
Sevoflurane pretreatment regulates abnormal expression of MicroRNAs associated with spinal cord ischemia/reperfusion injury in rats
Introduction
Spinal cord ischemia/reperfusion injury (SCII) is a serious spinal injury that generally arises from ischemia in spinal cord tissues (1). Severe complications can result from SCII, including paralysis or paraplegia, with the incidence of paraplegia being as high as 14% (2). To date, there are no effective interventions for the prevention and treatment of SCII given the multiple pathological factors, including oxidative stress, blood spinal cord barrier (BSCB), inflammation, apoptosis, and autophagy involved in the development of SCII (3,4). Most importantly, there are no effective drugs or therapeutics against SCII, highlighting the serious clinical challenge facing disease management. Thus, it is imperative to explore novel drugs to manage SCII and possibly their underlying working mechanisms.
MicroRNAs (miRNAs) are small (20–22 nt) non-coding RNAs (ncRNAs) (5), widely expressed in mammalian organisms. They mainly regulate post-transcriptional expression of genes by binding to the 3’-untranslated region (3’-UTR) of messenger RNA (mRNA) of the target gene (6,7). It has been suggested that miRNAs regulate numerous biological processes such as neurogenesis, inflammation, apoptosis, and autophagy, all of which participate in the pathogenesis of central nervous system (CNS) disorders, including SCII, cerebral ischemia/reperfusion (I/R) injury, spinal cord injury (SCI), and Parkinson’s disease (8-11). Moreover, the expression profile of miRNAs has been associated with different SCII development stages (12-14).
Sevoflurane, a common inhalational anesthetic, is routinely used for both induction and maintenance of anesthesia during surgeries (15). Increasing evidence indicates that sevoflurane exhibits neuroprotective properties (15-17). Our previous study demonstrated that preconditioning with 2.4% sevoflurane ameliorated SCII by inhibiting the secretion of matrix metallopeptidase 9 (MMP-9) and microglia recruitment which together modulated downstream neuronal apoptosis and inflammatory damage to BSCB (18). In a related study, sevoflurane preconditioning prevented the rapid development of SCII in rabbits, possibly via activation of the extracellular-signal-regulated kinase (ERK) pathway (19). Numerous studies have shown that sevoflurane reduces brain ischemia injury by regulating miRNAs’ expression (20,21). However, whether sevoflurane modulates SCII by also regulating the expression of miRNAs remains to be validated.
In this study, therefore, we explored the expression profile of miRNA in SCII rats pretreated with sevoflurane. Also, target genes for the differentially expressed miRNAs (DEmiRNAs) were uncovered to dissect further mechanisms underlying the modulation of SCII. The experimental design and flowchart in this study are shown in Figure 1. We present the following article in accordance with the ARRIVE reporting checklist (available at http://dx.doi.org/10.21037/atm-20-7864).
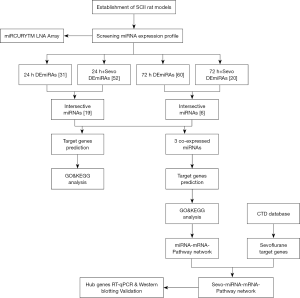
Methods
Experimental animals
Male adult Sprague-Dawley (SD) rats (200–250 g) were obtained from the Animal Center of China Medical University. All animals were housed for at least 1 week before the surgical operation. All experimental procedures were performed in line with the National Institutes of Health Guide’s guidelines for the Use and Care of Laboratory Animals (NIH Publication No.80-23, revised 1966). The Ethics Committee approved the protocol for this research of China Medical University (2020266).
Sevoflurane pretreatment and SCII rat models
Sevoflurane pretreatment was performed as previously described (18). In short, rats were exposed to a mixture of 2.4% sevoflurane, 33% oxygen, and air via spontaneous breathing through an unsealed facemask device. The end-tidal sevoflurane concentration was monitored continuously and maintained at 1 minimum alveolar concentration [(MAC) 2.4–2.7%] for 1 h, followed by 30 min washout. A cross-clamped aortic arch was applied to establish the SCII rat models as previously described (3,22) under pentobarbital sodium anesthesia (50 mg/kg) (Beyotime Biotechnology, Shanghai, China). Following 14 min of ischemia, the clips were removed, and after that, reperfusion was performed for 24 or 72 h. For the sham group, the same procedure was performed except occlusion. Before SCII induction, the SCII 24 h+sevo group and SCII 72 h+sevo groups first received sevoflurane pretreatment for 1 h.
Neurological function assessment
The neurological function of the rats was assessed at 24 or 72 h after reperfusion by independent investigators based on the Tarlov scoring system (23): here, 0 was awarded for mice with spastic paraplegia with no voluntary movement of lower limbs, 1 for spastic paraplegia and weak hind limb motor function, 2 for good antigravity strength with significant lower limb movement but the inability to stand, 3 for abnormal standing but unable to walk normally, and 4 for normal motor function.
