Abstract
Aim
This review aimed to provide an overview of the publications using metabolomics in research with physical exercises and to demonstrate how researchers have been applying this approach.
Methods
A systematic search in the databases Web of Science, SCOPUS and PubMed was performed, with the key words: "metabolomics" OR "metabonomics" and with "metabolomics" OR "metabonomics" AND "exercise" in the title or abstract of the articles. The search period was from 2000 to 2016. Forty-four original articles were selected. The studies found were separated into four categories: metabolic responses to physical exercise, supplementation and physical exercise, sports performance, and physical exercise related to diseases.
Results
It was possible to observe the exponential growth of the use of this approach in Sports and Health Sciences, and the four sub-fields towards which these researches involving exercise are directed, enabling a more comprehensive characterization of different metabolic profiles, as well as their study for identifying new biomarkers related to physical exercise.
Conclusions
The possibilities of using metabolomics approach are increasing in the fields of Health Sciences, Sports, and Physical Activity. The experimental design of the study is essential to take advantage of this tool and be able to answer questions in the metabolism comprehension.
Keywords
metabolomics; metabonomics; exercise; physical exercise
Introduction
Metabolomics
Metabolomics is known as a comprehensive analysis to identify, quantify and characterize molecules with low molecular weight called metabolites. This approach is gaining more and more space in several research fields, such as in the health, exact and biological sciences, comprising the quartet of the omics sciences (genomics, transcriptomics, proteomics, and metabolomics). These platforms together can provide relevant information for understanding the so-called Systems Biology11 Fernie AR, Trethewey RN, Krotzky AJ, Willmitzer L. Metabolite profiling: from diagnostics to systems biology. Nat Rev Mol Cell Biol. 2004;5(9):763-769.,22 Kitano H. Systems biology: A brief overview. Science. 2002;295(5560):1662-1664.,33 Jones OAH. Metabolomics and Systems Biology in Human Health and Medicine. 1 ed2014.,44 Roessner U, Bowne J. What is metabolomics all about? Biotechniques. 2009;46(5):363-365..
Metabolites are substrates and final products of cellular metabolism, that have essential roles in energy production, storage, signal transduction, apoptosis, in addition to providing an analysis of the physiological state of the organism55 Gieger C, Geistlinger L, Altmaier E, et al. Genetics meets metabolomics: a genome-wide association study of metabolite profiles in human serum. PLoS Genet. 2008;4(11):e1000282.,66 Johnson CH, Ivanisevic J, Siuzdak G. Metabolomics: beyond biomarkers and towards mechanisms. Nat Rev Mol Cell Biol. 2016;17(7):451-459.. Thus, the analysis of the metabolic profile by metabolomics has become a powerful tool, widely used for clinical diagnosis.
While genes and proteins are subject to epigenetic regulations, and post-transcriptional amendments, respectively, metabolites represent the direct signature of biochemical, cellular activity, and feature the same basic chemical structure regardless of species. Therefore, the metabolomic profile can be more easily correlated with phenotype, compared to genomic, transcriptomic and proteomic profiles77 Patti GJ, Yanes O, Siuzdak G. Innovation: Metabolomics: the apogee of the omics trilogy. Nat Rev Mol Cell Biol. 2012;13(4):263-269..
With the use of metabolomics, biochemical analysis can be performed at various times, in the same organism, to better understand the dynamic changes in metabolism, through the changes in the concentrations of these metabolites33 Jones OAH. Metabolomics and Systems Biology in Human Health and Medicine. 1 ed2014.. Metabolic concentrations can vary in response to disturbances in cellular homeostasis through genetic, nutritional and pathophysiological changes, and also through physical exercise88 Mo ML, Palsson BO, Herrgård MJ. Connecting extracellular metabolomic measurements to intracellular flux states in yeast. BMC Syst Biol. 2009;3:37..
Thus, in order to better understand the various applications and classification of metabolomic approaches, some of the key concepts are defined below:
Targeted Metabolomics: an analysis directed towards a specific group of metabolites that have already been listed, focusing on one or more metabolic pathways of interest, which one seeks to quantify for answering a hypothesis which has already been formulated77 Patti GJ, Yanes O, Siuzdak G. Innovation: Metabolomics: the apogee of the omics trilogy. Nat Rev Mol Cell Biol. 2012;13(4):263-269..
Untargeted Metabolomics: a global analysis which aims to measure the highest number of metabolites, at the same time, in biological samples77 Patti GJ, Yanes O, Siuzdak G. Innovation: Metabolomics: the apogee of the omics trilogy. Nat Rev Mol Cell Biol. 2012;13(4):263-269..
Metabolite Fingerprinting: an analysis in which the identification and quantification of metabolites are not the main purposes, but the distinction and classification of changes that have occurred in a biological sample. For this, an untargeted analysis is used99 Wolfender JL, Marti G, Thomas A, Bertrand S. Current approaches and challenges for the metabolite profiling of complex natural extracts. J Chromatogr A. 2015;1382:136-164..
Metabolite Profiling: an analysis that can be focused on a large group of metabolites, compounds related to a metabolic class or pathway, to understand their adjustment. It is a more focused kind of analysis compared to fingerprinting, as it identifies the metabolites11 Fernie AR, Trethewey RN, Krotzky AJ, Willmitzer L. Metabolite profiling: from diagnostics to systems biology. Nat Rev Mol Cell Biol. 2004;5(9):763-769.,99 Wolfender JL, Marti G, Thomas A, Bertrand S. Current approaches and challenges for the metabolite profiling of complex natural extracts. J Chromatogr A. 2015;1382:136-164..
Thus, these approaches have the purpose of describing in different ways the set of metabolites present in a certain metabolome1010 Fiehn O. Metabolomics--the link between genotypes and phenotypes. Plant Mol Biol. 2002;48(1-2):155-171.. Measuring the metabolome requires high-performance analytical techniques such as nuclear magnetic resonance and mass spectrometry.
Analytical Techniques used in Metabolomics
There are many combinations of techniques that can be used in metabolomics approaches, but the more widely used are nuclear magnetic resonance spectroscopy (NMR) and mass spectrometry (MS).
In NMR spectroscopy the absorption of electromagnetic radiation by the sample occurs. This absorption happens because of certain nuclei of the molecule, generating an NMR spectrum, which is a record of the frequencies of the absorption peaks against their intensities. The number of different orientations that a nucleus may take on when placed in a uniform magnetic field, called spin number, represents the angular momentum of a moving charge. This number can be determined by the mass and atomic number, 1H and 13C, being the ones most used in NMR, though 15N, 19F and 31P also exist. Radio frequency is given in megahertz (MHz)1111 Silverstein RM, et al. Spectrometric identification of organic compounds. John wiley & sons; 2014..
