
A blood-based 22-gene expression signature for hepatocellular carcinoma identification
Introduction
Hepatocellular carcinoma (HCC) accounts for 90% of liver cancer and is one of the most common and lethal malignancies. According to GLOBOCAN 2018, liver cancer has the sixth highest incidence rate and the fourth highest mortality rate among all cancers worldwide. It also ranks third in the causes of mortality in China (1). Liver cirrhosis (LC) with any cause, including Hepatitis B Virus (HBV) and Hepatitis C Virus (HCV) chronic infection or alcoholic cirrhosis, is the leading cause of HCC. Studies have indicated that the annual incidence rate of HCC in HBV- and HCV-associated LC patients is around 2–8% (2,3). Chronic HBV infection without cirrhosis is also a major risk factor for the development of HCC; it has an annual incidence rate of 0.5% (2,4). Due to China’s large population of individuals with the HBV infection, about half of all new liver cancer cases world wide occur in China each year (1). This makes liver cancer the fourth most common cancer in China, accounting for 9.53% of all cancers.
Liver ultrasonography (US) and the serum Alpha fetoprotein (AFP) level test are the most frequently applied HCC-monitoring methods in high-risk populations. A meta-analysis showed that US had a pooled sensitivity of 94% but was less effective when detecting early HCC, with a sensitivity of 63% (5). However, HCC is highly operative dependent and has a relatively low throughput. The AFP level is usually reported, but with a poor sensitivity of 40–73% and a specificity of 53.3–90% (6). Thus, a novel biomarker with superior performance in HCC screening is highly sought after.
The transcriptome of peripheral blood is a valuable source for biomarker studies. Due to its richness in information and development of microarray technology, many studies have assessed the peripheral blood transcriptome and its association with various diseases or drug responses (7-9). As one of the most effective interventions, whole blood transcriptome has been evaluated in several studies for its diagnostic potential for cancer in its early stages. Donati et al. identified a validated four-gene predictor set (ANKRD22, CLEC4D, VNN1, and IRAK3) that may prove useful in pancreatic ductal adenocarcinoma (PDAC) diagnosis (10). Aarøe et al. identified a diagnostic signature with high sensitivity (80.6%) and specificity (78.3%) for the early detection of breast cancer (11). In previous work in our laboratory, an 18-gene signature was identified for colorectal cancer diagnosis with a high sensitivity (84%) and specificity (88%). Functional analysis showed that most of the genes were associated with immune response (12).
In this study, we used an Affymetrix microarray for the expression profile of the whole blood transcriptome of HCC patients and the patients with high risk to develop HCC. Genes with diagnostic potential were selected and evaluated via qPCR. A 22-gene signature was finally generated with high accuracy in discriminating between the HCC group and the non-HCC control group.
Methods
Patients
A total of 316 patients (104 with CH, 112 with LC, and 100 with HCC) were enrolled at three hospitals (Ruijin and Renji Hospital of Shanghai Jiao Tong University School of Medicine, and the First Affiliated Hospital of Wenzhou Medical University). Approved by the ethics committee of above three hospitals, informed consents were obtained, and peripheral whole blood samples were collected in a PAX gene Blood RNA tube from patients with any etiology, including those of viral (e.g., HBV and HCV infection) or non-viral (alcohol and auto-immune hepatitis) origin. The study protocol conforms to the ethical guidelines of the 1975 Declaration of Helsinki (6th revision, 2008) as reflected in a priori approval by the institution’s human research committee. HCC patients were diagnosed using histological findings or based on typical imaging characteristics according to liver cancer guidelines. Samples were taken from these patients before any invasive intervention, including biopsy, surgery, or cancer treatments, such as chemotherapy or radiotherapy.
Data analysis and signature identification
Raw intensity data from microarray experiment were normalized using the robust multichip average (RMA) method and then filtered according to the median expression and standard deviation in all of the samples. Specifically, genes with a median expression value higher than 6 and/or standard deviation less than 0.5 were retained for downstream analysis.