RNA extraction and miRNA array
Total RNA of the L4-L6 segments of the spinal cord was extracted using TRIzol reagent (Takara Bio, Shiga, Japan) and the miRNeasy mini kit (Qiagen, West Sussex, UK) following the manufacturer’s instructions. The extracted RNAs were first labeled based on the miRCURYTM Hy3TM/HyTM Power labeling kit (Exiqon, Vedbaek, Denmark) and hybridized on a miRCURYTM LNA Array (version 18.0, Exiqon, Vedbaek, Denmark). After washing, the slides were scanned using an Axon GenePix 4000B microarray scanner (Axon Instruments, Foster City, CA, USA). The scanned images were imported into the GenePix Pro 6.0 platform (Axon Instruments) for grid alignments and analysis. Replicated miRNAs were averaged, and miRNAs with expression intensities ≥50 in all samples were used to calculate the normalized expression using the median normalization method. The DEmiRNAs were identified by volcano plot filtering. Finally, hierarchical clustering to identify differential expression of miRNAs among the samples was performed using the MEV software (Version 4.6; TIGR, Microarray Software Suite 4, Boston, MA, USA).
Prediction of target genes for miRNAs
The DEmiRNAs-mRNA interactions were predicted via using the miRDB (http://www.mirdb.org/), miRwalk (http://www.ma.uni-heidelberg.de/apps/zmf/mirwalk/), and TargetScan (http://www.targetscan.org/) database, with at least 2 databases included as the careening criteria.
Gene Ontology (GO) enrichment analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis
Metascape is a web-based portal designed to provide a comprehensive gene list annotation and analysis resource for experimental biologists (24). We used Metascape (http://metascape.org/) to perform GO and KEGG pathway analysis of target genes of DEmiRNAs (25).
Construction of the miRNA-mRNA-pathway network
The association among the DEmiRNAs, mRNAs, and the mRNA-related pathways were analyzed using the software Cytoscape 3.7.2 (https://cytoscape.org). Nodes of different shapes and colors represented the DEmiRNAs, mRNAs, and pathways in the network.
Screening of sevoflurane targets in SCII model and the construction of an integrative network
The key interactions among chemical-gene, chemical-disease, and gene-disease for predicting the significant associations and construction of the chemical-gene-disease networks were performed based on the comparative toxicogenomics database (CTD) (http://ctdbase.org/) (26). The same database was used to predict genes targeted by sevoflurane. The integrative network of sevo-miRNA-mRNA-pathway was then visualized using the Cytoscape 3.7.2 software.
Molecular docking
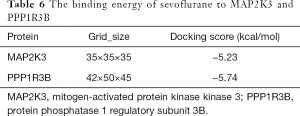
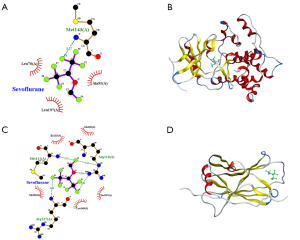
Molecular docking to identify the nature of binding between sevoflurane and its receptor proteins was performed using the AutoDock Tools (version 1.5.6, https://autodock.scripps.edu) (27). The crystal structures of target proteins, protein phosphatase 1 regulatory subunit 3B [(PPP2R3B) PDB ID: 3VN9] and MAP2K3: mitogen-activated protein kinase kinase 3 [(MAP2K3) PDB ID: 2EEF] were downloaded from the Research Collaboratory for Structural Bioinformatics Protein Data Bank (RCSB PDB) database, whereas the structures of the sevoflurane compound were obtained from the PubChem database. Before docking, the sevoflurane compound (ligands) and target proteins (receptors) were subjected to remove water molecules on the receptors, minimize energy, add polar hydrogen atoms, and set atom types (28).
Quantitative reverse transcription-polymerase chain reaction
The total RNA of the L4-L6 spinal cord section was extracted using TRIzol reagent (Takara Bio., Japan) following the manufacturer’s instructions. Complementary DNA (cDNA) was then synthesized through reverse transcription of the RNA using the Prime-Script RT reagent kit and gDNA Eraser (Takara) or MicroRNA Reverse Transcription Kit (Takara). Subsequently, a quantitative reverse transcription polymerase chain reaction (qRT-PCR) of the DNA transcript was then performed using the SYBR PremixEx TaqII Kit (Takara) in the ABI 7500 qRT-PCR system (Applied Biosystems). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) or U6 were used as the internal control. The expression levels of mRNA and miRNA were assessed based on the 2-ΔΔCt equation. The primers (Sangon Biotech, Shanghai, China) used in this study are shown in Table 1.

Full table
Western blotting
The L4-L6 segment of spinal cord tissues was lysed using radioimmunoprecipitation (RIPA) lysis buffer (R0010, Solarbio, Beijing, China) supplemented with phenylmethylsulfonyl fluoride [(PMSF) P0100, Solarbio] as previously described (9). The tissue homogenates were centrifuged at 12,000 rpm for 15 min at 4 °C to separate the lysis buffer’s proteins of interest. The protein concentrations were quantified using a bicinchoninic (BCA) protein assay kit (Beyotime Biotechnology). After that, 50 µg of total protein extracts were separated using 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE, KGP113K, KeyGen Biotech. Co. Ltd., Nanjing, China) and transferred on to polyvinylidene fluoride (PVDF) membranes (Millipore, Temecula, CA, USA). The membranes were blocked for 1.5 h using 5% nonfat dried milk dissolved in tris buffered saline with Tween 20 (TBST). The membranes were then incubated at 4 °C overnight with several primary antibodies; anti-MAP2K3 (Affinity, AF 6327; 1:1,000), anti-PPP1R3B (Abcam, ab 235049; 1:1,000), and anti-β-actin (ZSGB-BIO, China; 1:5,000). After 3 washes using TBST, the membranes were incubated for 2 h at room temperature with horseradish peroxidase (HRP)-conjugated goat anti-rabbit (Protein Tech, SA00001-2) or anti-mouse (Protein Tech, SA00001-1) secondary antibodies. Finally, proteins were then visualized using the electrochemiluminescence (ECL) kit (Bio-Rad, USA). The intensity of bands was measured using the Image Lab 5.0 software (https://www.bio-rad.com).