Among the advantages of the use of NMR in metabolomics is sample preparation, which requires minimum processes, being non-destructive or invasive, and widely used for biological fluids or solid biological materials, such as tissues. The main limitations of the technique include resolution and spectral sensitivity, which can improve with the use of high-intensity magnetic fields (greater than 500MHz). Another limitation is the number of identified metabolites that is smaller than about the use of other techniques such as mass spectrometry (MS)1212 Reo NV. NMR-based metabolomics. Drug Chem Toxicol. 2002;25(4):375-382..
In MS, a high mix of sensitivity and selectivity is offered. This technique provides highly specific chemical information, which is directly related to the chemical structure of the compound, such as mass precision, patterns of isotope distribution for the determination of the elemental formula, and the fragments of ions for structural elucidation or identification through the spectral combination of official compounds data. Another important factor is the high sensitivity of MS, which allows the detection and measurement from picomole to femtomole levels of primary and secondary metabolites. These advantages make MS a major analytical tool in metabolomics1313 Lei Z, Huhman DV, Sumner LW. Mass spectrometry strategies in metabolomics. J Biol Chem. 2011;286(29):25435-25442..
Within mass spectrometry, several different techniques can be applied and have different principles. Mass analysis can be direct, which is not effective in the identification of metabolites. It is necessary to associate with some chromatography for separation and identification of compounds, and for generating a complex analysis of the metabolome.. Among the most commonly used are gas chromatography (GC-MS), liquid chromatography (LC-MS), high-performance liquid chromatography (HPLC-MS), ultra-performance liquid chromatography (UPLC-MS) and separation through capillary electrophoresis (CE-MS)99 Wolfender JL, Marti G, Thomas A, Bertrand S. Current approaches and challenges for the metabolite profiling of complex natural extracts. J Chromatogr A. 2015;1382:136-164.,1313 Lei Z, Huhman DV, Sumner LW. Mass spectrometry strategies in metabolomics. J Biol Chem. 2011;286(29):25435-25442..
From these analysis platforms, spectra are generated and need to be processed and profiled for the data to be generated. With that, a large number of data is arranged in tables, requiring biostatistics tools for performing the reduction, analysis, and understanding of these results.
Data Analysis
Data analysis is one of the most important parts of metabolomics since a large number of data is generated by these platforms. From the obtaining of the data, it is necessary to use bioinformatics and chemometrics, which are mathematical, statistical and graphical applications to maximize the information that can be extracted from chemical or spectral data, for the data interpretation1414 Bollard ME, Stanley EG, Lindon JC, Nicholson JK, Holmes E. NMR-based metabonomic approaches for evaluating physiological influences on biofluid composition. NMR Biomed. 2005;18(3):143-162..
The first step is the standardization of such data to minimize their heteroscedasticity, due to biological variations, and variability of measurement techniques. The normalization of the samples can be made through the means, medians or an internal reference. These normalizations are commonly followed by non-linear conversions, such as logarithmic transformations, that helps to reduce the heteroscedasticity of the data enabling greater symmetry between the curves of data distribution, required for the application of linear techniques1515 van den Berg RA, Hoefsloot HC, Westerhuis JA, Smilde AK, van der Werf MJ. Centering, scaling, and transformations: improving the biological information content of metabolomics data. BMC Genomics. 2006;7:142.. Based on this, the scaling of the data is performed, in which each variable is divided by a scale factor of the variable itself, as a measure of data dispersion (such as the standard deviation) and another of the size of the emeasure (for example, the mean). The most commonly reported escalation techniques are auto-scaling and Pareto scaling, which aims to adjust differences in the concentrations of metabolites, adjusting them through a relative scale factor that makes them comparable with each other1515 van den Berg RA, Hoefsloot HC, Westerhuis JA, Smilde AK, van der Werf MJ. Centering, scaling, and transformations: improving the biological information content of metabolomics data. BMC Genomics. 2006;7:142..
Principal Component Analysis (PCA): an exploratory non-supervised analysis tool to reduce the dimensionality of the data set, increasing interpretability and minimizing the loss of information1616 Jolliffe IT, Cadima J. Principal component analysis: a review and recent developments. Philos Trans A Math Phys Eng Sci. 2016;374(2065):20150202.. This analysis is a technique for pattern recognition and not of classification, which has as goal replacing all correlated variables by a smaller number of non-correlated variables, usually referred to as main components1717 Sugimoto M, Kawakami M, Robert M, Soga T, Tomita M. Bioinformatics Tools for Mass Spectroscopy-Based Metabolomic Data Processing and Analysis. Curr Bioinform. 2012;7(1):96-108..
Partial Least Squares Discriminant Analysis (PLS-DA): a technique used to optimize the separation between samples of different groups. It is a supervised method, unlike PCA, used to show the variables (metabolites) that are most responsible for differences between the sample groups, through the calculation of principal components. The construction of a validated model is needed (cross-validation and permutations test) for it to be able to predict the classification of the variables involved in the model. Another analysis associated with PLS-DA is variable importance in projection (VIP), which shows a rank with the main metabolites participating in the segregation of the groups in the model, highlighting potential biomarkers33 Jones OAH. Metabolomics and Systems Biology in Human Health and Medicine. 1 ed2014.,1818 Xia J, Wishart DS. Web-based inference of biological patterns, functions and pathways from metabolomic data using MetaboAnalyst. Nat Protoc. 2011;6(6):743-760..
Partial Least Squares Orthogonal Discriminant Analysis (OPLS-DA): a supervised technique, which facilitates interpretation and transparency compared to PLS-DA. In OPLS-DA a regression between the multivariate and variable data which contain information from only one class is calculated. The advantage compared to PLS-DA is that a single component is used as a predictor of class, while the other components describe the orthogonal variation for the first predictive component1919 Westerhuis JA, van Velzen EJ, Hoefsloot HC, Smilde AK. Multivariate paired data analysis: multilevel PLSDA versus OPLSDA. Metabolomics. 2010;6(1):119-128.. For this model to be able to predict the classification of groups, the robustness of OPLS-DA is validated using two parameters, R2X which indicates the total variance explained in the data and Q2 (cum) which explains the degree of separation between classes, as well as the predictability of the model. For the observation of the main metabolites in the discrimination between the groups in the model, the S-Plot graph is used, which represents the contribution of spectral variables in the segregation of groups, showing data magnitude and reliability.
