulos D. The R Package JMbayes for Fitting Joint Models for Longitudinal and Time-to-Event Data Using MCMC. J Stat Softw 2016;72:1-46. [Crossref]
- Andrinopoulou ER, Clancy JP, Szczesniak RD. Multivariate joint modeling to identify markers of growth and lung function decline that predict cystic fibrosis pulmonary exacerbation onset. BMC Pulm Med 2020;20:142. [Crossref] [PubMed]
- Sing T, Sander O, Beerenwinkel N, et al. ROCR: visualizing classifier performance in R. Bioinformatics 2005;21:3940-1. [Crossref] [PubMed]
- Long JD, Mills JA. Joint modeling of multivariate longitudinal data and survival data in several observational studies of Huntington's disease. BMC Med Res Methodol 2018;18:138. [Crossref] [PubMed]
- Gustafson L, Jones R, Dufour-Zavala L, et al. Expert Elicitation Provides a Rapid Alternative to Formal Case-Control Study of an H7N9 Avian Influenza Outbreak in the United States. Avian Dis 2018;62:201-9. [Crossref] [PubMed]
- Burke SA, Trock SC. Use of Influenza Risk Assessment Tool for Prepandemic Preparedness. Emerg Infect Dis 2018;24:471-7. [Crossref] [PubMed]
- Wan DM, Kang XH, Bai W, et al. Zhonghua Jie He He Hu Xi Za Zhi 2019;42:750-4. [PubMed]
- Lu S, Li T, Xi X, et al. Prognosis of 18 H7N9 avian influenza patients in Shanghai. PLoS One 2014;9:e88728. [Crossref] [PubMed]
- Yang M, Gao H, Chen J, et al. Bacterial coinfection is associated with severity of avian influenza A (H7N9), and procalcitonin is a useful marker for early diagnosis. Diagn Microbiol Infect Dis 2016;84:165-9. [Crossref] [PubMed]
- Han J, Mou Y, Yan D, et al. Transient cardiac injury during H7N9 infection. Eur J Clin Invest 2015;45:117-25. [Crossref] [PubMed]
- Yu WQ, Ding MD, Dai GH, et al. Zhonghua Jie He He Hu Xi Za Zhi 2018;41:534-8. [PubMed]
- Zheng S, Wu J, Yu F, et al. Elevation of creatine kinase is linked to disease severity and predicts fatal outcomes in H7N9 infection. Clin Chem Lab Med 2017;55:e163-6. [Crossref] [PubMed]
- Cohen O, Leibovici L, Mor F, et al. Significance of elevated levels of serum creatine phosphokinase in febrile diseases: a prospective study. Rev Infect Dis 1991;13:237-42. [Crossref] [PubMed]
- Raimann JG, Calice-Silva V, Thijssen S, et al. Saliva Urea Nitrogen Continuously Reflects Blood Urea Nitrogen after Acute Kidney Injury Diagnosis and Management: Longitudinal Observational Data from a Collaborative, International, Prospective, Multicenter Study. Blood Purif 2016;42:64-72. [Crossref] [PubMed]
- Chen Y, Li X, Tian L, et al. Dynamic behavior of lymphocyte subgroups correlates with clinical outcomes in human H7N9 infection. J Infect 2014;69:358-65. [Crossref] [PubMed]


