Published online Apr 16, 2022. doi: 10.12998/wjcc.v10.i11.3352
Peer-review started: October 29, 2021
First decision: December 27, 2021
Revised: December 30, 2021
Accepted: February 12, 2022
Article in press: February 12, 2022
Published online: April 16, 2022
Colorectal cancer (CRC) imposes a tremendous burden on human health, with high morbidity and mortality. Circular ribonucleic acids (circRNAs), a new type of noncoding RNA, are considered to participate in cancer pathogenesis as microRNA (miRNA) sponges. However, the dysregulation and biological functions of circRNAs in CRC remain to be explored.
To identify potential circRNA biomarkers of CRC and explore their functions in CRC carcinogenesis.
CircRNAs and miRNAs differentially expressed in CRC tissues were identified by analyzing expression profiles from the Gene Expression Omnibus (GEO) database. Circ_0000375 and circ_0011536 were selected as CRC biomarker candidates. Quantitative real-time polymerase chain reaction was utilized to evaluate the expression of these 2 circRNAs in CRC tissues, serums and cell lines. Receiver operating characteristic curves were generated to assess the diagnostic performances of these 2 circRNAs. Then, functional experiments, including cell counting kit-8, wound healing and Transwell invasion assays, were performed after the overexpression of circ_0000375 and circ_0011536 in CRC cell lines. Furthermore, candidate target miRNAs of circ_0000375 and circ_0011536 were predicted via bioinformatics analysis. The expression levels of these miRNAs were explored in CRC cell lines and tissues from GEO datasets. A luciferase reporter assay was developed to examine the interactions between circRNAs and miRNAs. Based on the target miRNAs and downstream genes, functional enrichment analyses were applied to reveal the critical signaling pathways involved in CRC carcinogenesis.
Downregulated circ_0000375 and circ_0011536 expression was observed in CRC tissues in GSE126095, clinical CRC tissue and serum samples and CRC cell lines. The areas under the curve for circ_0000375 and circ_0011536 were 0.911 and 0.885 in CRC tissue and 0.976 and 0.982 in CRC serum, respectively. Moreover, the serum levels of these 2 circRNAs were higher in patients at 30 d postsurgery than in patients before surgery, suggesting that the serum expression of circ_0000375 and circ_0011536 is related to CRC tumorigenesis. Circ_0000375 and circ_0011536 overexpression inhibited the proliferation, migration and invasion of CRC cells. Furthermore, miR-1182 and miR-1246, which were overexpressed in CRC tissues in GSE41655, GSE49246 and GSE115513, were verified as target miRNAs of circ_0000375 and circ_0011536, respectively, by luciferase reporter assays. The downstream genes of miR-1182 and miR-1246 were enriched in some CRC-associated pathways, such as the Wnt signaling pathway.
Circ_0000375 and circ_0011536 may function as tumor suppressors in CRC progression, serving as novel biomarkers for CRC diagnosis and as promising candidates for therapeutic exploration.
Core Tip: The dysregulation and biological functions of circular ribonucleic acids (circRNAs) in colorectal cancer (CRC) have not been well elucidated. In this study, circ_0000375 and circ_0011536 were observed to be downregulated in CRC tissues, serum samples and cell lines. These 2 circRNAs were validated as biomarker candidates in tissues and serum samples with high diagnostic performance. Overexpression of circ_0000375 and circ_0011536 inhibited the growth, migration and invasion of CRC cells. Furthermore, miR-1182 and miR-1246 were upregulated in CRC tissues as target microRNA of circ_0000375 and circ_0011536, respectively. Overall, circ_0000375 and circ_0011536 may serve as diagnostic biomarkers and function as tumor suppressors in CRC.
- Citation: Yin TF, Du SY, Zhao DY, Sun XZ, Zhou YC, Wang QQ, Zhou GYJ, Yao SK. Identification of circ_0000375 and circ_0011536 as novel diagnostic biomarkers of colorectal cancer. World J Clin Cases 2022; 10(11): 3352-3368
- URL: https://www.wjgnet.com/2307-8960/full/v10/i11/3352.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v10.i11.3352
Colorectal cancer (CRC) is a common malignancy worldwide, with incidence and mortality rates ranking 3th and 2nd, respectively[1]. More than 50% of CRC patients are first diagnosed at an advanced stage, which creates enormous threats to human health and imposes a social burden[2]. The 5-year overall survival rate of CRC patients in stage Ⅰ can reach approximately 90%, while the rate is only 10% for stage Ⅳ patients[3], suggesting that the prognoses of patients with advanced CRC are notably poorer. The great challenges related to early diagnosis and successful therapy are attributed to the high potential for metastasis or relapse, treatment resistance, high heterogeneity and multistep initiation of CRC. Conventional tumor markers widely used in clinical practice, such as glycoprotein carcinoembryonic antigen, exhibit unsatisfactory specificity and sensitivity[4]. Thus, there is an urgent need to discover innovative and reliable diagnostic biomarkers to improve predictive accuracy, guide individualized treatment and assess clinical outcome.
Recently, an increasing number of circular ribonucleic acids (circRNAs) have been identified through bioinformatic analysis and high-throughput sequencing technology. CircRNAs have drawn widespread attention as a class of single-stranded noncoding RNA molecules with a circular structure lacking 5’ and 3’ ends that are more stable than linear RNAs and resistant to RNA exonucleases[5]. CircRNAs are highly expressed in eukaryotes and exert diverse functions, including sponging microRNAs (miRNAs), regulating transcription and splicing in the nucleus, interacting with proteins and undergoing translation in the cytoplasm[6,7]. Accumulating evidence has demonstrated that circRNAs contain adequate miRNA-binding elements to sponge and sequester corresponding miRNAs, acting as competing endogenous RNAs at the posttranscriptional level[8,9]. Some circRNAs have been reported to be not only potential diagnostic and prognostic indicators for CRC but also regulators of carcinogenesis via their interactions with miRNAs and their target genes. For example, hsa_circ_0005273 (circPTK2) in CRC tissue is associated with tumor growth and metastasis as well as prognosis and might serve as a therapeutic target for CRC metastasis[10]. Serum exosomal hsa_circ_0004771 can act as a diagnostic biomarker of CRC[11]. Hsa_circ_0001955 and hsa_circ_0000977 were found to mediate the miRNA-mRNA regulatory network in CRC[12]. Collectively, circRNAs could play vital roles in the tumorigenesis of CRC and have potential as novel biomarkers for diagnosis and clinical outcome prediction.