There are still other secondary analyses that are typically conducted, such as Pathway Analysis, Enrichment Analysis, and Integrated Pathway Analysis to study the metabolic pathways related to the metabolites found2020 Xia J, Sinelnikov IV, Han B, Wishart DS. MetaboAnalyst 3.0--making metabolomics more meaningful. Nucleic Acids Res. 2015;43(W1):W251-257.. In addition, there are also clustering analyses such as K-means clustering, Hierarchical clustering and Self-organizing map, which aim to identify the groups in the same set of data with significant intra-group similarities and differences between groups2121 Ren S, Hinzman AA, Kang EL, Szczesniak V, Lu LJ. Computational and statistical analysis of metabolomics data. Metabolomics. 2015;11:1492-1513..
Use of Metabolomics in Research fields
Several research fields have been using metabolomics approaches to understanding better the mechanisms of action, metabolic pathways involved in response to homeostasis disturbances, and for the identification of new biomarkers related to diseases in human beings, plants or animals2222 Griffiths WJ, Koal T, Wang Y, Kohl M, Enot DP, Deigner HP. Targeted metabolomics for biomarker discovery. Angew Chem Int Ed Engl. 2010;49(32):5426-5445.. Currently, the Medical Sciences have been using metabolomics for elucidation on cardiovascular diseases2323 Kordalewska M, Markuszewski MJ. Metabolomics in cardiovascular diseases. J Pharm Biomed Anal. 2015;113:121-136., cancer2424 Vermeersch KA, Styczynski MP. Applications of metabolomics in cancer research. J Carcinog. 2013;12:9., obesity2525 Rauschert S, Uhl O, Koletzko B, Hellmuth C. Metabolomic biomarkers for obesity in humans: a short review. Ann Nutr Metab. 2014;64(3-4):314-324.,2626 Xie B, Waters MJ, Schirra HJ. Investigating potential mechanisms of obesity by metabolomics. J Biomed Biotechnol. 2012;2012:805683., diabetes2727 Friedrich N. Metabolomics in diabetes research. J Endocrinol. 2012;215(1):29-42., among others since most of the causes of these diseases are related to some metabolic dysregulation2828 Vinayavekhin N, Homan EA, Saghatelian A. Exploring disease through metabolomics. ACS Chem Biol. 2010;5(1):91-103..
Other fields of Science have also been using the technique in studies with plants2929 Bhalla R, Narasimhan K, Swarup S. Metabolomics and its role in understanding cellular responses in plants. Plant Cell Rep. 2005;24(10):562-571.,3030 Sumner LW, Mendes P, Dixon RA. Plant metabolomics: large-scale phytochemistry in the functional genomics era. Phytochemistry. 2003;62(6):817-836., regulation of metabolic pathways3131 Zhang GF, Sadhukhan S, Tochtrop GP, Brunengraber H. Metabolomics, pathway regulation, and pathway discovery. J Biol Chem. 2011;286(27):23631-23635., environment3232 Bundy JG, Davey MP, Viant MR. Environmental metabolomics: a critical review and future perspectives. Metabolomics. 2009;5(1):18., nutrition3333 WISHART DS. Metabolomics: applications to food science and nutrition research. Trends in Food Science and Technology. 2008;19(9):482., and physical exercise3434 Lewis GD, Farrell L, Wood MJ, et al. Metabolic signatures of exercise in human plasma. Sci Transl Med. 2010;2(33):33ra37.,3535 Viant MR, Bearden DW, Bundy JG, et al. International NMR-based environmental metabolomics intercomparison exercise. Environ Sci Technol. 2009;43(1):219-225..
Physical exercise is characterized as something which disturbs metabolic homeostasis. When carried out in different populations, intensities, protocols, and pathophysiological conditions generate a metabolic imbalance, which can be measured through the change in the concentration of metabolites by metabolomics, to try to understand the mechanisms of metabolic responses better. Thus, this review is aimed to provide an overview of the publications using metabolomics in researches with physical exercises, focusing on how researchers have been applying this new approach.
Methods
For the conducting of this review, firstly a systematic search was carried out in 3 databases: Web of Science, SCOPUS and PubMed with the keywords: "metabolomics" OR "metabonomics" in the title or abstract of the articles. The search period was from 2000 to 2016. Then, in these same databases and the same period, the following keywords were used: "metabolomics" OR "metabonomics" AND "exercise".
In the second step a search in these three databases was conducted, in the same period, using the keywords: "metabolomics" OR "metabonomics" AND "exercise" OR "physical exercise" OR "physical activity" in the title or abstract of the articles.
For the second search, only articles which met the following criteria were included in the analysis: human studies; studies with physical exercise described in detail in the methodology, studies with the use of metabolomics, studies published in English in the period from 2000 to 2016. The studies that did not fit the criteria were excluded. By the end, 44 articles had been selected (Figure 1).
Results
Publications with Metabolomics
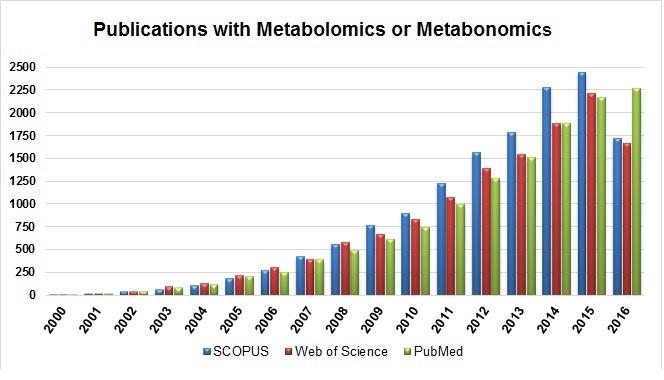
The first search resulted in all publications with the keywords metabolomics or metabonomics found in the three databases. In Figure 2 it is possible to see the number of publications per year. From this search, an exponential increase in the number of publications was observed from 2000 to 2016, showing how much this field of research has been growing each year.
In Figure 3 the number of publications per year with metabolomics or metabonomics is shown, including the keyword "exercise." It is possible to see that the first publications with exercise appeared only in 2007, a period of about seven years without any related research having been observed. Comparing both pictures, it seems that there has been an increase in the number of publications each year, but looking at the total number of publications, the field of metabolomics applied to physical exercise still appears much lower in quantity.
Number of publications per year using the key terms "metabolomics" or "metabonomics" and "exercise".