After data preprocessing, two feature selection algorithms, mRMR and Lasso, were combined with the support vector machine (SVM) classification model to identify signatures with the best performance in cancer and noncancer discrimination.
Genes selected via the two strategies were combined and validated using the qPCR method.
Five candidate reference genes (CSNK1G2, PPIB, FPGS, DECR1, and CRY2) that were reported to be stably expressed in human whole blood were evaluated (13). Four statistical approaches were used for the evaluation: geNorm, normFinder, bestKeeper, and delta-Ct (dCt). Three genes (CSNK1G2, PPIB, and FPGS) were finally selected as the reference gene set for data normalization. The Ct geometric mean of the three reference genes was used for normalization and subtracted by Ct of each gene that was validated.
The same Lasso-SVM algorithm implementation was used for the qPCR validation study. A 10-fold cross-validation accuracy metric was used for model selection.
Differentially expressed genes were identified via the significant analysis of microarray (SAM) method. Annotation and function analysis were performed via The Database for Annotation, Visualization, and Integrated Discovery (DAVID), which is an online annotation tool for transcriptome study.
Results
Patient characteristics
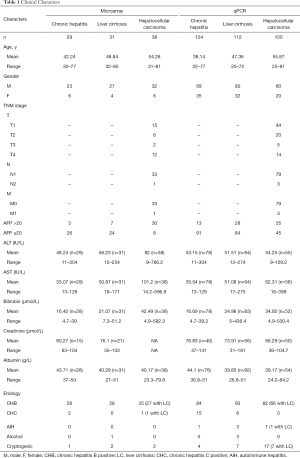
Ninety-eight samples (29 CH, 31 LC, and 38 HCC) were processed to a microarray. Of these, 89 (90.82%) were infected with the hepatitis B virus, and 3 (3.06%) were infected with the hepatitis C virus. Moreover, 36 of the 38 HCC patients had LC. Patients were stratified according to their serological AFP level. A total of 3 (10.34%) CH patients, 7 (22.58%) LC patients, and 30 (78.95%) HCC patients were AFP-positive (>20 ng/mL). Their tumor sizes were either measured using imaging technology such as an US/CT scan or determined after surgery. The longest axis of the largest tumor (if there were multiple nodules) was defined as the diameter of the nodule. The tumor sizes of the 27 HCC patients were recorded: 9 were less than 3 cm, 10 were between 3 and 5 cm, 3 were between 5 and 10 cm, and 5 were larger than 10 cm.
The 316 samples (104 CH, 112 LC, and 100 HCC) were processed to qPCR. Of these, 259 (81.96%) were infected with the hepatitis B virus, and 21 (6.65%) were infected with hepatitis C. In addition, 74 of the 100 HCC patients had LC as a background disease. Moreover, 13 (12.5%) CH patients, 28 (25.0%) LC patients, and 55 (55.0%) HCC patients were AFP-positive (>20 ng/mL). The longest axis of the largest tumor (if there were multiple nodules) was defined as the diameter of the nodule. The tumor sizes of 80 HCC patients were recorded; 33 were less than 3 cm, 14 were between 3 and 5 cm, 17 were between 5 and 10 cm, and 16 were larger than 10 cm (Table 1).
Full table
Gene selection from microarray data
For genes represented by more than one probe set, the probe set with the highest mean value across all of the samples was chosen for further analysis. Genes with a median expression less than 6 and a standard deviation less than 0.5 across all of the samples were removed. This preprocessing procedure reduced the number of genes to 7,127.
Two strategies were then used for feature selection: (I) minimal redundancy maximal relevance (mRMR), which was developed in 2005 by Ding and Peng (14). The method selects genes with a minimum correlation with each other and a maximum relevance with the target phenotype; (II) least Absolute Shrinkage and Selection Operator (Lasso), proposed by Tibshirani, shrinks some coefficients and sets others to 0. Hence, the method aims to retain the good features of both subset selection and ridge regression (15). The SVM classification model was subsequently used for signature identification.