Statistical analysis
Data were analyzed using the Graph-Pad Prism 8 software (GraphPad Software, Inc., San Diego, CA, USA). Continuous data were expressed as mean ± standard deviation (SD). The differences between groups were analyzed using the 2-tailed Student’s unpaired t-test. Statistical significance was set at P<0.05. All experiments were performed at least 3 times.
Results
Neurological evaluation
The rats’ neurological function was evaluated at 24 and 72 h after reperfusion based on the Tarlov scores (Figure 2A). The SCII group was found to show significantly lower Tarlov scores. Compared to controls, sevoflurane pretreatment generated a markedly improved neurological function at each time point (P<0.05).
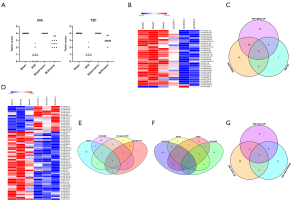
Sevoflurane pretreatment altered the expression of multiple miRNAs
Compared to sham rats, we identified 31 DEmiRNAs 24 h after SCII induction, 30 of which were downregulated, and 1 was upregulated (Figure 2B and Table 2). However, sevoflurane pretreated SCII rats (24 h) exhibited over-expression of 52 miRNAs, with no miRNA shown to be downregulated (https://cdn.amegroups.cn/static/public/atm-20-7864-1.xlsx). Among the identified DEmiRNAs, 19 (Figure 2C and Table 3) were intersected. At 72 h post-reperfusion, we uncovered 60 DEmiRNAs, 43 of which were downregulated, whereas 17 were upregulated (Figure 2D and Table 4). Compared to controls, 11 and 9 miRNAs were over and under-expressed, respectively, in SCII sevoflurane pre-treatment rats (72 h) (https://cdn.amegroups.cn/static/public/atm-20-7864-2.xlsx). Among DEmiRNAs in the above 2 analyses, 6 intersected genes (4 upregulated and 2 downregulated) were obtained (Figure 2E and Table 5). Also, there were 20 overlapped DEmiRNAs both at 24 and 72 h after reperfusion (Figure 2F). Interestingly, rno-miR-221-3p, rno-miR-181a-1-3p, and rno-miR-21-5p were all upregulated at both 24 and 72 h after sevoflurane pretreatment (Figure 2G).
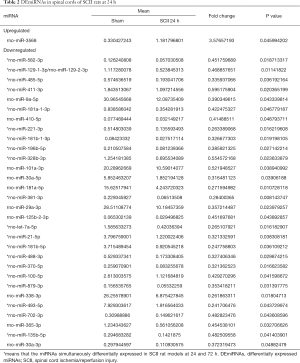
Full table
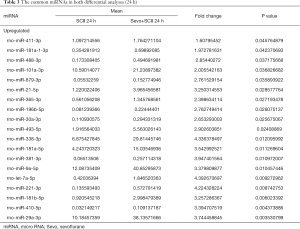
Full table
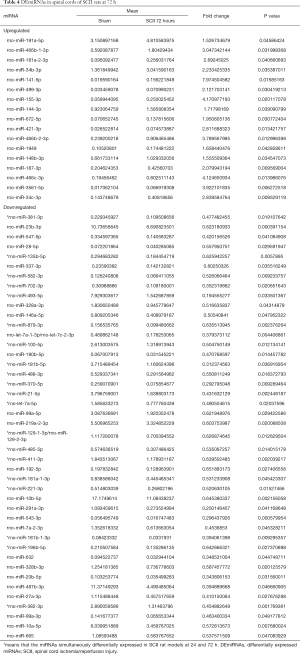
Full table
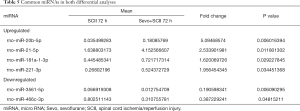
Full table
Validation of DEmiRNAs
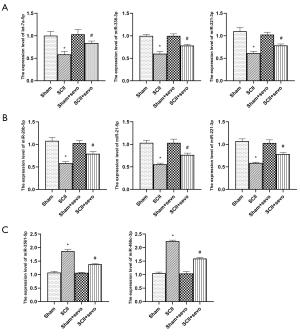
The qRT-PCR for 8 selected DEmiRNAs (top 3 upregulated miRNAs at 24 h after sevoflurane pretreatment, top 3 upregulated miRNAs at 72 h following sevoflurane pre-treatment, and 2 downregulated miRNAs at 72 h following sevoflurane pretreatment) revealed that the expression of rno-let-7a-5p, miR-338-3p, and miR-221-3p was downregulated at 24 h in SCII. However, sevoflurane pre-treatment up-regulated the expression of the 3 miRNAs. Meanwhile, the expression of miR-20b-5p, miR-21-5p, and miR-221-3p was also downregulated at 72 h in the SCII group, whereas the sevoflurane pretreated group displayed up-regulated expression of the 3 miRNAs. Furthermore, miR-3561-5p and miR-466c-3p were over-expressed in the SCII group at 72 h, but sevoflurane pre-treatment modulated the expression of these miRNAs (Figure 3). In general, qRT-PCR and miRNA array analyses revealed comparable findings. Therefore, our miRNA array profile was deemed highly reliable, revealing miRNAs strongly associated with SCII pathogenesis.