MiRNAs are small single-stranded noncoding RNA molecules that are 19-25 nucleotides in size and mediate the posttranscriptional silencing of target genes by combining with the 3’ untranslated region of a mRNA transcript to contribute to translational inhibition and mRNA destabilization[13,14]. The miRNAs in tissue samples and cell-free biological fluids, have been reported as crucial gene regulators, promising candidate biomarkers and therapeutic targets[14,15]. Considerable miRNAs have been observed to regulate various biological and pathological processes in many diseases, especially cancer. For instance, many miRNAs differentially expressed in CRC can influence the growth, progression, autophagy, apoptosis and even microbiome of CRC cells[16-18], and act as noninvasive biomarkers of CRC[19].
The aim of this study was to identify promising circRNA biomarkers for CRC diagnosis and obtain insights into their functions in modulating the development of CRC. In the present study, we investigated the expression levels of 2 circRNAs, circ_0000375 and circ_0011536, in tissue and serum samples as well as in CRC cell lines. The biological functions of the 2 circRNAs were explored by evaluating the proliferation, migration and invasion of CRC cell lines. The expression of target miRNAs and their interactions with circRNAs were also investigated in CRC. Collectively, our findings revealed the circRNAs and their miRNA interactions that are critically important to the tumorigenesis of CRC, providing potential biomarkers for diagnosis and therapeutic options.
The expression profiles of circRNAs and miRNAs of CRC patients and negative controls (NCs) were obtained from the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo). The GSE126095 (10 CRCs vs 10 NCs) dataset was utilized for circRNA expression analysis, while GSE41655 (33 CRCs vs 15 NCs), GSE49246 (40 CRCs vs 40 NCs) and GSE115513 (750 CRCs vs 649 NCs) were used for miRNA analysis. The platform and series matrix files of the datasets were downloaded from the GEO database, and then the probe matrix was converted into the corresponding RNA name referring to the annotation from the platform files. The “limma” R package was used for normalization of raw data and identification of the differentially expressed circRNAs and miRNAs between CRC and NC samples. The target miRNAs were predicted with the circular RNA interactome database (CircInteractome, https://circinteractome.nia.nih.gov/). Gene Ontology and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses were employed to reveal the vital signaling pathways of miRNAs involved in the tumorigenesis of CRC using the “clusterProfiler” R package.
In this study, patients with CRC and healthy control (HC) participants were enrolled from May 2019 to May 2021 at China-Japan Friendship Hospital. All patients included in our study were newly diagnosed with colorectal adenocarcinoma confirmed by postoperative pathological examination. Patients who had already received radiotherapy, chemotherapy or other molecular targeted therapy before surgery were screened out. Age- and sex-matched HC participants were recruited from a population of individuals without major abnormalities and with negative results after complete colonoscopy. In total, 34 tumor tissue samples from CRC patients and 24 colon tissue samples from HCs were obtained. Serum samples were collected from HCs before colonoscopy (n = 28), from CRC patients before curative surgery (n = 56) and from CRC patients at 30 d after surgery (n = 14). Demographic data and clinicopathological features, including age, gender, TNM stage, tumor size, tumor location and lymph node metastasis, were collected. Tissue samples taken from CRC patients or HCs were frozen in liquid nitrogen immediately and stored at -80 °C until further analysis. Serum specimens were immediately centrifuged at 4000 g for 10 min at 4 °C, and 1 mL of supernatant was stored at -80 °C. This study was approved by the Ethics Committee of the China-Japan Friendship Hospital (No. 2018-116-K85-1), and all participants signed written informed consent forms.
Three CRC cell lines (HCT, RKO, and SW480) and a human normal colonic epithelial cell line (FHC) were obtained from the American Type Culture Collection. All cell lines except RKO were cultured in Roswell Park Memorial Institute-1640 medium (RPMI-1640; Invitrogen, Carlsbad, United States), and RKO cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Invitrogen). All the cell lines were supplemented with 100 IU/mL penicillin, 100 μg/mL streptomycin and 10% fetal bovine serum (FBS) and cultured at 37 °C in a humidified atmosphere with 5% CO2.
An empty pcDNA3.1 vector and overexpression plasmids for circRNAs were synthesized by GenePharma (Shanghai, China). Then, Lipofectamine 3000 Transfection Reagent (Invitrogen) was utilized to transfect HCT116 and SW480 cells with the above plasmids according to the manufacturer’s instructions. 24 h later, CRC cells were collected for subsequent experiments.
Total RNA was isolated from tissues, serums and cells using the RNAprep Pure Cell Kit (Solarbio, Beijing, China) following the manufacturer’s protocol. Then, the isolated RNA was reverse transcribed into cDNA using the Hifair® Ⅱ 1st Strand cDNA Synthesis Kit (YESEN, Shanghai, China). Then, quantitative reverse transcription-polymerase chain reaction (qRT-PCR) was conducted using Hieff® qPCR SYBR® Green Master Mix (No Rox) (YESEN). The relative expression levels of circRNAs and miRNAs were quantified by the 2-ΔΔCt method, using GAPDH and U6 as internal reference genes, respectively. The primers for qRT-PCR were synthesized by Sangon Biotech and are listed in Supplementary Table 1. To further ascertain the back-splicing and junction sequence of each circRNA, Sanger sequencing of the amplification products of circRNAs was performed by Tsingke (Beijing, China).
To determine the stability characteristics of circRNAs, 2 circRNAs and their corresponding linear mRNAs were digested with RNase R. A 5-μg aliquot of total RNA was incubated with or without 1 μL of 10-fold reaction buffer and 0.5 μL of RNase R (20 U/μL) for 30 min at 37 °C. Then, the expression levels of the circRNAs and their corresponding linear mRNAs were quantified by PCR.
A total of 1500 SW480 or HCT116 cells in 100 μL of medium were seeded in each well of a 96-well plate. After 24 h, the CRC cells were transfected with oe-circRNA plasmid or vector. Then, at 0 h, 24 h, 48 h and 72 h, 10 μL of cell counting kit-8 (CCK-8) reagent (Solarbio) and 90 μL of medium were added after the previous medium was removed. Then, the cells were incubated for 1 h at 37 °C with 5% CO2, and the optical density (OD) was detected at 450 nm using a microplate reader (BioTek Instruments, United States).
SW480 or HCT116 cells were cultured in 6-well plates for 24 h, and then, cell transfection was performed as described above. After incubation for 48 h, a wound was scratched with a 200 µL sterile pipette tip. Then, the cells were washed with PBS 3 times and incubated in serum-free medium. Representative images were captured at 0 h and 24 h after scratching. The total wound area was measured using ImageJ software, and the relative migration rate was calculated.