For the mRMR-SVM process, the detailed implementation was as follows:
(I) The whole process began with an external iterative Leave One Out Cross Validation (LOOCV) procedure. In each iteration, only one sample was left out as an external validation sample, and all of the remaining samples were used in a training dataset.
(II) In each iteration, 30 runs of gene selection and model training were performed, each with a different number (ranging from 1 to 50) of genes to be selected. Each run consisted of two steps: mRMR gene selection and SVM model training with a 10-fold cross-validation procedure both applied on the training set.
(III) For gene selection, the mRMR algorithm was applied to the external training dataset to search for subsets of n genes that had a maximum relevance with the clinical status and a minimum redundancy within the gene sets. Once gene selection was completed, the external training set was further split into 10 folds to initiate an internal 10-fold cross-validation procedure to train an SVM classification model using the selected genes as input features. The trained SVM model was then used to classify the external validation sample.
(IV) The external LOOCV procedure was repeated in such a way that each sample function was an external validation sample only once. The performance of the SVM models with certain numbers of genes and parameters was reported as the external LOOCV validation metric of accuracy, which was then used to determine the optimal number of gene numbers. The best model was thus selected and applied to the whole dataset, and the resulting signature was deemed to be the final gene signature (Figure 1A).

(V) We performed a grid search to get the optimal number of genes. Thirty-two genes reached the highest accuracy for distinguishing non-cancers from cancers. Thirty-two genes were also the optimal amount for the sensitivity and specificity.
The procedure of Lasso-SVM followed a slightly different principle (Figure 1B). In this procedure, Lasso feature selection and SVM classification were sequentially combined and trained in a repeated cross-validation process to select the best combination of the Lasso parameter, lamda, and the SVM parameter, C. Twenty-two genes were finally selected as the optimal signature according to the accuracy metric.
Genes were found overlapped between the two procedures. Thus, we generated a combined set with 43 genes for the qPCR study.
qPCR validation and signature identification
For the qPCR validation experiment, 43 target genes and five reference genes (CSNK1G2, PPIB, FPGS, DECR1, and CRY2) were tested. Three genes (CSNK1G2, PPIB, and FPGS) were finally selected to be used as reference genes for data normalization according to the algorithms described in methods.
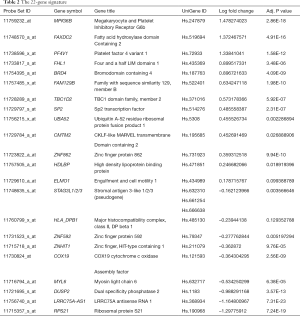
The Lasso-SVM procedure was applied to the normalized qPCR data. We achieved the optimal classification performance when lambda equaled 0.01, which corresponded to a signature of 22 genes (Table 2). In the end, six genes existed in the results of both model training procedures for the microarray; 12 genes were from mRMR-SVM alone, and 4 genes were from Lasso-SVM (Figure 1C).
Full table
Function and pathway analysis of Differential Expressed Genes and Diagnostic Signature
Differential Expressed Genes (DEGs) were identified using the limma package of R. Probe sets, which fulfilled the criteria of LogFC >1 or <–1 and P value lower than 0.05. They were selected and submitted to DAVID for further analysis. Nighty three probe sets representing 64 genes were found to be up-regulated, and 422 probe sets associated with 284 genes were down-regulated in the HCC group.
The most significant biological process among the up-regulated genes was platelet degranulation (10 genes), with adjusted P value =2.49E-08 (Benjamini), and blood coagulation (7 genes), with adjusted P value =7.9E-3. In the KEGG pathway analysis, platelet activation was also listed as one of the top most significant pathways, with five genes enriched (adjusted P value =0.184).
In the down-regulated genes, the most significantly enriched pathways were ribosome (44 genes, with Benjamini adjusted P value =1.09E-43) and oxidative phosphorylation (12 genes, with adjusted P value =5.12E-04). The top-ranked biological process GO clusters also mainly comprised associated genes (Figure 2).