Prediction of target genes
Based on the miRDB, miRwalk, and TargetScan databases, at 24 h of sevoflurane pretreatment, the DEmiRNAs targeted 2,117 genes (https://cdn.amegroups.cn/static/public/atm-20-7864-3.xlsx), while at 72 h, the DEmiRNAs targeted 1,251 genes (https://cdn.amegroups.cn/static/public/atm-20-7864-4.xlsx). Meanwhile, the 3 DEmiRNAs over-expressed at 24, and 72 h targeted 187 genes.
Functional enrichment analysis of the DEmiRNAs target genes
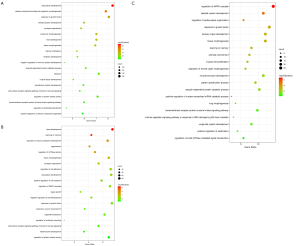
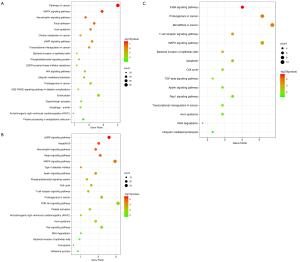
The GO enrichment analysis and KEGG pathway analysis revealed a significant difference in the activated and modulated pathways between the 24 and 72 h post sevoflurane treatment (Figures 4,5). For instance, GO analysis demonstrated that the cell surface receptor signaling pathway involved in cell-cell signaling and protein kinase activity regulation were important biological processes both at 24 and 72 h in the SCII with sevoflurane pretreatment groups (Figure 4A,B). Functional enrichment analysis of top the 20 most significant genes based on GO revealed the target genes for the 3 DEmiRNAs which were overexpressed both at 24 and 48 h post sevoflurane pretreatment mainly participated in the regulation of mitogen-activated protein kinase (MAPK) cascade, skeletal system development, and post-synapse organization and growth (Figure 4C). The KEGG pathway enrichment analysis revealed that the MAPK, neurotrophin, and cyclic adenosine monophosphate (cAMP) signaling were the most significantly upregulated pathways both at 24 and 72 h in the SCII with sevoflurane pretreatment groups (Figure 5A,B). The KEGG analysis further revealed the 3 co-upregulated DEmiRNAs at 24 and 72 h SCII post sevoflurane pretreatment participated in MAPK, ErbB, and apoptosis regulation, and transforming growth factor-beta (TGF-beta) signaling pathways (Figure 5C). These findings implied that sevoflurane regulates SCII at both 24 and 72 h via several pathways.
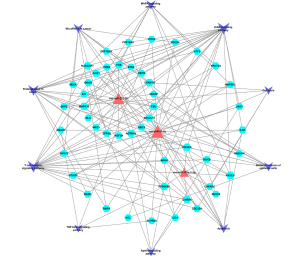
Construction of the miRNA-mRNA-pathway network
The miRNA-mRNA-pathway network (Figure 6) demonstrated a significant interconnection among miRNA and between mRNAs and multiple signaling pathways. For instance, MAPK10, FOS, and FASLG regulated the ErbB, apoptosis, MAPK, and T cell receptor signaling pathways, whereas the PIK3CA regulated the ErbB, apoptosis, and T cell receptor signaling pathways. Interestingly, MAPK10 and FOS were target genes for rno-miR-221-3p, whereas FASLG was a target for rno-miR-21-5p.
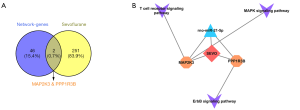
The sevo-miRNA-mRNA-pathway integrative network
Bioinformatics analysis of the CTD database identified 253 sevoflurane target genes (https://cdn.amegroups.cn/static/public/atm-20-7864-5.xlsx). Interestingly, genes regulating the expression of mitogen-activated protein kinase kinase 3 (MAP2K3) and protein phosphatase 1 regulatory subunit 3B (PPP1R3B) (Figure 7A) were all targeted by both sevoflurane and DEmiRNAs. The integrative network (Figure 7B) further revealed that MAP2K3 and PPP1R3B genes were both targets for rno-miR-21-5p and sevoflurane. Interestingly, MAP2K3 regulates the MAPK and T cell receptor signaling pathways, whereas PPP1R3B regulates the ErbB signaling pathway. These findings demonstrated that sevoflurane regulates SCII via the rno-miR-21-5p, MAP2K3, PPP1R3B, and several other key signaling pathways.
Molecular docking
The bioinformatics analysis uncovered MAP2K3 and PPP1R3B as the most critical proteins associated with SCII modulation upon sevoflurane treatment. A ligand and a protein were displayed by a ball-and-stick and a cartoon chain, respectively. The Vina score describes the strength of binding between a protein and a compound more often than not. The more negative the Vina value is, the stronger the binding between a compound and a protein (28). The strength of binding between sevoflurane and MAP2K3 as well as PPP1R3B are shown in Table 6. Here, the Vina score for the binding between sevoflurane and MAP2K3 and PPP1R3B were all negative, but less than −5, implying that sevoflurane exhibits good binding with both MAP2K3 and PPP1R3B (Figure 8).