CRC cells were suspended in 200 μL of serum-free medium and seeded in the upper chambers coated with matrigel (Corning, United States). The bottom 24-well plates were filled with 600 μL of medium containing 10% FBS as a chemoattractant. After incubation for 72 h, the cells remaining in the upper chamber were gently removed with cotton swabs, and the cells on the lower surface were photographed and counted after being fixed with 4% paraformaldehyde and stained with 0.1% crystal violet.
Fragments of circ_0000375 and circ_0011536 and corresponding mutant sequences were designed and inserted into a luciferase reporter vector (GP-miRGLO) to produce circ_0000375-WT, circ_0011536-WT, circ_0000375-MUT and circ_0011536-MUT, respectively (GenePharma, China). HEK293T cells obtained from the American Type Culture Collection were seeded in 12-well plates and then cotransfected with a mixture of these plasmids and miRNA mimics or mimics NC. After incubation for 24 h, firefly and Renilla luciferase activities were examined with a Dual-Luciferase Reporter Assay System (Promega, United States) in line with the manufacturer’s instructions.
Statistical Program for Social Science version 26.0 (SPSS 26.0), R software (version 4.0.3) and GraphPad Prism (version 9) were used to perform statistical analysis. Quantitative data with a normal distribution are presented as the mean ± SD and were analyzed using Student’s t-test or one-way analysis of variance, while variables with a nonnormal distribution are presented as the median and interquartile range and were compared using a Mann-Whitney U test or Kruskal-Wallis test. Moreover, categorical variables were analyzed by the chi-square test or a nonparametric test. Receiver operating characteristic (ROC) curves were constructed to examine the diagnostic efficiency of RNA molecules by assessing the area under the curve (AUC). A two-tailed P < 0.05 was considered statistically significant.
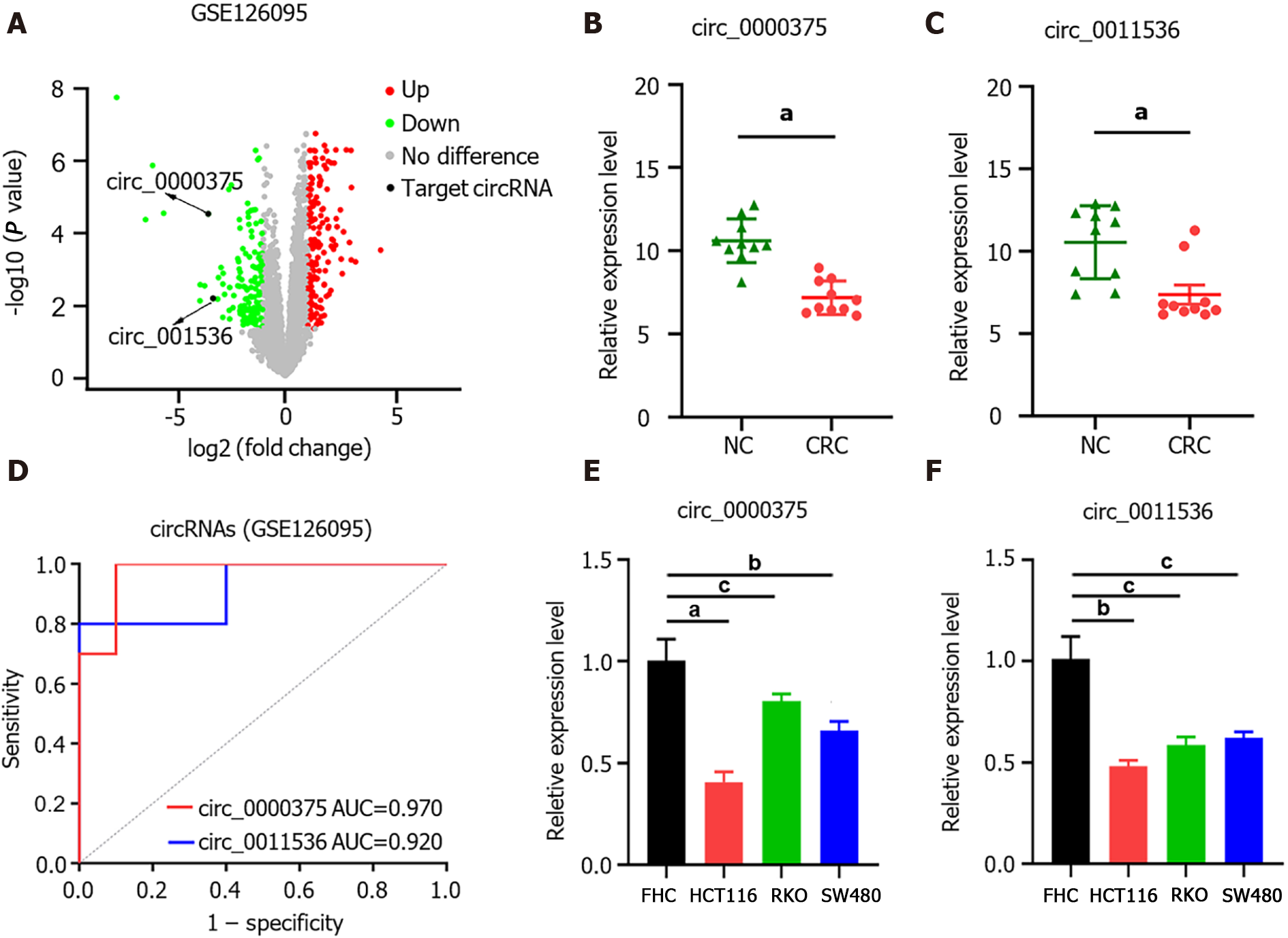
In GSE126095, the expression levels of circRNAs were compared between CRC (n = 10) and NC (n = 10) samples. The circRNAs differentially expressed between CRC and NC were obtained with the criteria |log2 fold change| > 1 and P value < 0.05 (Figure 1A). Based on the circRNA regulatory network identified in our previous study[20], we selected 2 circRNAs, namely, hsa_circ_0000375 (circ_0000375) and hsa_circ_0011536 (circ_0011536), which were downregulated in CRC (Figure 1B and C). The AUCs of circ_0000375 and circ_0011536 were 0.970 and 0.920, respectively (Figure 1D), suggesting that these 2 circRNAs were useful as CRC biomarkers in the GSE126095 dataset. To further confirm the value of the 2 circRNAs in CRC, we investigated the expression levels of the 2 circRNAs in cell lines. Compared with the normal colonic epithelial cell line FHC, CRC cell lines (HCT, RKO and SW480) showed significantly decreased expression levels of circ_0000375 and circ_0011536 (Figure 1E and F).
