Abstract
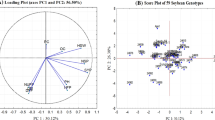
The development of expressed sequence tag-derived simple sequence repeats (EST-SSRs) provided a useful tool for investigating plant genetic diversity. In the present study, 22 polymorphic EST-SSRs from grain soybean were identified and used to assess the genetic diversity in 48 vegetable soybean accessions. Among the 22 EST-SSR loci, tri-nucleotides were the most abundant repeats, accounting for 50.00% of the total motifs. GAA was the most common motif among tri-nucleotide repeats, with a frequency of 18.18%. Polymorphic analysis identified a total of 71 alleles, with an average of 3.23 per locus. The polymorphism information content (PIC) values ranged from 0.144 to 0.630, with a mean of 0.386. Observed heterozygosity (H o) values varied from 0.0196 to 1.0000, with an average of 0.6092, while the expected heterozygosity (H e) values ranged from 0.1502 to 0.6840, with a mean value of 0.4616. Principal coordinate analysis and phylogenetic tree analysis indicated that the accessions could be assigned to different groups based to a large extent on their geographic distribution, and most accessions from China were clustered into the same groups. These results suggest that Chinese vegetable soybean accessions have a narrow genetic base. The results of this study indicate that EST-SSRs from grain soybean have high transferability to vegetable soybean, and that these new markers would be helpful in taxonomy, molecular breeding, and comparative mapping studies of vegetable soybean in the future.
Similar content being viewed by others
References
Anderson, J.A., Churchill, G.A., Autrique, J.E., Tanksley, S.D., Sorrells, M.E., 1993. Optimizing parental selection for genetic linkage maps. Genome, 36(1):181–186. [doi:10. 1139/g93-024]
Arikit, S., Yoshihashi, T., Wanchana, S., Tanya, P., Juwattanasomran, R., Srinives, P., Vanavichit, A., 2011. A PCR-based marker for a locus conferring aroma in vegetable soybean (Glycine max L.). Theor. Appl. Genet., 122(2):311–316. [doi:10.1007/s00122-010-1446-y]
Choudhary, S., Sethy, N.K., Shokeen, B., Bhatia, S., 2009. Development of chickpea EST-SSR markers and analysis of allelic variation across related species. Theor. Appl. Genet., 118(3):591–608. [doi:10.1007/s00122-008-0923-z]
Cornelious, B.K., Sneller, C.H., 2002. Yield and molecular diversity of soybean lines derived from crosses of northern and southern elite parents. Crop Sci., 42(2):642–647. [doi:10.2135/cropsci2002.0642]
Gong, Y.M., Xu, S.C., Mao, W.H., Hu, Q.Z., Zhang, G.W., Ding, J., Li, Y.D., 2010a. Developing new SSR markers from ESTs of pea (Pisum sativum L.). J. Zhejiang Univ.-Sci. B (Biomed. & Biotechnol.), 11(9):702–707. [doi:10.1631/jzus.B1000004]
Gong, Y.M., Xu, S.C., Mao, W.H., Hu, Q.Z., Zhang, G.W., Ding, J., Li, Z.Y., 2010b. Generation and characterization of 11 novel EST derived microsatellites from Vicia faba (Fabaceae). Am. J. Bot., 97(7):69–71. [doi:10.3732/ajb. 1000166]
Gong, Y.M., Xu, S.H., Mao, W.H., Li, Z.Y., Hu, Q.Z., Zhang, G.W., Ding, J., 2011. Genetic diversity analysis of faba Bean (Vicia faba L.) based on EST-SSR markers. Agric. Sci. China, 10(6):838–844. [doi:10.1016/S1671-2927 (11)60069-2]
Hisano, H., Sato, S., Isobe, S., Sasamoto, S., Wada, T., Matsuno, A., Fujishiro, T., Yamada, M., Nakayama, S., Nakamura, Y., et al., 2007. Characterization of the soybean genome using EST-derived microsatellite markers. DNA Res., 14(6):271–281. [doi:10.1093/dnares/dsm025]