Full table
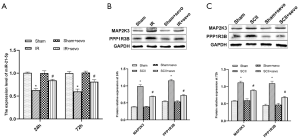
Validation of critical miRNAs and proteins in SCII
The qRT-PCR and western blot analysis revealed that compared with controls, SCII induced significant under-expression of rno-miR-21-5p but up-regulated the expression of PPP1R3B and MAP2K3 proteins. However, compared with the SCII group, the sevoflurane pretreatment group exhibited over-expression of rno-miR-21-5p but underexpression of PPP1R3B and MAP2K3 proteins (Figure 9).
Discussion
As one of the most serious spinal cord complications, SCII generally results from spinal cord injuries or thoracoabdominal aortic surgery and may lead to paralysis or paraplegia and neurologic deficits [1, 2]. However, the molecular mechanisms underlying the development of SCII are not well understood. Elucidating these processes may uncover novel targets for the prevention and treatment of SCII.
The miRNAs have been shown to regulate genes associated with the pathological mechanism of SCII (13,14,29). For example, miR-30c protects PC12 cells against oxygen-glucose deprivation (OGD)-caused apoptosis and inflammatory response and inhibits the development of SCII by modulating the expression of SIRT1 (30). Specifically, miR-21 regulates reactive astrocyte’s polarization and promotes nerites growth and synapsis formation following acute SCII (31). Meanwhile, overexpression of miR-136 ameliorates cell apoptosis induced by SCII via the TIMP3 signaling pathway (32). In this study, we uncovered 31 DEmiRNAs (30 downregulated miRNAs and 1 upregulated) after 24 h of SCII and 60 DEmiRNAs (43 downregulated and 17 upregulated) following 72 h of SCII. Interestingly, 20 miRNAs were dysregulated at both 24 and 72 h after SCII.
The protective effect of sevoflurane against SCII has been demonstrated in numerous studies (19,33). Particularly, post-treatment sevoflurane ameliorates SCII via free radicals, which up-regulates antioxidant enzymes’ expression (33). Moreover, accumulating evidence indicates that sevoflurane preconditioning might reduce ischemic injury in the brain, kidney, and heart and relieve hypoxic and ischemic injury via miRNAs (34-36). A related study revealed that sevoflurane preconditioning protected against cerebral I/R injury both in vitro and in vivo, possibly by inhibiting the expression of miR-181a while promoting that of X-linked inhibitor of apoptosis (XIAP) (34). Meanwhile, sevoflurane protects against renal and ischemic I/R injury in rats via miR-17-3p and miR-27a (35). It has also been shown that miR-374 could mitigate myocardial I/R damage in rat models following sevoflurane pretreatment by targeting SP1 via activating the PI3K/Akt pathway (36). This study found 19 and 6 miRNAs were differently expressed at 24 and 72 h, respectively, across sham, SCII, and SCII-sevoflurane pretreatment groups. Sevoflurane pretreatment reversed the abnormal expression pattern of the 19 (24 h after SCII) and 6 miRNAs (72 h after SCII) induced by SCII. This suggests that sevoflurane exerts its therapeutic effects against SCII by restoring the normal expression of miRNAs. Also, rno-miR-221-3p, rno-miR-181a-1-3p, and rno-miR-21-5p were all over-expressed both at 24 and 72 h after sevoflurane pretreatment of SCII rats. Combined with our previous study (18), these findings strengthen the hypothesis that sevoflurane modulates SCII in part by regulating the abnormal expression of certain miRNAs. The expression of the target genes of the 3 DEmiRNAs over-expressed at 24 and 72 h after SCII was upregulated and regulated in several pathways including MAPK, ErbB, Apoptosis, 'T cell receptor activation, and TGF-beta signaling pathways.