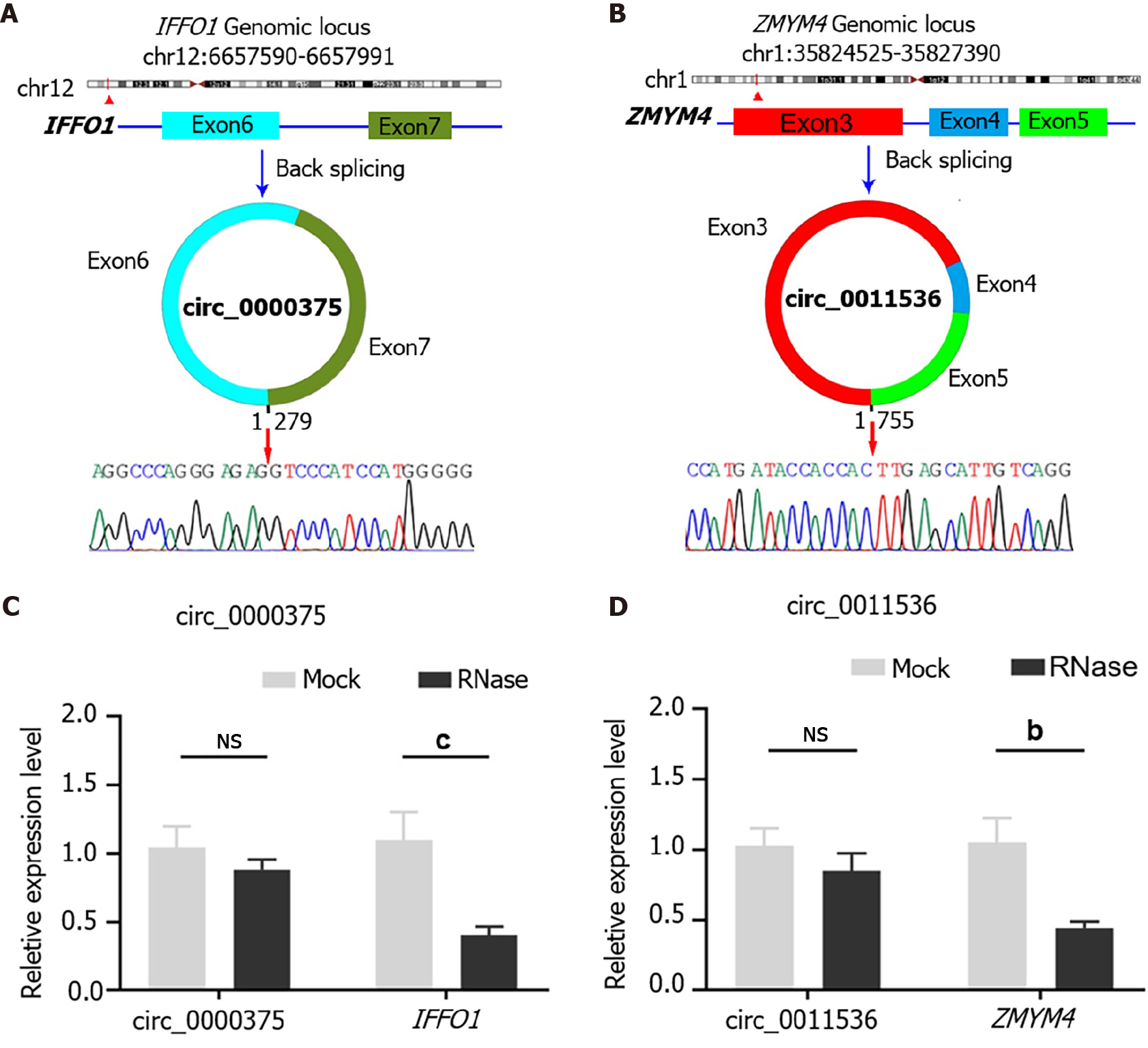
Circ_0000375 originates from head-to-tail splicing of exon 6 and exon 7 in the linear mRNA IFFO1, which is located on chromosome 12. According to the Sanger sequencing results for PCR products, the target sequence of circ_0000375 was verified to be the head-to-tail splicing site (Figure 2A). For circ_0011536, which is generated by back-splicing of exon 3 to exon 5 of the ZMYM4 gene, our primer covered the splicing site, so the inverse complementary sequence of the amplification products of circ_0011536 was consistent with that in the circBase database (Figure 2B). Subsequently, the expression level of circ_0000375 did not show an obvious decline after CRC tissues were degraded with RNase R, but its corresponding linear mRNA, IFFO1, was significantly downregulated (Figure 2C). Similar results were obtained for circ_0011536 and ZMYM4 (Figure 2D). These results revealed that circ_0000375 and circ_0011536 were more resistant to RNase R digestion than the corresponding linear mRNAs.