Metabolomics and Physical Exercise
The second search conducted in the Scopus, Web of Science and PubMed databases with the keywords "metabolomics" or "metabonomics", "physical exercise" or "physical activity" resulted in a total of 81 articles. Next, the titles and abstracts of all articles were analyzed, and 37 of them were excluded, according to the flowchart shown in Figure 1, resulting in 44 selected articles. The articles were also divided into four categories: 1 - metabolic responses to exercise, 2 - supplementation and exercise, 3 - sports performance and 4 --exercise related to diseases (Figure 4).
Percentages for the number of publications by category of classification regarding the subject of investigation.
The complete list of selected articles with exercise and metabolomics can be seen in Table 1, which are separated by year of publication, authors, published a periodical, the methodology used, exercise protocol, type of exercise and type of intervention and category.
Discussion
This review was conducted to provide an overview of how metabolomics approach has been explored in research in the field of Sport Science and Physical Education. As well as show the ways that researchers have chosen to better understand physical exercise using this new method of analysis. To facilitate comparisons and results, we divide the articles into subtopics, which fit the most, and some of them are discussed below based on the information contained in Table 1.
Metabolic Response to Exercise
About 46% of the articles with metabolomics and physical exercise seek to investigate, using different protocols and intensities, the metabolic response to exercise in healthy young individuals (mostly). As can be seen in Table 1, 77% of the protocols presented were acute and 82% aerobic, only two of them were chronic and one acute chronic. Analyzing the exercise protocols used in those studies, 25% of them use high-intensity interval training (HIIT), and 25% continuous aerobic training protocols in an attempt to promote metabolic changes.
The study by Zafeiridis et al.3636 Zafeiridis A, Chatziioannou AC, Sarivasiliou H, et al. Global Metabolic Stress of Isoeffort Continuous and High Intensity Interval Aerobic Exercise: A Comparative (1)H NMR Metabonomic Study. J Proteome Res. 2016;15(12):4452-4463. compared three aerobic exercise protocols of the same volume, with them being continuous, short HIIT (30 s) and long HIIT (3 min). The three protocols were performed by volunteers with a two-week interval between them, and the volumes of training were all similar. The PCA and PLS-DA graphics showed segregation between the moments before and after exercise, but there was no segregation between the training sessions. Therefore, the set of metabolic responses during aerobic exercise appear to depend more on global effort and work performed than on the specific features of each training session, because although different, they have generated similar metabolic changes. The metabolic pathways related to bioenergetics (glycolysis and the citric acid cycle) appear the ones which most changed.
In Danaher et al.3737 Danaher J, Gerber T, Wellard RM, Stathis CG, Cooke MB. The use of metabolomics to monitor simultaneous changes in metabolic variables following supramaximal low volume high intensity exercise. Metabolomics. 2016;12(1):12., on the other hand, comparisons were made between two HIIT protocols with different intensities, the first of them being at 150% of the VO2max: thirty 20s sprints and 40s rest. The second protocol had an intensity of 300% of the VO2max and consisted of thirty 10s sprints and 50s rest. Segregation was observed between the metabolic response of the protocols in the PLS-DA chart, and major disturbances were also found in the metabolism of fatty acids, lipids, and glycolysis in the group with 300% of VO2max. The article by Kuehnbaum et al.3838 Kuehnbaum NL, Gillen JB, Kormendi A, et al. Multiplexed separations for biomarker discovery in metabolomics: Elucidating adaptive responses to exercise training. Electrophoresis. 2015. included a 6-week chronic HIIT intervention in overweight/obese and sedentary young women, with two sessions per week, consisting of 10 60s sprints at 90% of the VO2max. There was a difference in metabolic response before and after the intervention; also, exercise-induced a positive adjustment in the expression of plasmatic o-acetylcarnitine, regarded as an improvement in muscle oxidative capacity.
Other articles performed the comparison between HIIT and continuous moderate exercise in a stationary bicycle. In Enea et al.3939 Enea C, Seguin F, Petitpas-Mulliez J, et al. (1)H NMR-based metabolomics approach for exploring urinary metabolome modifications after acute and chronic physical exercise. Anal Bioanal Chem. 2010;396(3):1167-1176., sprinting at top speed was performed for 30 s with a 90 min break in a semi-lying position. The continuous protocol was conducted at 75% of the VO2max until exhaustion and 90 min of recovery in a semi-lying position. There was no difference in the metabolic profile, and the study showed that the intensity of the protocol is important, because it promotes greater changes in metabolic response. In Peake et al.4040 Peake JM, Tan SJ, Markworth JF, Broadbent JA, Skinner TL, Cameron-Smith D. Metabolic and hormonal responses to isoenergetic high-intensity interval exercise and continuous moderate-intensity exercise. Am J Physiol Endocrinol Metab. 2014;307(7):E539-552., ten 4 min sprints at 80% of the VO2max and 2 min of recovery at 50 watts were performed. The continuous moderate protocol was conducted at 65% of the VO2max, and the duration was given about HIIT's volume, so both protocols had the same volume. Changes occurred in the metabolism of lipids, in the citric acid cycle, and in proteins for the group that performed high-intensity interval exercise.
The studies by Pechlivanis (2010, 2013, and 2015)4141 Pechlivanis A, Kostidis S, Saraslanidis P, et al. (1)H NMR-based metabonomic investigation of the effect of two different exercise sessions on the metabolic fingerprint of human urine. J Proteome Res. 2010;9(12):6405-6416.
42 Pechlivanis A, Kostidis S, Saraslanidis P, et al. 1H NMR study on the short- and long-term impact of two training programs of sprint running on the metabolic fingerprint of human serum. J Proteome Res. 2013;12(1):470-480.-4343 Pechlivanis A, Papaioannou KG, Tsalis G, Saraslanidis P, Mougios V, Theodoridis GA. Monitoring the Response of the Human Urinary Metabolome to Brief Maximal Exercise by a Combination of RP-UPLC-MS and (1)H NMR Spectroscopy. J Proteome Res. 2015;14(11):4610-4622. assessed the acute and chronic metabolic response of the HIIT of 80 m sprints in maximum speed, but with running instead of with a stationary bicycle. The 2010 study was performed on urine samples, using three blocks 80m sprints with 10 s and 1 min intervals between them. A 20 min break was taken between the blocks and the collection held 35 min after. The 2013 study was conducted on blood samples and was a continuation of the previous survey. The same protocol of training three times a week for eight weeks was performed. There was greater segregation between the groups with 10-s interval than between the groups with a 1-minute interval in the PCA chart. In the 2015 study, three 80 m sprints with 10 min interval between the first and second and a 10 s interval between the second and the third were performed. Blood collections were performed after 1 h, 1,5 h, and 2 h. The results were presented in OPLS-DA graphs comparing the times of collection. The greatest discriminators between rest and exercise were hypoxanthine and inosine. These metabolites are part of the cycle of purines and high concentrations after exercise indicate a high rate of breakage and synthesis of ATP in muscles. Another novelty introduced by the authors changed in the metabolites of the intestinal microbiota, such as 2-hydroxy isobutyrate, TMAO, and formate.