Moreover, the integrative network for the underlying sevoflurane-related protective mechanism of SCII further demonstrated that sevoflurane modulates SCII by regulating the expression of rno-miR-21-5p, MAP2K3, PPP1R3B, and key associated signaling pathways including MAPK, T cell receptor signaling, and ErbB. Molecular docking analysis, on its part, demonstrated that sevoflurane exhibits good binding activity with MAP2K3 and PPP1R3B, the most critical proteins in the development/pathogenesis of SCII. The role of miR-21-5p in the brain, spinal cord, and heart injury has been widely reported (37-39). In one study, a combination of gastrodin and rhynchophylline inhibited the activation of inflammasomes induced by cerebral ischemia via upregulating the expression of miR-21-5p and miR-331-5p (40). In a related study, miR-21-5p modulated apoptosis of neurons after SCI via the miR-21-5p/PDCD4/caspase-3 pathway (34). In a separate study, silencing miR-21-5p induced polarization of astrocytes to the A2 phenotype and promoted the formation of synapses by targeting glypican 6 via the signal transducer and activator of the transcription-3 pathway after acute ischemic SCI (31). Research has shown that the MAPK signaling pathway participates in SCI and traumatic brain injury (TMI) (41,42). Inhibiting the MAPK signaling pathway represses the expression levels of tumor necrosis factor β (TNF-β), interleukin 6 (IL-6), and IL-1β in SCI (41). Metformin, a first line medication against type 2 diabetes, modulates microglial cells' activation and inhibits the release of several pro-inflammatory cytokines, including IL-6, IL-1β, and TNF-α, partly via the MAPK signaling pathway, which improves neuro-behavioral function following TMI (42). A separate study demonstrated that miR-21-5p exerted its protective effect against blood-brain barrier (BBB) disruption by inhibiting the expression of MAP2K3 protein secreted via the MAPK signaling pathway (37). Additionally, miR-21-5p inhibits the growth and metastasis of melanoma cells by down-regulating the expression of MAP2K3 (43). Microglia activation has been implicated in the development/pathogenesis of SCII (44). Interestingly, sevoflurane inhibits microglia activation by suppressing the MAPK and nuclear factor-kB (NF-kB) signaling pathways (45). In combination with propofol, Sevoflurane protects myocardial cells against SCII induced damage by modulating the expression of MAP2K3. They also and regulate cell apoptosis via the Bcl-2/Bax pathway (46). The T cell receptor signaling pathway has also been implicated in developing CNS diseases and associated pathology (47). For example, T cell receptor signaling was shown to be activated in CD8+ TEMRA cells in the cerebrospinal fluid of patients with Alzheimer’s disease (48). The ErbB signaling pathway on its part has been associated with traumatic brain injury, SCI, and mechanical allodynia (49-51). As such, inhibition of the ErbB signaling in parvalbumin-positive interneurons protects against ethanol-induced TMI (50). The NRG1-ErbB signaling mediates the incision-induced mechanical allodynia and microglia activation. Consequently, pharmacological blocking of NRG1-ErbB signaling suppresses the expression of NRG1 types I and II by repressing the small interfering RNA (siRNA) incision-induced microglia activation and modulated mechanical allodynia (51). Polymorphisms of PPP1R3B are associated with ischemic stroke (IS), increased risk of developing coronary artery disease (CAD), and high serum lipid levels (52). However, the roles of miR-21-5p, MAP2K3, PPP1R3B, and sevoflurane in SCII have not been studied. This study established that compared with the sham group, SCII significantly downregulated the expression of rno-miR-21-5p, but upregulated that of PPP1R3B and MAP2K3 proteins. However, sevoflurane pretreatment up-regulated the expression of miR-21-5p but down-regulated those of PPP1R3B and MAP2K3 proteins. Nonetheless, the effect of miR-21-5p, MAP2K3, PPP1R3B, and sevoflurane on SCII still required further investigation.
In conclusion, sevoflurane ameliorates SCII by regulating the abnormal expression of miRNAs, particularly the miR-21-5p, its target genes (MAP2K3 and PPP1R3B), and the related signaling pathways.
Acknowledgments
Funding: This study was supported financially by the National Natural Science Foundation of China (81971152).
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-7864
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-7864
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-7864). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The protocol for this research was approved by the Ethics Committee of China Medical University (2020266), and conducted according to the 3R principle for animal experiments. All experimental procedures were performed in line with the guidelines of the National Institutes of Health Guide for the Use and Care of Laboratory Animals (NIH Publication No.80-23, revised 1966).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Fang B, Li XQ, Bi B, et al. Dexmedetomidine attenuates blood-spinal cord barrier disruption induced by spinal cord ischemia reperfusion injury in rats. Cell Physiol Biochem 2015;36:373-83. [Crossref] [PubMed]