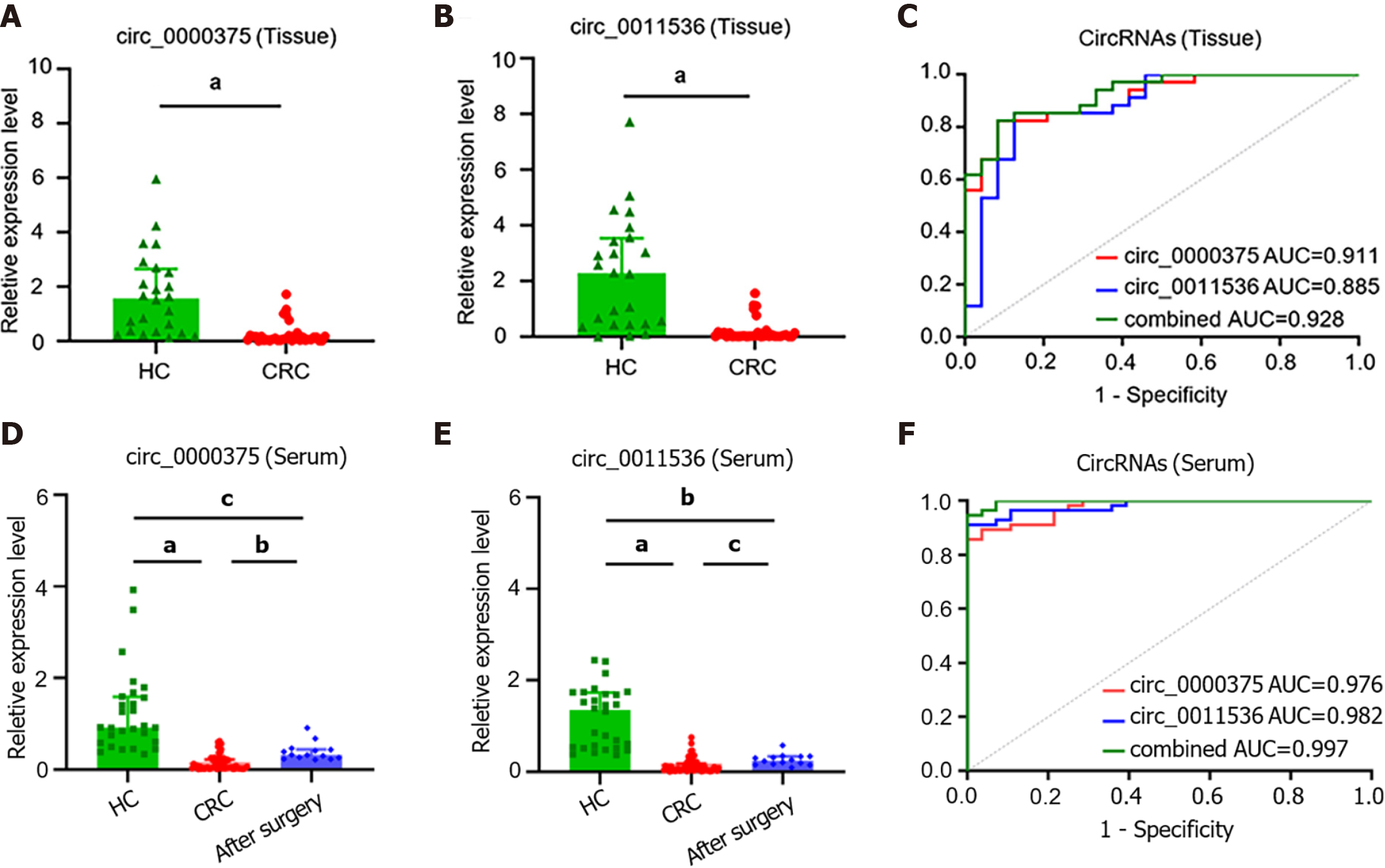
The clinicopathological features of CRC patients and HCs are listed in Table 1. Downregulation of circ_0000375 and circ_0011536 was validated in CRC tissues compared with HC tissues (Figure 3A and B). ROC curve analysis revealed that the AUCs of circ_0000375 and circ_0011536 were 0.911 and 0.885, respectively. Combined with both circRNAs, the AUC was 0.928, indicating possible better performance based on these 2 circRNAs (Figure 3C). Furthermore, the median expression levels of circ_0000375 and circ_0011536 in tissues were used to divide the CRC patients into low and high expression groups, which were used for subgroup analysis of the clinicopathological characteristics of the CRC patients (Table 2). The results indicated that patients with lymph node metastasis or advanced TNM stage tended to exhibit high expression of circ_0011536. However, the expression of circ_0000375 in tissue was not related to clinicopathological indexes.
| Tissues | Serums | ||||
| HC (n = 24) | CRC (n = 34) | HC (n = 28) | CRC (n = 56) | AS (n = 14) | |
| Age (yr) (M) (P25, P75) | 62.0 (55.0, 69.0) | 64.5 (58.8, 73.0) | 63.5 (56.3, 71.8) | 66.0 (60.3, 75.0) | 61.0 (49.5, 68.3) |
| Gender | |||||
| Male | 17 | 23 | 15 | 39 | 9 |
| Female | 7 | 11 | 13 | 17 | 5 |
| TNM stage | |||||
| Ⅰ | 9 | 9 | 2 | ||
| Ⅱ | 9 | 16 | 4 | ||
| Ⅲ | 10 | 21 | 6 | ||
| Ⅳ | 6 | 10 | 2 | ||
| Tumor size | |||||
| ≤ 5 cm | 20 | 29 | 9 | ||
| > 5 cm | 14 | 27 | 5 | ||
| Tumor location | |||||
| Left colon | 6 | 12 | 3 | ||
| Right colon | 5 | 9 | 2 | ||
| Rectum | 23 | 35 | 9 | ||
| Lymph node metastasis | |||||
| No | 19 | 35 | 6 | ||
| Yes | 15 | 21 | 8 | ||
| n | Circ_0000375 expression | P value | Circ_0011536 expression | P value | ||||
| Low | High | Low | High | |||||
| Age (yr) | 0.730 | 0.300 | ||||||
| ≤ 65 | 19 | 9 | 10 | 8 | 11 | |||
| > 65 | 15 | 8 | 7 | 9 | 6 | |||
| Gender | 0.714 | |||||||
| Female | 11 | 6 | 5 | 5 | 6 | 0.714 | ||
| Male | 23 | 11 | 12 | 12 | 11 | |||
| TNM stage | 0.169 | 0.039a | ||||||
| Ⅰ-Ⅱ | 18 | 11 | 7 | 12 | 6 | |||
| Ⅲ-Ⅳ | 16 | 6 | 10 | 5 | 11 | |||
| Tumor size | 0.486 | 1.000 | ||||||
| ≤ 5 cm | 20 | 9 | 11 | 10 | 10 | |||
| > 5 cm | 14 | 8 | 6 | 7 | 7 | |||
| Tumor location | 0.271 | 0.714 | ||||||
| Colon | 11 | 4 | 7 | 5 | 6 | |||
| Rectum | 23 | 13 | 10 | 12 | 11 | |||
| Lymph node metastasis | 0.084 | 0.016a | ||||||
| No | 19 | 12 | 7 | 13 | 6 | |||
| Yes | 15 | 5 | 10 | 4 | 11 | |||
Furthermore, the expression levels of the 2 circRNAs in serum samples from CRC patients were lower than those in serum samples of HCs (Figure 3D and E). For CRC patients after surgery, the levels of circ_0000375 and circ_0011536 in the serum were elevated compared with the levels in preoperative patients but were still significantly lower than the levels in HCs significantly, indicating that circ_0000375 and circ_0011536 in the serum were closely related to CRC tumorigenesis. The AUCs of circ_0000375, circ_0011536 and combined circRNAs in CRC serum were 0.976, 0.982 and 0.997, respectively (Figure 3F), indicating that these 2 circRNAs could serve as new biomarkers for CRC diagnosis and that both circRNAs might yield better performance. The expression levels of circ_0000375 and circ_0011536 in the serum were not correlated with the clinicopathological indexes of CRC patients, which suggested the robustness of the expression of these 2 circRNAs among different subtypes of CRC patients (Supplementary Table 2).