Keatinge, J.D.H., Easdown, W.J., Yang, R.Y., Chadha, M.L., Shanmugasundaram, S., 2011. Overcoming chronic malnutrition in a future warming world: the key importance of mungbean and vegetable soybean. Euphytica, 180(1):129–141. [doi:10.1007/s10681-011-0401-6]
Kuroda, Y., Tomooka, N., Kaga, A., Wanigadeva, S.M.S.W., Vaughan, D.A., 2009. Genetic diversity of wild soybean (Glycine soja Sieb. et Zucc.) and Japanese cultivated soybeans [G. max (L.) Merr.] based on microsatellite (SSR) analysis and the selection of a core collection. Genet. Resour. Crop Evol., 56(8):1045–1055. [doi:10. 1007/s10722-009-9425-3]
Li, A.Q., Zhao, C.Z., Wang, X.J., Liu, Z.J., Zhang, L.F., Song, G.Q., Yin, J., Li, C.S., Xia, H., Bi, Y.P., 2010. Identification of SSR markers using soybean (Glycine max) ESTs from globular stageembryos. Electron. J. Biotechnol., 13(5):1–11. [doi:10.2225/vol13-issue5-fulltext-5]
Li, G., Ra, W.H., Park, J.W., Kwon, S.W., Lee, J.H., Park, C.B., Park, Y.J., 2011. Developing EST-SSR markers to study molecular diversity in Liriope and Ophiopogon. Biochem. Syst. Ecol., 39(4–6):241–252. [doi:10.1016/j.bse.2011.08.012]
Li, Y.H., Guan, R.X., Liu, Z.X., Ma, Y.S., Wang, L.X., Li, L.H., Lin, F.Y., Luan, W.J., Chen, P.Y., Yan, Z., et al., 2008. Genetic structure and diversity of cultivated soybean (Glycine max (L.) Merr.) landraces in China. Theor. Appl. Genet., 117(6):857–871. [doi:10.1007/s00122-008-0825-0]
Liu, Y.L., Li, Y.H., Zhou, G.A., Uzokwe, N., Chang, R.Z., Chen, S.Y., Qiu, L.J., 2010. Development of soybean EST-SSR markers and their use to assess genetic diversity in the Subgenus soja. Agric. Sci. China, 9(10):1423–1429. [doi:10.1016/S1671-2927(09)60233-9]
Lu, Y.B., Yang, Y.W., Wu, P.D., 2006. Separation of phosphatidylcholine from soybean phospholipids by simulated moving bed. J. Zhejiang Univ.-Sci. B, 7(7):559–564. [doi:10.1631/jzus.2006.B0559]
Metzgar, D., Bytof, J., Wills, C., 2000. Selection against frameshift mutations limits microsatellite expansion in coding DNA. Genome Res., 10(1):72–80.
Mimura, M., Coyne, C.J., Bambuck, M.W., Lumpkin, T.A., 2007. SSR diversity of vegetable soybean [Glycine max (L.) Merr.]. Genet. Resour. Crop Evol., 54(3):497–508. [doi:10.1007/s10722-006-0006-4]
Moe, K.T., Zhao, W.G., Song, H.S., Kim, Y.H., Chung, J.W., Cho, Y.I., Park, P.H., Park, H.S., Chae, S.C., Park, Y.J., 2010. Development of SSR markers to study diversity in the genus Cymbidium. Biochem. Syst. Ecol., 38(4): 585–594. [doi:10.1016/j.bse.2010.07.004]
Morgante, M., Hanafey, M., Powell, W., 2002. Microsatellites are preferentially associated with non-repetitive DNA in plant genomes. Nat. Genet., 30(2):194–200. [doi:10.1038/ng822]
Roy, J.K., Lakshmikumaran, M.S., Balyan, H.S., Gupta, P.K., 2004. AFLP-based genetic diversity and its comparison with diversity based on SSR, SAMPL, and phenotypic traits in bread wheat. Biochem. Genet., 42(1/2):43–59. [doi:10.1023/B:BIGI.0000012143.48298.71]
Saldivar, X., Wang, Y.J., Chen, P., Mauromoustakos, A., 2010. Effects of blanching and storage conditions on soluble sugar contents in vegetable soybean. LWT Food Sci. Technol., 43(9):1368–1372. [doi:10.1016/j.lwt.2010.04.017]