- Fu J, Sun H, Wei H, et al. Astaxanthin alleviates spinal cord ischemia-reperfusion injury via activation of PI3K/Akt/GSK-3β pathway in rats. J Orthop Surg Res 2020;15:275. [Crossref] [PubMed]
- Chen F, Li X, Li Z, et al. The roles of chemokine (C-X-C motif) ligand 13 in spinal cord ischemia-reperfusion injury in rats. Brain Res 2020;1727:146489 [Crossref] [PubMed]
- Li XQ, Fang B, Tan WF, et al. miR-320a affects spinal cord edema through negatively regulating aquaporin-1 of blood-spinal cord barrier during bimodal stage after ischemia reperfusion injury in rats. BMC Neurosci 2016;17:10. [Crossref] [PubMed]
- Chandran R, Mehta SL, Vemuganti R. Non-coding RNAs and neuroprotection after acute CNS injuries. Neurochem Int 2017;111:12-22. [Crossref] [PubMed]
- Zhai F, Zhang X, Guan Y, et al. Expression profiles of microRNAs after focal cerebral ischemia/reperfusion injury in rats. Neural Regen Res 2012;7:917-23. [PubMed]
- Balsam LB. Spinal cord ischemia-reperfusion injury: MicroRNAs and mitophagy at a crossroads. J Thorac Cardiovasc Surg 2017;154:1509-10. [Crossref] [PubMed]
- Bhalala OG, Srikanth M, Kessler JA. The emerging roles of microRNAs in CNS injuries. Nat Rev Neurol 2013;9:328-39. [Crossref] [PubMed]
- Li XQ, Yu Q, Tan WF, et al. MicroRNA-125b mimic inhibits ischemia reperfusion-induced neuroinflammation and aberrant p53 apoptotic signalling activation through targeting TP53INP1. Brain Behav Immun 2018;74:154-65. [Crossref] [PubMed]
- Li X, Lou X, Xu S, et al. Knockdown of miR-372 Inhibits Nerve Cell Apoptosis Induced by Spinal Cord Ischemia/Reperfusion Injury via Enhancing Autophagy by Up-regulating Beclin-1. J Mol Neurosci 2018;66:437-44. [Crossref] [PubMed]
- Wang R, Yang Y, Wang H, et al. MiR-29c protects against inflammation and apoptosis in Parkinson's disease model in vivo and in vitro by targeting SP1. Clin Exp Pharmacol Physiol 2020;47:372-82. [Crossref] [PubMed]
- Liu ZG, Li Y, Jiao JH, et al. MicroRNA regulatory pattern in spinal cord ischemia-reperfusion injury. Neural Regen Res 2020;15:2123-30. [Crossref] [PubMed]
- Li JA, Zan CF, Xia P, et al. Key genes expressed in different stages of spinal cord ischemia/reperfusion injury. Neural Regen Res 2016;11:1824-9. [Crossref] [PubMed]
- Hu JR, Lv GH, Yin BL. Altered microRNA expression in the ischemic-reperfusion spinal cord with atorvastatin therapy. J Pharmacol Sci 2013;121:343-6. [Crossref] [PubMed]
- Yu Q, Li L, Liang WM. Effect of sevoflurane preconditioning on astrocytic dynamics and neural network formation after cerebral ischemia and reperfusion in rats. Neural Regen Res 2019;14:265-71. [Crossref] [PubMed]
- Shi CX, Jin J, Wang XQ, et al. Sevoflurane attenuates brain damage through inhibiting autophagy and apoptosis in cerebral ischemia-reperfusion rats. Mol Med Rep 2020;21:123-30. [PubMed]
- Yu F, Tong LJ, Cai DS. Sevoflurane inhibits neuronal apoptosis and expressions of HIF-1 and HSP70 in brain tissues of rats with cerebral ischemia/reperfusion injury. Eur Rev Med Pharmacol Sci 2020;24:5082-90. [PubMed]
- Li XQ, Cao XZ, Wang J, et al. Sevoflurane preconditioning ameliorates neuronal deficits by inhibiting microglial MMP-9 expression after spinal cord ischemia/reperfusion in rats. Mol Brain 2014;7:69. [Crossref] [PubMed]
- Ding Q, Wang Q, Deng J, et al. Sevoflurane preconditioning induces rapid ischemic tolerance against spinal cord ischemia/reperfusion through activation of extracellular signal-regulated kinase in rabbits. Anesth Analg 2009;109:1263-72. [Crossref] [PubMed]
- Shi H, Sun BL, Zhang J, et al. miR-15b suppression of Bcl-2 contributes to cerebral ischemic injury and is reversed by sevoflurane preconditioning. CNS Neurol Disord Drug Targets 2013;12:381-91. [Crossref] [PubMed]
- Zhang Y, Shan Z, Zhao Y, et al. Sevoflurane prevents miR-181a-induced cerebral ischemia/reperfusion injury. Chem Biol Interact 2019;308:332-8. [Crossref] [PubMed]
- Li XQ, Lv HW, Tan WF, et al. Role of the TLR4 pathway in blood-spinal cord barrier dysfunction during the bimodal stage after ischemia/reperfusion injury in rats. J Neuroinflammation 2014;11:62. [Crossref] [PubMed]
- Aslan A, Cemek M, Eser O, et al. Does dexmedetomidine reduce secondary damage after spinal cord injury? An experimental study. Eur Spine J 2009;18:336-44. [Crossref] [PubMed]
- Zhou Y, Zhou B, Pache L, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun 2019;10:1523. [Crossref] [PubMed]
- Zhou J, Hui X, Mao Y, et al. Identification of novel genes associated with a poor prognosis in pancreatic ductal adenocarcinoma via a bioinformatics analysis. Biosci Rep 2019;39:BSR20190625 [Crossref] [PubMed]
- Davis AP, Grondin CJ, Johnson RJ, et al. The Comparative Toxicogenomics Database: update 2019. Nucleic Acids Res 2019;47:D948-54. [Crossref] [PubMed]
- Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 2010;31:455-61. [PubMed]
- Guo W, Wang Y, Fan M, et al. Integrating metabolomics and network pharmacology to explore the protective effect of gross saponins of Tribulus terrestris L. fruit against ischemic stroke in rat. J Ethnopharmacol 2020;263:113202 [Crossref] [PubMed]