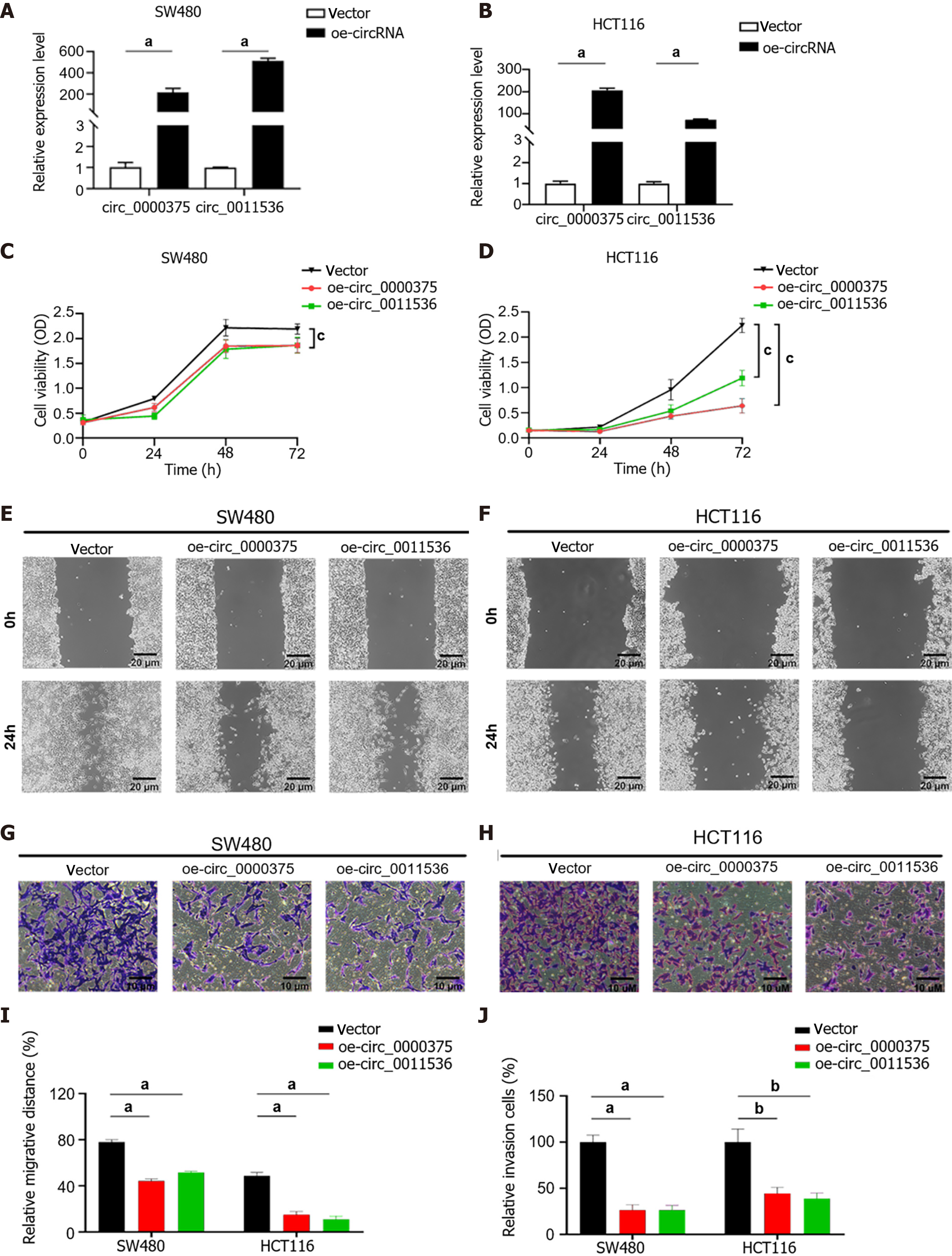
To investigate the biological effects of circ_0000375 and circ_0011536 on CRC cells, SW480 and HCT116 cells were transfected with overexpression vectors for these 2 circRNAs (Figure 4A and B). CCK-8 assays revealed that upregulation of circ_0000375 or circ_0011536 remarkably inhibited the proliferation of CRC cells (Figure 4C and D), indicating that these 2 circRNAs inhibited the proliferation of CRC cells. Wound healing assays were utilized to verify the function of the 2 circRNAs related to the migration of CRC cells, suggesting that the migratory abilities of CRC cells were significantly restrained by overexpression of circ_0000375 or circ_0011536 (Figure 4E, F and I). Transwell assay results suggested that these 2 circRNAs also affected the invasion of CRC cells. Upregulation of circ_0000375 or circ_0011536 suppressed CRC cell invasion capability (Figure 4G, H and J). Therefore, circ_0000375 and circ_0011536 suppressed cell proliferation, migration and invasion in CRC cells.