Among the few articles that have used strength exercise protocols, Berton et al.4444 Berton R, Conceição MS, Libardi CA, et al. Metabolic time-course response after resistance exercise: A metabolomics approach. J Sports Sci. 2016:1-8. conducted a metabolic response time-course after an acute session of strength exercise in young people. A 4 series protocol with ten repetitions at 70% of 1-RM and blood collections 60 min before, before exercise, 5, 15, 30 and 60 min after exercise were conducted. Metabolites were divided between rapid response metabolites, expressed 5 min after the end of the session, intermediate response metabolites expressed 15 and 30 min after, and slow response metabolites changed only after 60 min.
Only two studies used combined training protocols (strength and aerobic). The first of these was the one by Huffman et al.4545 Huffman KM, Koves TR, Hubal MJ, et al. Metabolite signatures of exercise training in human skeletal muscle relate to mitochondrial remodelling and cardiometabolic fitness. Diabetologia. 2014;57(11):2282-2295., who investigated subjects of both genders, age between 18 and 70 years old, eutrophic or overweight and obese. The subjects were divided into several groups, with aerobic and strength exercise protocols. A combined group performed aerobic training (AT) 3 times a week at a percentage from 65 to 80 % of the VO2max, and strength training (ST) 3 times a week, three series of 8 to 12 repetitions in exercise equipment, with exercises for the lower and upper members. The combined protocol promoted improvements in the VO2max and insulin sensitivity, in addition to changes in metabolites from the family of acylcarnitines, which were considered markers of trainability and related to resistance and insulin sensitivity. In the study by Glynn et al.4646 Glynn EL, Piner LW, Huffman KM, et al. Impact of combined resistance and aerobic exercise training on branched-chain amino acid turnover, glycine metabolism and insulin sensitivity in overweight humans. Diabetologia. 2015;58(10):2324-2335. overweight, sedentary, and insulin resistant subjects were invited to participate in a 6-month combined training, with 3 to 4 sessions of AT at 60-85% of the VO2max and ST in 8 different exercise equipment. The best predictor of improvement in insulin sensitivity was considered.
In general, it should be noted that the use of exercise as a disturber of homeostasis and promoter of changes in metabolic response has increased over the years and that high-intensity interval training still seems to be the most widely used as exercise stimulus to detect possible biomarkers in metabolomics field.
Supplementation and Physical Exercise
The second category of classification is supplementation and physical exercise. It is possible to see in Table 1 that 89% of exercise interventions are acute and 78% are aerobic exercises. However, only two articles use HIIT (22%), a relatively small number compared with the number of studies seeking changes in metabolic response. Most studies used continuous moderate exercise and the intake of some kind of supplement after or during exercise.
In Kirwan et al.4747 Kirwan GM, Coffey VG, Niere JO, Hawley JA, Adams MJ. Spectroscopic correlation analysis of NMR-based metabonomics in exercise science. Anal Chim Acta. 2009;652(1-2):173-179., the protocol consisted of performing an intermittent session in a stationary bicycle, followed by a low-carbohydrate meal the night before the protocol. Subjects should do ten hours fast, go back to the lab in the morning and cycle at 70 % of the VO2max to exhaustion. Immediately after the session, and 60, 120 and 180 minutes after it, 4 g of carbohydrate were ingested. In addition, 6 mg of caffeine were ingested immediately after the exercise and 2 h after it. The authors demonstrated new information about the dynamic changes in the concentration of metabolites in plasma, in response to exercise, fatigue and nutrient supplementation during recovery, in human beings.
In the study by Jacobs et al.4848 Jacobs DM, Hodgson AB, Randell RK, et al. Metabolic response to decaffeinated green tea extract during rest and moderate-intensity exercise. J Agric Food Chem. 2014;62(40):9936-9943., volunteers ingested two capsules of decaffeinated green tea extract (156 ± 3 mg of green tea extract, 284 ± 6 mg of catechins, and 3 mg of caffeine) with 200 ml of water or placebo (273 ± 25 mg of cellulose) and rested for 2 h while sitting. After this period, they exercised for 30 min in a stationary bicycle at 55% of the VO2max. These volunteers ingested, during 28 days, four capsules of the extract, two before breakfast and two before dinner. Exercise sessions took place on the first day, after seven days and after 28 days. Among the results, it was suggested that decaffeinated green tea extract increased the metabolic markers of lipid oxidation during rest after seven days, but not during exercise. After 28 days, these markers continued to increase during rest, but others also increased, suggesting an improvement in the oxidation of fatty acids in mitochondria.
The work by Yde et al.4949 Yde CC, Ditlev DB, Reitelseder S, Bertram HC. Metabonomic Response to Milk Proteins after a Single Bout of Heavy Resistance Exercise Elucidated by 1H Nuclear Magnetic Resonance Spectroscopy. Metabolites. 2013;3(1):33-46., is the only one that used an exclusive strength protocol. Ten series of 8 repetitions each at 80 % of 1RM and a 3 min recovery period between series were performed. After finishing, the volunteers ingested the following drinks: water, whey protein or calcium caseinate (0.3 g/kg lean body mass). Blood collections were made after 70, 220 and 370 min. The authors found differences between the groups that ingested milk proteins compared to the group that ingested water, but among the groups that ingested whey and caseinate, there was no difference in the segregation of groups.
In the study by Miccheli et al.5050 Miccheli A, Marini F, Capuani G, et al. The influence of a sports drink on the postexercise metabolism of elite athletes as investigated by NMR-based metabolomics. J Am Coll Nutr. 2009;28(5):553-564. rowing athletes from the Italian Olympic team were used. The protocol was performed on a rowing ergometer, consisting of 20 min of warm-up, 1000 m in maximum speed, followed by 50 min of submaximal exercise to cause dehydration. Then the groups were rehydrated with an electrolytic beverage with green tea extract (Isotè Coop). Group B was rehydrated with oligomineral water, ingesting 25 ml every 5 min of recovery, until reaching 500 ml. A week later the groups repeated the protocol and rehydration were swapped between the groups. Blood and urine samples were collected before, immediately after the exercise and 120 min after it. There was a clear segregation in the PLS-DA models between the groups that ingested water and the electrolyte drink. Moreover, the group that ingested the drink seemed to have a better metabolic recovery than the group that ingested only water.