In the 22-gene signature list, 13 genes were up-regulated in the HCC group, and 9 genes were down-regulated as a result of qPCR. However, after submission to DAVID, no biological process or pathways was significantly enriched. Five signature genes with logFC >1 or <–1 were also included in the DEG list. The up-regulated genes, MPIG6B and PF4V1, were associated with the function of the platelet FAXDC2, which is related to oxidoreductase activity, as described in the Gene Ontology annotation. The down-regulated gene, RPS21, was a ribosomal protein, and the other one was a non-coding RNA with an unclear function.
Signature performance
As classification output, the probability value that a sample was HCC was used for the analysis of model performance.
Compared with the serological AFP level, the performance of our signature was much better. The AUC reached 0.94 (95% CI, 0.908−0.964) when AFP got 0.684 (95% CI, 0.629−0.735). In the samples with an AFP less than 20 ng/mL, the AUC of the signature was 0.93 (0.888−0.960). At the optimal cutoff, the 22-gene signature had an 88% sensitivity and an 88.3% specificity in all of the samples; it had a 91.3% sensitivity and an 83.24% specificity in the AFP-negative group (Figure 3A,B).
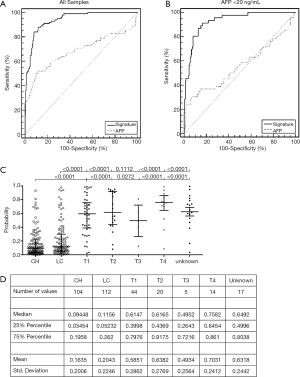
The probability values of the CH, LC, and HCC groups with different tumor T stages were plotted (Figure 3C,D). As shown in the dot-plot, there was a significant difference between the non-HCC and the HCC group, but not between the CH and LC groups and not among the tumor T stage subgroups.
Discussion
Peripheral blood is one of the most useful biomarkers for various diseases. Its circulating nature provides peripheral blood cells the opportunity to communicate and interact with diseased organs. Specific molecules released from a diseased organ could increase in peripheral blood. Change in host immune status with the development of the disease is another interesting phenomenon that could be utilized as a potential biomarker resource. With the development of microarray technology, the transcriptome of peripheral whole blood could easily be profiled in an accurate and reproducible way. Due to the richness in gene expression information, a whole blood transcriptome is an attractive field of biomarker study for various diseases, such as infectious disease (7,9), neurodegenerative disease (8,16), and cancers (17,18).
In the current work, patients with HCC and patients under high risk of developing HCC were enrolled. The total RNA of the peripheral blood from 98 patients was purified and processed to a microarray using a standard procedure. Two strategies, mRMR-SVM and Lasso-SVM, were applied for feature selection. Selected genes were combined for further validation through qPCR. Then, qPCR was performed in an enlarged population with 316 samples and with a combined gene list. Twenty-two genes were selected through the Lasso-SVM method and generated a good result in discriminating between HCC and non-HCC samples. The AUC reached 0.94 in all of the samples and 0.93 in AFP-negative samples and the small tumor group. The signature generated had a largely similar distribution of probability scores among subgroups of different tumor sizes, which indicated that the common biological behavior in HCC was captured by the genes selected in this study.
In the DEG list generated from the microarray, more genes were down-regulated. Ribosome and oxidative phosphorylation were the two most obviously enriched pathways; both were involved with a significant number of genes. These genes were more likely associated with lymphocytes, according to a previous study that reported on their cell-type-specific gene expression profile (19). In the up-regulation group, genes associated with platelet activation were quite significant in our data. The association between platelets and cancer was reported in several papers. Over a century ago, thrombocytosis was found to be associated with solid tumors. In another study, platelet count of peripheral blood was found to be an indicator of the existence of occult cancer (20). In ovarian cancer, tumor-derived IL6 stimulated thrombopoietin production by the liver, thereby stimulating megakaryopoiesis and thrombocytosis (21,22).