Shultz, J.L., Kazi, S., Bashir, R., Afzal, J.A., Lightfoot, D.A., 2007. The development of BAC-end sequence-based microsatellite markers and placement in the physical and genetic maps of soybean. Theor. Appl. Genet., 114(6): 1081–1090. [doi:10.1007/s00122-007-0501-9]
Song, Q.J., Marek, L.F., Shoemaker, R.C., Lark, K.G., Concibido, V.C., Delannay, X., Specht, J.E., Cregan, P.B., 2004. A new integrated genetic linkage map of the soybean. Theor. Appl. Genet., 109(1):122–128. [doi:10.1007/s00122-004-1602-3]
Temnykh, S., Park, W.D., Ayres, N., Cartinhour, S., Hauck, N., Lipovich, L., Cho, Y.G., Ishii, T., McCouch, S.R., 2000. Mapping and genome organization of microsatellite sequences in rice (Oryza sativa L.). Theor. Appl. Genet., 100(5):697–712. [doi:10.1007/s001220051342]
Varshney, R.K., Graner, A., Sorrells, M.E., 2005. Genic microsatellite markers in plants: features and applications. Trends Biotechnol., 23(1):48–55. [doi:10.1016/j.tibtech. 2004.11.005]
Wang, X.B., Mulock, B., Guus, B., McCallum, B.T., 2010. Development of EST-derived simple sequence repeat markers for wheat leaf rust fungus, Puccinia triticina Eriks. Can. J. Plant Pathol., 32(1):98–107. [doi:10.1080/07060661003594133]
Wen, Z.X., Ding, Y.L., Zhao, T.J., Gai, J.Y., 2009. Genetic diversity and peculiarity of annual wild soybean (G. soja Sieb. et Zucc.) from various eco-regions in China. Theor. Appl. Genet., 119(2):371–381. [doi:10.1007/s00122-009-1045-y]
Xia, Z.J., Tsubokura, Y., Hoshi, M., Hanawa, M., Yano, C., Okamura, K., Ahmed, T., Anai, T., Watanabe, S., Hayashi, M., et al., 2007. An integrated high-density linkage map of soybean with RFLP, SSR, STS, and AFLP markers using a single F2 population. DNA Res., 14(6): 257–269. [doi:10.1093/dnares/dsm027]
Yinbo, G., Peoples, M.B., Rerkasem, B., 1997. The effect of N fertilizer strategy on N2 fixation, growth and yield of vegetable soybean. Field Crop Res., 51(3):221–229. [doi:10.1016/S0378-4290(96)03464-8]
Young, G., Mebrahtu, T., Johnson, J., 2000. Acceptability of green soybeans as a vegetable entity. Plant Food Hum. Nutr., 55(4):323–333. [doi:10.1023/A:1008164925103]
Zhou, X., Carter, J.T.E., Cui, Z., Miyazaki, S., Burton, J.W., 2002. Genetic diversity patterns in Japanese soybean cultivars based on coefficient of parentage. Crop Sci., 42(4):1331–1342. [doi:10.2135/cropsci2002.1331]
Author information
Authors and Affiliations
Corresponding author
Additional information
The two authors contributed equally to this work
Project supported by the National Natural Science Foundation of China (Nos. 31101538, 31000942, and 31000676), the Grand Science and Technology Special Project of Zhejiang Province (Nos. 2010 C02006, 2012C12903-4-1, and 2012C12903-6-3), the Zhejiang Provincial Natural Science Foundation of China (No. LY12C15004), the Public Welfare Project of Zhejiang Province (No. 2011C22011), and the Shaoxing Important Science and Technology Projects (No. 2012A22008), China
Rights and permissions
About this article
Cite this article
Zhang, Gw., Xu, Sc., Mao, Wh. et al. Determination of the genetic diversity of vegetable soybean [Glycine max (L.) Merr.] using EST-SSR markers. J. Zhejiang Univ. Sci. B 14, 279–288 (2013). https://doi.org/10.1631/jzus.B1200243
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1631/jzus.B1200243
Key words
- Expressed sequence tag (EST)
- Simple sequence repeat (SSR)
- Genetic diversity
- Microsatellites
- Vegetable soybean