Based on these articles and considering the others described in Table 1, it is noted that the use of metabolomics, coupled with exercise and supplements, would help to better understanding the recovery period post-exercise, using different types of exercise protocols and supplementation on the metabolic expression analysis.
Sports Performance
In what concerns the third category of selected articles, about 20% are related to sports performance. In these studies, most volunteers are athletes of some sports modality, and the exercise protocols are related to the type of training of such modalities, such as cyclists, runners, and soccer players, among others.
The study by Yan et al.5151 Yan B, A J, Wang G, et al. Metabolomic investigation into variation of endogenous metabolites in professional athletes subject to strength-endurance training. J Appl Physiol (1985). 2009;106(2):531-538., used senior class rowers with over seven years experience and junior class rowers with only three years experience in China. A two-week training period before a national championship was analyzed, with a focus on speed and aerobic capacity increase. 30 hours of training were carried out, with 11 sessions a week. The collections were made before training in the first and second week and three days after the last session and the control group did the same collections, without training. Based on the PLS-DA graph presented, it was possible to observe a large segregation between both groups with different profiles. In the second chart, it was possible to observe a different metabolic response between the basal, first and second week of training.
In Ra et al.5252 Ra SG, Maeda S, Higashino R, Imai T, Miyakawa S. Metabolomics of salivary fatigue markers in soccer players after consecutive games. Appl Physiol Nutr Metab. 2014;39(10):1120-1126. intercollegiate soccer players were analyzed to find fatigue markers in saliva. The players had three consecutive games (90 min) in 3 days. Collections were made before and after the three days. Some salivary metabolites increased after three consecutive days, and the authors suggested them as important to be studied as markers of fatigue. About the saliva markers, the study by Santone et al.5353 Santone C, Dinallo V, Paci M, D'Ottavio S, Barbato G, Bernardini S. Saliva metabolomics by NMR for the evaluation of sport performance. J Pharm Biomed Anal. 2014;88:441-446. used the Yo-Yo test in soccer players, for performance evaluation. The athletes were divided into two groups, those who had good results in the trial and those who had bad results. The collections were made before and after the three days of games. There was no difference in the metabolic profile of the athletes who responded well and badly to the test, and there were changes in some metabolites after the test. The authors claim that saliva can be a promising biofluid for the measurement of performance in athletes.
Another example is the study by Nieman et al.5454 Nieman DC, Shanely RA, Luo B, Meaney MP, Dew DA, Pappan KL. Metabolomics approach to assessing plasma 13- and 9-hydroxy-octadecadienoic acid and linoleic acid metabolite responses to 75-km cycling. Am J Physiol Regul Integr Comp Physiol. 2014;307(1):R68-74. which was carried out with cyclists who participate in road tests. The athletes performed a 75 km protocol in their competition bikes in CompuTrainer Pro model 8001 at the laboratory. The course was hilly and of medium difficulty. Blood samples were taken before the exercise while fasting, immediately after it, 1,5 h and 21 h after it. It was possible to observe the metabolic changes that occurred from the moment before to the time after the protocol, as well as for 1,5 h and 21 h after it. The biggest changes took place in the metabolites of the metabolism of lipids and carnitine.
Unlike other studies with physical exercise, those related to sports performance did not use pre-made training protocols, but the athletes' training protocols, for the metabolic research. Diet control was far more rigorous, and the stimuli, both under load, as in duration, were much greater due to the physiological condition of these athletes.
Exercise Related to Diseases
The last category of classification of articles is the one, which relates physical exercise to diseases. These studies used therapeutic exercise supporter of diseases, such as insulin resistance, diabetes, and obesity, as noted in Table 1. Thus, the types of intervention are mostly chronic, unlike the others, which have been presented in this review. The kind of exercise is not only aerobic, but the combination of strength and aerobic training, or combined training.
In the study by Kuhl et al.5555 Kuhl J, Moritz T, Wagner H, et al. Metabolomics as a tool to evaluate exercise-induced improvements in insulin sensitivity. Metabolomics. 2008;4(3):9., a 50-minute protocol was performed, three times a week for 12 weeks, combining aerobic and strength exercises in the same session by healthy people and people with type 2 diabetes. In the OPLS-DA models, it was possible to note a segregation between healthy individuals and the control group, in addition to a large segregation between trained and the control group, healthy and with diabetes, showing that exercise was a great way to improve insulin sensitivity and that metabolomics may be used as a diagnostic tool.
The study by Kuehnbaum et al. 20145656 Kuehnbaum NL, Gillen JB, Gibala MJ, Britz-McKibbin P. Personalized metabolomics for predicting glucose tolerance changes in sedentary women after high-intensity interval training. Sci Rep. 2014;4:6166., on the other hand, recruited overweight and obese sedentary women to perform a 6-week HIIT protocol, three times a week, for a total of 18 sessions. Each session was composed of ten 60 s sprints at 90 % of the HRmax and 1-minute interval between sprints. The authors suggested that the use of metabolomics, to find biomar-kers which act as predictors of changes in glucose tolerance, in sedentary women, after interval training, is an efficient tool.
In Reinehr et al.5757 Reinehr T, Wolters B, Knop C, et al. Changes in the serum metabolite profile in obese children with weight loss. Eur J Nutr. 2015;54(2):173-181. a lifestyle intervention was performed in overweight and obese children, for weight loss. The intervention consisted of nutritional, psychological, medical guidance and physical exercises during one year. The physical exercises were mostly aerobic and included different sorts of games since the target population was overweight and obese children. The results showed that some metabolites affected by obesity had their effect reversed due to weight loss, but some showed irreversible changes and remained the same after the changes in lifestyle.
By looking at these examples of studies, it may be noted that the focus of these studies is not to show a metabolic response to exercise, but to use exercise as a metabolic change and metabolic homeostasis control tool, associated with morbidities such as DM2, obesity, and others.
Final Considerations and Future Prospects
With this review, it was possible to show what kinds of research are being conducted with metabolomics in the Health Sciences and Physical Activity fields. It was possible to note the exponential increase of the use of metabolomics in researches over the last 16 years. The earliest records of the use of metabolomics in research with physical exercise were around 2007, and have been growing consistently, albeit in a slight fraction compared with the total number of metabolomics studies.