The final selected signature had quite a different gene composition with the DEG list, largely because the de-redundancy step-through removal of highly correlated genes was applied in feature selection. The possible inter-patient heterogeneity could be another reason for the huge difference between the DEGs and the signature. More up-regulated genes were enrolled in the signature, including MPIG6B, a platelet surface receptor that plays an inhibition role in platelet activation (23). Then, there was PF4V1, also known as CXCL4L1, which has only three amino acids different from CXCL4, is released from thrombin-stimulated human platelets, and affects angiogenesis (24). Finally, there was FAXDC2, a member of the fatty acid hydroxylase superfamily. It not only upregulated but also enhanced the process of megakaryocytic maturation, which participates in platelet production (25). In the down-regulated genes, the ribosomal protein S21 was reported to be more associated with lymphocytes (19).
Other genes in the signature list were more ubiquitously expressed and involved in various complex biological function associated to cancer. BRD4 is a transcriptional and epigenetic regulator that plays a pivotal role during embryogenesis and cancer development (10). UBA52 was found participated in the degradation of CCNB1, and was critical in cell cycle progression and proliferation of NSCLC cell lines (26). CMTM2 expression could predict the prognostic outcomes of diffuse gastric cancer (27). Downregulation of Elmo1 was found suppressed the migration and invasion of TNBC epithelial cells (28). Some of these genes underwent a relatively small fold change between the HCC and non-HCC group, which represented a fine-tuning of the model.
Conclusions
For this study, we analyzed genes that were differentially expressed between HCC patients and patients with a high risk of developing HCC (e.g., had CH and LC). Platelet activation and a decrease in lymphocyte function were the two main biological phenomena observed. Our signature was identified through qPCR in an enlarged cohort of samples. A good performance was achieved in the AFP-negative samples and patients with small tumors. More validation is necessary to further confirm the performance of the signature.
Acknowledgments
Funding: None.
Footnote
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ferlay J, Ervik M, Lam F, et al. Global Cancer Observatory: Cancer Today. Lyon, France: International Agency for Research on Cancer. accessed [20/10/2018]. Available online: https://gco.iarc.fr/today
- The NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines™) Hepatobiliary Cancers. (Version 1.2018). © 2018. Available online: www.NCCN.org
- Goodgame B, Haheen NJ, Galanko J, et al. The risk of end stage liver disease and hepatocellular carcinoma among persons infected with hepatitis C virus: publication bias? Am J Gastroenterol 2003;98:2535-42. [Crossref] [PubMed]
- Beasley RP, Lin CC, Hwang LY, et al. Hepatocellular carcinoma and hepatitis B virus: a prospective study of 22 707 men in Taiwan. Lancet 1981;2:1129-33. [Crossref] [PubMed]
- Singal A, Volk ML, Waljee A, et al. Meta-analysis: surveillance with ultrasound for early-stage hepatocellular carcinoma in patients with cirrhosis. Alimentary pharmacology & therapeutics 2009;30:37-47. [Crossref] [PubMed]
- Behne T, Copur MS. Biomarkers for hepatocellular carcinoma. Int J Hepatol 2012;2012:859076. [Crossref] [PubMed]
- Nikolayeva I, Bost P, Casademont I, et al. A blood RNA signature detecting severe disease in young dengue patients at hospital arrival. J Infect Dis 2018;217:1690-98. [Crossref] [PubMed]
- Shamir R, Klein C, Amar D, et al. Analysis of blood-based gene expression in idiopathic Parkinson disease. Neurology 2017;89:1676-83. [Crossref] [PubMed]
- Sambarey A, Devaprasad A, Mohan A, et al. Unbiased identification of blood-based biomarkers for pulmonary tuberculosis by modeling and mining molecular interaction networks. EBioMedicine 2017;15:112-26. [Crossref] [PubMed]
- Donati B, Lorenzini E, Ciarrocchi A. BRD4 and Cancer: going beyond transcriptional regulation. Mol Cancer 2018;17:164. [Crossref] [PubMed]
- Aarøe J, Lindahl T, Dumeaux V, et al. Gene expression profiling of peripheral blood cells for early detection of breast cancer. Breast Cancer Res 2010;12:R7. [Crossref] [PubMed]
- Xu Y, Xu Q, Yang L, et al. Identification and validation of a blood-based 18-gene expression signature in colorectal cancer. Clin Cancer Res 2013;19:3039-49. [Crossref] [PubMed]
- Stamova BS, Apperson M, Walker WL, et al. Identification and validation of suitable endogenous reference genes for gene expression studies in human peripheral blood. BMC Med Genomics 2009;2:49. [Crossref] [PubMed]
- Ding C, Peng H. Minimum redundancy feature selection from microarray gene expression data. J Bioinform Comput Biol 2005;3:185-205. [Crossref] [PubMed]
- Tibshirani R. Regression shrinkage and selection via the lasso. J R Stat Soc Series B Stat Methodol 1996;8:267-88.
