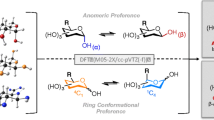
Abstract
D-glucosamine (GluN) is a vital amino monosaccharide present in bio-polymers like chitin and chitosan. Using density functional theory (DFT), we examined its molecular and structural properties in different conformations and protonation states. Results revealed the β-form as more stable than the α-form when neutral. Protonation led to shorter hydrogen bond distances with improved interactions. Frontier molecular orbital analysis showed the amino group's significant role in influencing HOMO and LUMO orbitals based on protonation. The calculated energy gap (ΔE) indicated protonated D-glucosamine’s higher stability and lower reactivity compared to neutral isomers. These findings enhance our comprehension of D-glucosamine’s in bio-polymers applications.
Graphical abstract





Similar content being viewed by others
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
J.W. Anderson, R.J. Nicolosi, J.F. Borzelleca, Glucosamine effects in humans: a review of effects on glucose metabolism, side effects, safety considerations and efficacy. Food Chem. Toxicol. 43(2), 187–201 (2005). https://doi.org/10.1016/j.fct.2004.11.006
D.L. Bertuzzi et al., General protocol to obtain D-glucosamine from biomass residues: shrimp shells, cicada sloughs and cockroaches. Global Chall. 2(11), 1800046 (2018). https://doi.org/10.1002/gch2.201800046
C. Virués et al., Formulation of anomerization and protonation in d-glucosamine, based on 1H NMR. Carbohydr. Res. 490, 107952 (2020). https://doi.org/10.1016/j.carres.2020.107952
H. Amiri et al., Chitin and chitosan derived from crustacean waste valorization streams can support food systems and the UN sustainable development goals. Nat Food 3(10), 822–828 (2022). https://doi.org/10.1038/s43016-022-00591-y
K. Piekarska, M. Sikora, M. Owczarek, J. Jóźwik-Pruska, M. Wiśniewska-Wrona, Chitin and chitosan as polymers of the future—obtaining, modification, life cycle assessment and main directions of application. Polymers (Basel) 15(4), 793 (2023). https://doi.org/10.3390/polym15040793
K. Ogawa, T. Yui, K. Okuyama, Three D structures of chitosan. Int. J. Biol. Macromol. 34(1–2), 1–8 (2004). https://doi.org/10.1016/j.ijbiomac.2003.11.002
R.A. Muzzarelli, M. Mattioli-Belmonte, A. Pugnaloni, G. Biagini, Biochemistry, histology and clinical uses of chitins and chitosans in wound healing. EXS 87, 251–264 (1999). https://doi.org/10.1007/978-3-0348-8757-1_18
I.M. van der Lubben, J.C. Verhoef, G. Borchard, H.E. Junginger, Chitosan and its derivatives in mucosal drug and vaccine delivery. Eur. J. Pharm. Sci. 14(3), 201–207 (2001). https://doi.org/10.1016/S0928-0987(01)00172-5
R. Hejazi, M. Amiji, Chitosan-based gastrointestinal delivery systems. J. Control. Release 89(2), 151–165 (2003). https://doi.org/10.1016/S0168-3659(03)00126-3
M. Prabaharan, J.F. Mano, Chitosan-based particles as controlled drug delivery systems. Drug Deliv. 12(1), 41–57 (2004). https://doi.org/10.1080/10717540590889781
V.R. Sinha et al., Chitosan microspheres as a potential carrier for drugs. Int. J. Pharm. 274(1–2), 1–33 (2004). https://doi.org/10.1016/j.ijpharm.2003.12.026
J.-K. Francis Suh, H.W.T. Matthew, Application of chitosan-based polysaccharide biomaterials in cartilage tissue engineering: a review. Biomaterials 21(24), 2589–2598 (2000). https://doi.org/10.1016/S0142-9612(00)00126-5
M.G. Cascone, N. Barbani, C.C.P. Giusti, G. Ciardelli, L. Lazzeri, Bioartificial polymeric materials based on polysaccharides. J. Biomater. Sci. Polym. Ed. 12(3), 267–281 (2001). https://doi.org/10.1163/156856201750180807
M.C. Neuffer, J. McDivitt, D. Rose, K. King, C.C. Cloonan, J.S. Vayer, Hemostatic dressings for the first responder: a review. Mil. Med. 169(9), 716–720 (2004). https://doi.org/10.7205/MILMED.169.9.716
T.H. Fischer, A.P. Bode, M. Demcheva, J.N. Vournakis, Hemostatic properties of glucosamine-based materials. J. Biomed. Mater. Res. A 80A(1), 167–174 (2007). https://doi.org/10.1002/jbm.a.30877