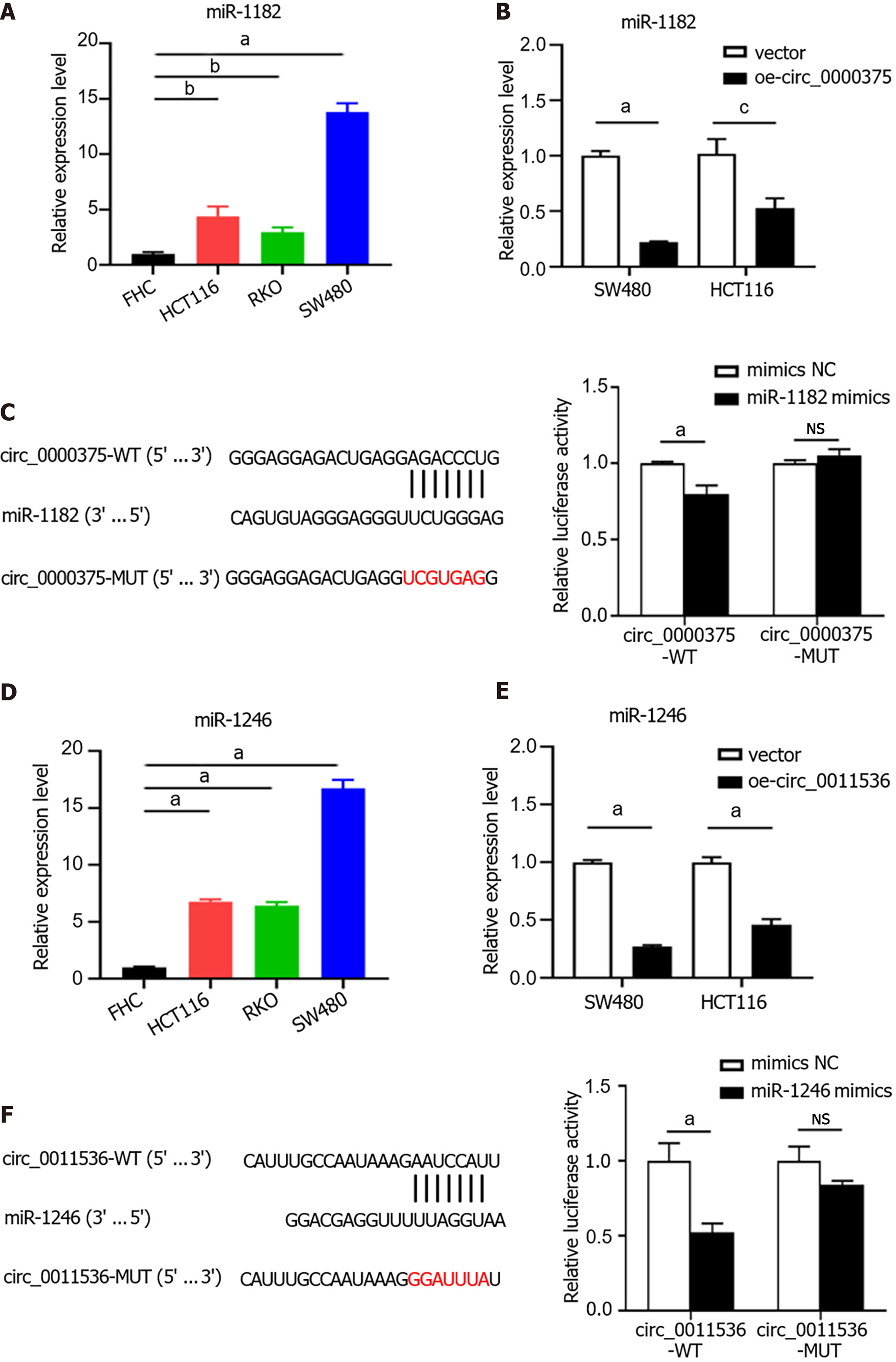
Given that mounting evidence has demonstrated that circRNAs serve as miRNA sponges, the miRNA binding abilities of circ_0000375 and circ_0011536 in CRC were explored using the CircInteractome database. We identified miR-1182 and miR-1246 as potential targets of circ_0000375 and circ_0011536, respectively, and then examined the interaction of these miRNAs with the circRNAs. The expression of miR-1182 was elevated in CRC cell lines compared with the FHC cell line (Figure 5A). MiR-1182 was downregulated in circ_0000375-overexpressing CRC cells (Figure 5B). Moreover, to examine the direct interaction between circ_0000375 and miR-1182, we conducted a dual-luciferase reporter assay. The results showed that the luciferase activity of circ_0000375-WT was significantly reduced in the miR-1182 mimics group compared with the mimics NC group, while no difference was observed with circ_0000375-MUT (Figure 5C). Similarly, miR-1246 was also upregulated in CRC cell lines (Figure 5D) but downregulated after overexpression of circ_0011536 (Figure 5E). MiR-1246 mimics led to decreased luciferase activity of circ_0011536-WT, but no change was found for circ_0011536-MUT (Figure 5F). Overall, circ_0000375 and circ_0011536 were confirmed to serve as sponges for miR-1182 and miR-1246, respectively, and the binding sites were uncovered in the present study.
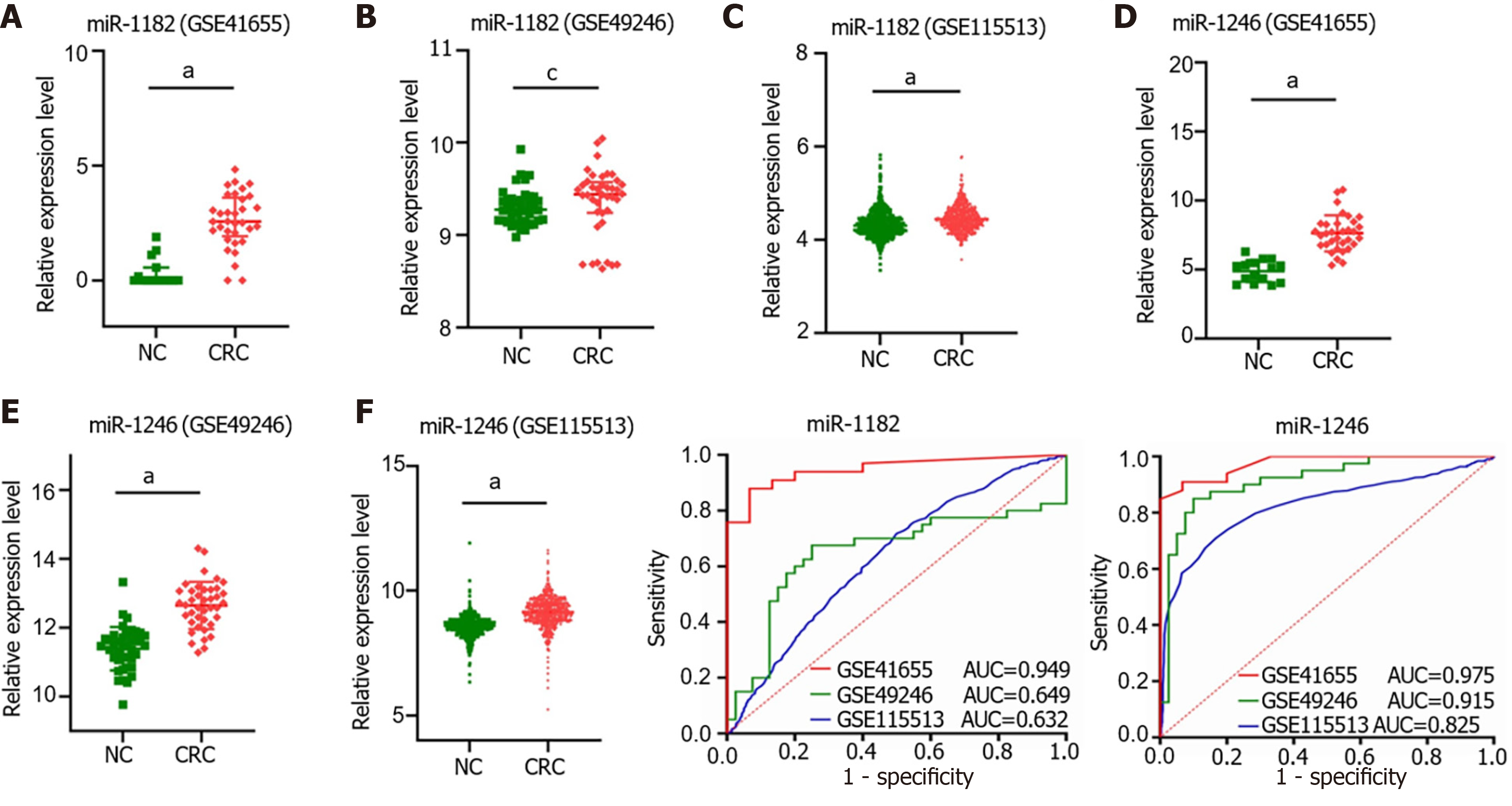
We compared the miR-1182 and miR-1246 expression levels among 3 GEO datasets, GSE41655 (33 CRCs vs 15 NCs), GSE49246 (40 CRCs vs 40 NCs) and GSE115513 (750 CRCs vs 649 NCs). The results suggested that miR-1182 was overexpressed in CRC tissues in GSE41655, GSE49246 and GSE115513 (Figure 6A-C), supporting our discovery in vitro. Consistently, the expression of miR-1246 was also increased in CRC tissues in GSE41655, GSE49246 and GSE115513 (Figure 6D-F). The diagnostic performances of miR-1182 and miR-1246 in these 3 GEO datasets are shown in Figure 6G and H, suggesting that these miRNAs could serve as CRC indicators. These results revealed that miR-1182 and miR-1246, predicted as target miRNAs for circ_0000375 and circ_0011536 in our study, are overexpressed in CRC tissues and might act as potential biomarkers for CRC diagnosis.