- Chikina MD, Gerald CP, Li X, et al. Low-variance RNAs identify Parkinson's disease molecular signature in blood. Movement Disorders 2015;30:813-21. [Crossref] [PubMed]
- Gross ME. Blood-based gene expression profiling in castrate-resistant prostate cancer. BMC Med 2015;13:219. [Crossref] [PubMed]
- Isaksson HS, Sorbe B, Nilsson TK. Whole blood RNA expression profiles in ovarian cancer patients with or without residual tumors after primary cytoreductive surgery. Oncol Rep 2012;27:1331-5. [PubMed]
- Palmer C, Diehn M, Alizadeh AA, et al. Cell-type specific gene expression profiles of leukocytes in human peripheral blood. BMC Genomics 2006;7:115. [Crossref] [PubMed]
- Bailey SE, Ukoumunne OC, Shephard E, et al. How useful is thrombocytosis in predicting an underlying cancer in primary care? a systematic review. Fam Pract 2017;34:4-10. [Crossref] [PubMed]
- Stone RL, Nick AM, McNeish IA, et al. Paraneoplastic thrombocytosis in ovarian cancer. N Engl J Med 2012;366:610-8. [Crossref] [PubMed]
- Haemmerle M, Stone RL, Menter DG, et al. The platelet lifeline to cancer: Challenges and opportunities. Cancer Cell 2018;33:965-83. [Crossref] [PubMed]
- Newland SA, Macaulay IC, Floto AR, et al. The novel inhibitory receptor G6B is expressed on the surface of platelets and attenuates platelet function in vitro. Blood 2007;109:4806-9. [Crossref] [PubMed]
- Struyf S, Burdick M D, Proost P, et al. Platelets Release Cxcl4l1, a Nonallelic Variant of the Chemokine Platelet Factor-4/cxcl4 and Potent Inhibitor of Angiogenesis. Circ Res 2004;95:855-7. [Crossref] [PubMed]
- Machlus KR, Italiano JE. The incredible journey: From megakaryocyte development to platelet formation. J Cell Biol 2013;201:785-796. [Crossref] [PubMed]
- Wang F, Chen X, Yu X, et al. Degradation of CCNB1 mediated by APC11 through UBA52 ubiquitination promotes cell cycle progression and proliferation of non-small cell lung cancer cells. Am J Transl Res 2019;11:7166-85. [PubMed]
- Choi JH, Kim YB, Ahn JM, et al. Identification of genomic aberrations associated with lymph node metastasis in diffuse-type gastric cancer. Exp Mol Med 2018;50:6. [Crossref] [PubMed]
- Liang Y, Wang S, Zhang Y. Downregulation of Dock1 and Elmo1 suppresses the migration and invasion of triple-negative breast cancer epithelial cells through the RhoA/Rac1 pathway. Oncol Lett 2018;16:3481-8. [PubMed]