J.N. Vournakis, M. Demcheva, A. Whitson, R. Guirca, E.R. Pariser, Isolation, purification, and characterization of Poly-N-Acetyl glucosamine use as a hemostatic agent. J. Trauma: Injury, Infection Critical Care 57(1), S2–S6 (2004). https://doi.org/10.1097/01.TA.0000136741.66698.9D
D. Bálint, L. Jäntschi, Comparison of molecular geometry optimization methods based on molecular descriptors. Mathematics 9(22), 2855 (2021). https://doi.org/10.3390/math9222855
M. Manathunga, A.W. Götz, K.M. Merz, Computer-aided drug design, quantum-mechanical methods for biological problems. Curr. Opin. Struct. Biol. 75, 102417 (2022). https://doi.org/10.1016/j.sbi.2022.102417
M.J. Frisch et al., Gaussian 09, Revision A.02 (Gaussian Inc, Wallingford CT, 2016)
A.D. Becke, Density-functional thermochemistry. V. Systematic optimization of exchange-correlation functionals. J Chem Phys 107(20), 8554–8560 (1997). https://doi.org/10.1063/1.475007
P. Singla, M. Riyaz, S. Singhal, N. Goel, Theoretical study of adsorption of amino acids on graphene and BN sheet in gas and aqueous phase with empirical DFT dispersion correction. Phys. Chem. Chem. Phys. 18(7), 5597–5604 (2016). https://doi.org/10.1039/C5CP07078C
H.T. Larijani, M. Jahanshahi, M.D. Ganji, M.H. Kiani, Computational studies on the interactions of glycine amino acid with graphene, h-BN and h-SiC monolayers. Phys. Chem. Chem. Phys. 19(3), 1896–1908 (2017). https://doi.org/10.1039/C6CP06672K
A. Shokuhi Rad, M. Esfahanian, S. Maleki, G. Gharati, Application of carbon nanostructures toward SO2 and SO3 adsorption: a comparison between pristine graphene and N-doped graphene by DFT calculations”. J. Sulfur Chem. 37(2), 176–188 (2016). https://doi.org/10.1080/17415993.2015.1116536
A.S. Rad, Al-doped graphene as a new nanostructure adsorbent for some halomethane compounds: DFT calculations. Surf Sci 645, 6–12 (2016). https://doi.org/10.1016/j.susc.2015.10.036
A.R. Katritzky, N.G. Akhmedov, J. Doskocz, P.P. Mohapatra, C.D. Hall, A. Güven, NMR spectra, GIAO and charge density calculations of five-membered aromatic heterocycles. Magn. Reson. Chem. 45(7), 532–543 (2007). https://doi.org/10.1002/mrc.1967
K. Momma, F. Izumi, VESTA 3 for three-dimensional visualization of crystal, volumetric and morphology data. J. Appl. Crystallogr. 44(6), 1272–1276 (2011). https://doi.org/10.1107/S0021889811038970
G. Schaftenaar, E. Vlieg, G. Vriend, Molden 2.0: quantum chemistry meets proteins. J. Comput. Aided Mol. Des. 31(9), 789–800 (2017). https://doi.org/10.1007/s10822-017-0042-5
C.H. Suresh, G.S. Remya, P.K. Anjalikrishna, Molecular electrostatic potential analysis: a powerful tool to interpret and predict chemical reactivity. WIREs Comput. Mol. Sci. (2022). https://doi.org/10.1002/wcms.1601
A. Suvitha, S. Periandy, P. Gayathri, NBO, HOMO–LUMO, UV, NLO, NMR and vibrational analysis of veratrole using FT-IR, FT-Raman, FT-NMR spectra and HF–DFT computational methods. Spectrochim Acta A Mol. Biomol. Spectrosc. 138, 357–369 (2015). https://doi.org/10.1016/j.saa.2014.11.011
M. Uzzaman, M. JabedulHoque, Physiochemical, molecular docking, and pharmacokinetic studies of Naproxen and its modified derivatives based on DFT. Int. J. Sci. Res. Manag. (2018). https://doi.org/10.18535/ijsrm/v6i9.c01
J. Aihara, Reduced HOMO−LUMO gap as an index of kinetic stability for polycyclic aromatic hydrocarbons. J. Phys. Chem. A 103(37), 7487–7495 (1999). https://doi.org/10.1021/jp990092i
Acknowledgments
To Roberto López Rendón†, your legacy will live on through the years. The author is gratefully for the computing time granted by the Supercomputer Hybrid Cluster “Xiuhcoatl” at General Coordination of Information and Communications Technologies (CGSTIC) of Cinvestav-IPN. This research/thesis was partially supported by the NLHPC supercomputing infrastructure (ECM-02).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The author declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ávila-Avilés, R.D. Quantum chemical analysis of isomerization and protonation of amino group in D-glucosamine. MRS Communications 13, 1303–1308 (2023). https://doi.org/10.1557/s43579-023-00455-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1557/s43579-023-00455-x