This review showed the four sub-fields towards which these researches involving exercise are directed: metabolic responses of physical exercise in different protocols and intensities. Physical exercise combined with supplementation to understand the organism's metabolic changes and the recovery process. Sports performance, with analysis of metabolic changes in athletes in their training procedures, and exercise related to diseases, using it as a form of non-pharmacological therapy in the treatment of comorbidities, and as an attenuator of metabolic disturbances caused by diseases in the organism's homeostasis.
With this, we can expect that the use of the metabolomics on sports science will continue growing. The current studies still have an exploratory character, in the understanding of metabolism as a separated compartment. However, in the near future, it is believed that the focus of research will be directed towards a better understanding of particular groups of metabolites that have been exploited, and are expressed in certain conditions (protocols, intensities), to be able to find metabolites which act as predictors of changes in metabolic profiles and integrated systems. They should be measured quickly, or even that metabolites are expressed based on certain exercise protocols, and that they may be ingested in an exogenous manner, to improve the performance of a person in terms of aerobic capacity, strength, lean mass gain, fat mass loss, among many other beneficial effects of physical exercise in relation to the treatment of comorbidities.
The paths to be followed and the possibilities of using this approach are numerous. The techniques, methods of data analysis and interpretation are also numerous. It is up to the researchers to find the best experimental design as possible to take advantage of this tool and to be able to answer their questions, making Science further progress in the understanding of metabolism
References
-
1Fernie AR, Trethewey RN, Krotzky AJ, Willmitzer L. Metabolite profiling: from diagnostics to systems biology. Nat Rev Mol Cell Biol. 2004;5(9):763-769.
-
2Kitano H. Systems biology: A brief overview. Science. 2002;295(5560):1662-1664.
-
3Jones OAH. Metabolomics and Systems Biology in Human Health and Medicine. 1 ed2014.
-
4Roessner U, Bowne J. What is metabolomics all about? Biotechniques. 2009;46(5):363-365.
-
5Gieger C, Geistlinger L, Altmaier E, et al. Genetics meets metabolomics: a genome-wide association study of metabolite profiles in human serum. PLoS Genet. 2008;4(11):e1000282.
-
6Johnson CH, Ivanisevic J, Siuzdak G. Metabolomics: beyond biomarkers and towards mechanisms. Nat Rev Mol Cell Biol. 2016;17(7):451-459.
-
7Patti GJ, Yanes O, Siuzdak G. Innovation: Metabolomics: the apogee of the omics trilogy. Nat Rev Mol Cell Biol. 2012;13(4):263-269.
-
8Mo ML, Palsson BO, Herrgård MJ. Connecting extracellular metabolomic measurements to intracellular flux states in yeast. BMC Syst Biol. 2009;3:37.
-
9Wolfender JL, Marti G, Thomas A, Bertrand S. Current approaches and challenges for the metabolite profiling of complex natural extracts. J Chromatogr A. 2015;1382:136-164.
-
10Fiehn O. Metabolomics--the link between genotypes and phenotypes. Plant Mol Biol. 2002;48(1-2):155-171.
-
11Silverstein RM, et al. Spectrometric identification of organic compounds. John wiley & sons; 2014.
-
12Reo NV. NMR-based metabolomics. Drug Chem Toxicol. 2002;25(4):375-382.
-
13Lei Z, Huhman DV, Sumner LW. Mass spectrometry strategies in metabolomics. J Biol Chem. 2011;286(29):25435-25442.
-
14Bollard ME, Stanley EG, Lindon JC, Nicholson JK, Holmes E. NMR-based metabonomic approaches for evaluating physiological influences on biofluid composition. NMR Biomed. 2005;18(3):143-162.
-
15van den Berg RA, Hoefsloot HC, Westerhuis JA, Smilde AK, van der Werf MJ. Centering, scaling, and transformations: improving the biological information content of metabolomics data. BMC Genomics. 2006;7:142.
-
16Jolliffe IT, Cadima J. Principal component analysis: a review and recent developments. Philos Trans A Math Phys Eng Sci. 2016;374(2065):20150202.
-
17Sugimoto M, Kawakami M, Robert M, Soga T, Tomita M. Bioinformatics Tools for Mass Spectroscopy-Based Metabolomic Data Processing and Analysis. Curr Bioinform. 2012;7(1):96-108.
-
18Xia J, Wishart DS. Web-based inference of biological patterns, functions and pathways from metabolomic data using MetaboAnalyst. Nat Protoc. 2011;6(6):743-760.
-
19Westerhuis JA, van Velzen EJ, Hoefsloot HC, Smilde AK. Multivariate paired data analysis: multilevel PLSDA versus OPLSDA. Metabolomics. 2010;6(1):119-128.
-
20Xia J, Sinelnikov IV, Han B, Wishart DS. MetaboAnalyst 3.0--making metabolomics more meaningful. Nucleic Acids Res. 2015;43(W1):W251-257.
-
21Ren S, Hinzman AA, Kang EL, Szczesniak V, Lu LJ. Computational and statistical analysis of metabolomics data. Metabolomics. 2015;11:1492-1513.
-
22Griffiths WJ, Koal T, Wang Y, Kohl M, Enot DP, Deigner HP. Targeted metabolomics for biomarker discovery. Angew Chem Int Ed Engl. 2010;49(32):5426-5445.
-
23Kordalewska M, Markuszewski MJ. Metabolomics in cardiovascular diseases. J Pharm Biomed Anal. 2015;113:121-136.
-
24Vermeersch KA, Styczynski MP. Applications of metabolomics in cancer research. J Carcinog. 2013;12:9.
-
25Rauschert S, Uhl O, Koletzko B, Hellmuth C. Metabolomic biomarkers for obesity in humans: a short review. Ann Nutr Metab. 2014;64(3-4):314-324.
-
26Xie B, Waters MJ, Schirra HJ. Investigating potential mechanisms of obesity by metabolomics. J Biomed Biotechnol. 2012;2012:805683.
-
27Friedrich N. Metabolomics in diabetes research. J Endocrinol. 2012;215(1):29-42.
-
28Vinayavekhin N, Homan EA, Saghatelian A. Exploring disease through metabolomics. ACS Chem Biol. 2010;5(1):91-103.
-
29Bhalla R, Narasimhan K, Swarup S. Metabolomics and its role in understanding cellular responses in plants. Plant Cell Rep. 2005;24(10):562-571.