- Zhou Z, Han B, Jin H, et al. Changes in long non-coding RNA transcriptomic profiles after ischemia-reperfusion injury in rat spinal cord. PeerJ 2020;8:e8293 [Crossref] [PubMed]
- Wang X, Su X, Gong F, et al. MicroRNA-30c abrogation protects against spinal cord ischemia reperfusion injury through modulating SIRT1. Eur J Pharmacol 2019;851:80-7. [Crossref] [PubMed]
- Su Y, Chen Z, Du H, et al. Silencing miR-21 induces polarization of astrocytes to the A2 phenotype and improves the formation of synapses by targeting glypican 6 via the signal transducer and activator of transcription-3 pathway after acute ischemic spinal cord injury. FASEB J 2019;33:10859-71. [Crossref] [PubMed]
- Jin R, Xu S, Lin X, et al. MiR-136 controls neurocytes apoptosis by regulating Tissue Inhibitor of Metalloproteinases-3 in spinal cord ischemic injury. Biomed Pharmacother 2017;94:47-54. [Crossref] [PubMed]
- Wang Q, Chen Q, Ding Q, et al. Sevoflurane postconditioning attenuates spinal cord reperfusion injury through free radicals-mediated up-regulation of antioxidant enzymes in rabbits. J Surg Res 2011;169:292-300. [Crossref] [PubMed]
- Zhang T, Ni S, Luo Z, et al. The protective effect of microRNA-21 in neurons after spinal cord injury. Spinal Cord 2019;57:141-9. [Crossref] [PubMed]
- Yamamoto M, Morita T, Ishikawa M, et al. Specific microRNAs are involved in the reno-protective effects of sevoflurane preconditioning and ischemic preconditioning against ischemia reperfusion injury in rats. Int J Mol Med 2020;45:1141-9. [Crossref] [PubMed]
- Zhang SB, Liu TJ, Pu GH, et al. MicroRNA-374 Exerts Protective Effects by Inhibiting SP1 Through Activating the PI3K/Akt Pathway in Rat Models of Myocardial Ischemia-Reperfusion After Sevoflurane Preconditioning. Cell Physiol Biochem 2018;46:1455-70. [Crossref] [PubMed]
- Yao X, Wang Y, Zhang D. microRNA-21 Confers Neuroprotection Against Cerebral Ischemia-Reperfusion Injury and Alleviates Blood-Brain Barrier Disruption in Rats via the MAPK Signaling Pathway. J Mol Neurosci 2018;65:43-53. [Crossref] [PubMed]
- Chen R, Tai Y, Zhang Y, et al. MicroRNA-21 attenuates oxygen and glucose deprivation induced apoptotic death in human neural stem cells with inhibition of JNK and p38 MAPK signaling. Neurosci Lett 2019;690:11-6. [Crossref] [PubMed]
- Tang M, Pan H, Zheng Z, et al. Prostaglandin E1 protects cardiomyocytes against hypoxia-reperfusion induced injury via the miR-21-5p/FASLG axis. Biosci Rep 2019;39:BSR20190597 [Crossref] [PubMed]
- Zhang HS, Liu MF, Ji XY, et al. Gastrodin combined with rhynchophylline inhibits cerebral ischaemia-induced inflammasome activation via upregulating miR-21-5p and miR-331-5p. Life Sci 2019;239:116935 [Crossref] [PubMed]
- Ye J, Xue R, Ji ZY, et al. Effect of NT-3 on repair of spinal cord injury through the MAPK signaling pathway. Eur Rev Med Pharmacol Sci 2020;24:2165-72. [PubMed]
- Tao L, Li D, Liu H, et al. Neuroprotective effects of metformin on traumatic brain injury in rats associated with NF-κB and MAPK signaling pathway. Brain Res Bull 2018;140:154-61. [Crossref] [PubMed]
- Zhou M, Yu X, Jing Z, et al. Overexpression of microRNA-21 inhibits the growth and metastasis of melanoma cells by targeting MKK3. Mol Med Rep 2019;20:1797-807. [Crossref] [PubMed]
- Ta Na HS, An M, Zhang T, et al. Dexmedetomidine inhibits microglial activation through SNHG14/HMGB1 pathway in spinal cord ischemia-reperfusion injury mice. Int J Neurosci 2020; Epub ahead of print. [Crossref] [PubMed]
- Yu X, Zhang F, Shi J. Effect of sevoflurane treatment on microglia activation, NF-kB and MAPK activities. Immunobiology 2019;224:638-44. [Crossref] [PubMed]
- Liu Y, Shi L, Liu C, et al. Effect of combination therapy of propofol and sevoflurane on MAP2K3 level and myocardial apoptosis induced by ischemia-reperfusion in rats. Int J Clin Exp Med 2015;8:6427-35. [PubMed]
- He X, Fan L, Wu Z, et al. Gene expression profiles reveal key pathways and genes associated with neuropathic pain in patients with spinal cord injury. Mol Med Rep 2017;15:2120-8. [Crossref] [PubMed]
- Gate D, Saligrama N, Leventhal O, et al. Clonally expanded CD8 T cells patrol the cerebrospinal fluid in Alzheimer's disease. Nature 2020;577:399-404. [Crossref] [PubMed]
- Wu Y, Chen H, Tan Z, et al. Therapeutic effects of Erbin inhibitor on spinal cord contusion in mice. Am J Transl Res 2019;11:2570-9. [PubMed]
- Chandrasekar A, Olde Heuvel F, Wepler M, et al. The Neuroprotective Effect of Ethanol Intoxication in Traumatic Brain Injury Is Associated with the Suppression of ErbB Signaling in Parvalbumin-Positive Interneurons. J Neurotrauma 2018;35:2718-35. [Crossref] [PubMed]
- Xiang Y, Liu T, Yang H, et al. NRG1-ErbB signalling promotes microglia activation contributing to incision-induced mechanical allodynia. Eur J Pain 2015;19:686-94. [Crossref] [PubMed]
- Li WJ, Yin RX, Huang JH, et al. Association between the PPP1R3B polymorphisms and serum lipid traits, the risk of coronary artery disease and ischemic stroke in a southern Chinese Han population. Nutr Metab (Lond) 2018;15:27. [Crossref] [PubMed]