-
30Sumner LW, Mendes P, Dixon RA. Plant metabolomics: large-scale phytochemistry in the functional genomics era. Phytochemistry. 2003;62(6):817-836.
-
31Zhang GF, Sadhukhan S, Tochtrop GP, Brunengraber H. Metabolomics, pathway regulation, and pathway discovery. J Biol Chem. 2011;286(27):23631-23635.
-
32Bundy JG, Davey MP, Viant MR. Environmental metabolomics: a critical review and future perspectives. Metabolomics. 2009;5(1):18.
-
33WISHART DS. Metabolomics: applications to food science and nutrition research. Trends in Food Science and Technology. 2008;19(9):482.
-
34Lewis GD, Farrell L, Wood MJ, et al. Metabolic signatures of exercise in human plasma. Sci Transl Med. 2010;2(33):33ra37.
-
35Viant MR, Bearden DW, Bundy JG, et al. International NMR-based environmental metabolomics intercomparison exercise. Environ Sci Technol. 2009;43(1):219-225.
-
36Zafeiridis A, Chatziioannou AC, Sarivasiliou H, et al. Global Metabolic Stress of Isoeffort Continuous and High Intensity Interval Aerobic Exercise: A Comparative (1)H NMR Metabonomic Study. J Proteome Res. 2016;15(12):4452-4463.
-
37Danaher J, Gerber T, Wellard RM, Stathis CG, Cooke MB. The use of metabolomics to monitor simultaneous changes in metabolic variables following supramaximal low volume high intensity exercise. Metabolomics. 2016;12(1):12.
-
38Kuehnbaum NL, Gillen JB, Kormendi A, et al. Multiplexed separations for biomarker discovery in metabolomics: Elucidating adaptive responses to exercise training. Electrophoresis. 2015.
-
39Enea C, Seguin F, Petitpas-Mulliez J, et al. (1)H NMR-based metabolomics approach for exploring urinary metabolome modifications after acute and chronic physical exercise. Anal Bioanal Chem. 2010;396(3):1167-1176.
-
40Peake JM, Tan SJ, Markworth JF, Broadbent JA, Skinner TL, Cameron-Smith D. Metabolic and hormonal responses to isoenergetic high-intensity interval exercise and continuous moderate-intensity exercise. Am J Physiol Endocrinol Metab. 2014;307(7):E539-552.
-
41Pechlivanis A, Kostidis S, Saraslanidis P, et al. (1)H NMR-based metabonomic investigation of the effect of two different exercise sessions on the metabolic fingerprint of human urine. J Proteome Res. 2010;9(12):6405-6416.
-
42Pechlivanis A, Kostidis S, Saraslanidis P, et al. 1H NMR study on the short- and long-term impact of two training programs of sprint running on the metabolic fingerprint of human serum. J Proteome Res. 2013;12(1):470-480.
-
43Pechlivanis A, Papaioannou KG, Tsalis G, Saraslanidis P, Mougios V, Theodoridis GA. Monitoring the Response of the Human Urinary Metabolome to Brief Maximal Exercise by a Combination of RP-UPLC-MS and (1)H NMR Spectroscopy. J Proteome Res. 2015;14(11):4610-4622.
-
44Berton R, Conceição MS, Libardi CA, et al. Metabolic time-course response after resistance exercise: A metabolomics approach. J Sports Sci. 2016:1-8.
-
45Huffman KM, Koves TR, Hubal MJ, et al. Metabolite signatures of exercise training in human skeletal muscle relate to mitochondrial remodelling and cardiometabolic fitness. Diabetologia. 2014;57(11):2282-2295.
-
46Glynn EL, Piner LW, Huffman KM, et al. Impact of combined resistance and aerobic exercise training on branched-chain amino acid turnover, glycine metabolism and insulin sensitivity in overweight humans. Diabetologia. 2015;58(10):2324-2335.
-
47Kirwan GM, Coffey VG, Niere JO, Hawley JA, Adams MJ. Spectroscopic correlation analysis of NMR-based metabonomics in exercise science. Anal Chim Acta. 2009;652(1-2):173-179.
-
48Jacobs DM, Hodgson AB, Randell RK, et al. Metabolic response to decaffeinated green tea extract during rest and moderate-intensity exercise. J Agric Food Chem. 2014;62(40):9936-9943.
-
49Yde CC, Ditlev DB, Reitelseder S, Bertram HC. Metabonomic Response to Milk Proteins after a Single Bout of Heavy Resistance Exercise Elucidated by 1H Nuclear Magnetic Resonance Spectroscopy. Metabolites. 2013;3(1):33-46.
-
50Miccheli A, Marini F, Capuani G, et al. The influence of a sports drink on the postexercise metabolism of elite athletes as investigated by NMR-based metabolomics. J Am Coll Nutr. 2009;28(5):553-564.
-
51Yan B, A J, Wang G, et al. Metabolomic investigation into variation of endogenous metabolites in professional athletes subject to strength-endurance training. J Appl Physiol (1985). 2009;106(2):531-538.
-
52Ra SG, Maeda S, Higashino R, Imai T, Miyakawa S. Metabolomics of salivary fatigue markers in soccer players after consecutive games. Appl Physiol Nutr Metab. 2014;39(10):1120-1126.
-
53Santone C, Dinallo V, Paci M, D'Ottavio S, Barbato G, Bernardini S. Saliva metabolomics by NMR for the evaluation of sport performance. J Pharm Biomed Anal. 2014;88:441-446.
-
54Nieman DC, Shanely RA, Luo B, Meaney MP, Dew DA, Pappan KL. Metabolomics approach to assessing plasma 13- and 9-hydroxy-octadecadienoic acid and linoleic acid metabolite responses to 75-km cycling. Am J Physiol Regul Integr Comp Physiol. 2014;307(1):R68-74.
-
55 Kuhl J, Moritz T, Wagner H, et al. Metabolomics as a tool to evaluate exercise-induced improvements in insulin sensitivity. Metabolomics. 2008;4(3):9.
-
56Kuehnbaum NL, Gillen JB, Gibala MJ, Britz-McKibbin P. Personalized metabolomics for predicting glucose tolerance changes in sedentary women after high-intensity interval training. Sci Rep. 2014;4:6166.
-
57Reinehr T, Wolters B, Knop C, et al. Changes in the serum metabolite profile in obese children with weight loss. Eur J Nutr. 2015;54(2):173-181.
Publication Dates
-
Publication in this collection
2017
History
-
Received
16 Mar 2016 -
Accepted
04 Apr 2016