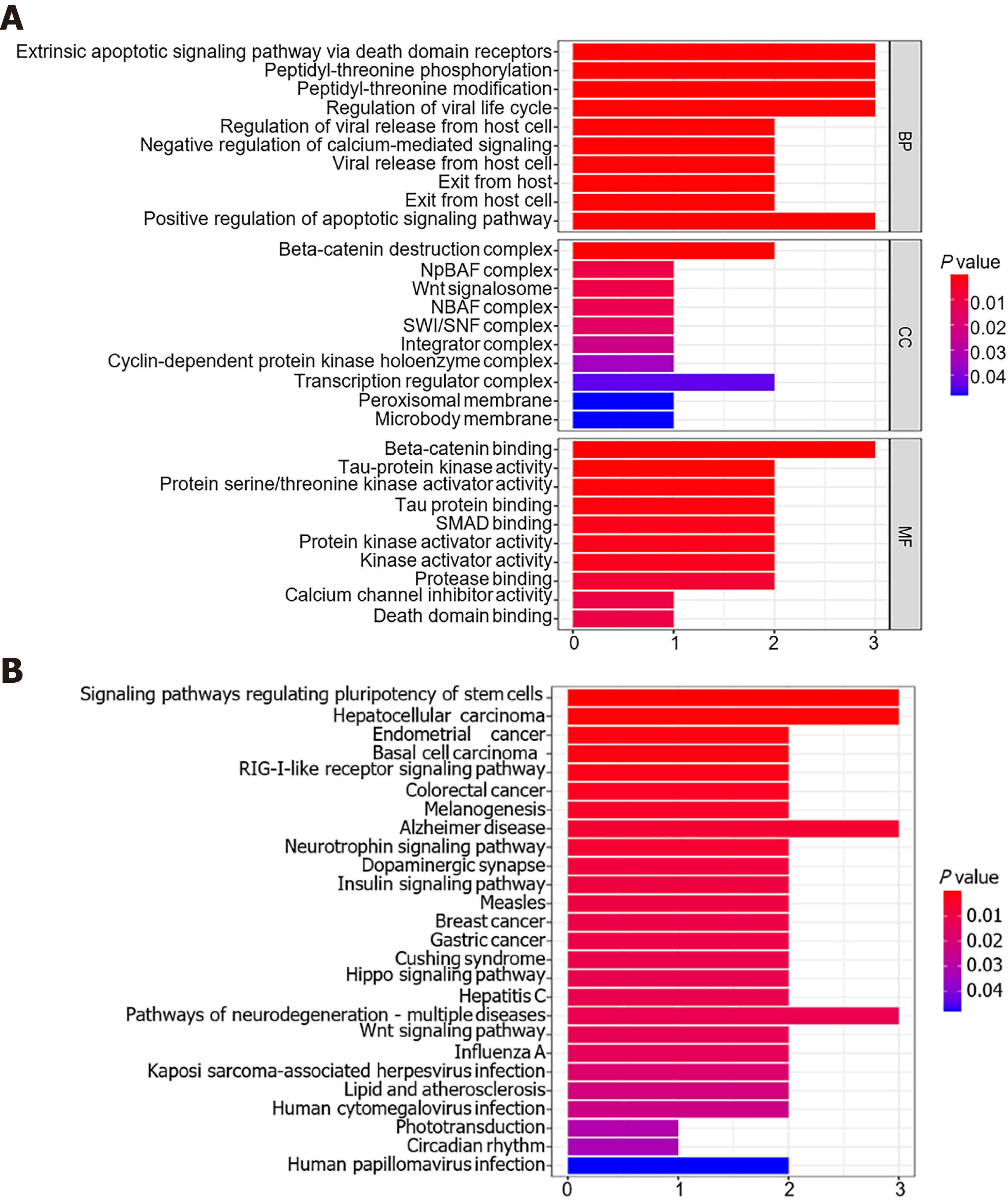
A total of 19 genes were predicted to be potential targets of miR-1182 and miR-1246 using the TargetScan, miRDB and miRTarBase databases. MTRNR2L8, TRIM25, DIRC2, ZNF609, GTF3C4, ARID1A, DARS and MAVS were recognized as downstream genes of miR-1182, and DYRK1A, GSK3B, AXIN2, CRADD, KIAA0895, GPATCH11, CALM2, CKS2, SKIL, RORA and ZNF502 were identified as targets for miR-1246 (Supplementary Table 3). Based on these 19 genes, the top 10 enrichment terms in GO analysis were revealed, including the apoptotic signaling pathway for biological processes, Wnt/beta-catenin signaling pathway and SWI/SNF complex for cellular components, beta-catenin binding and SMAD binding for molecular functions (Figure 7A). Meanwhile, KEGG analysis revealed that these genes were enriched in some tumorigenesis-associated pathways, including the signaling pathways regulating pluripotency of stem cells, the RIG-Ⅰ-like receptor signaling pathway, the Hippo signaling pathway and the Wnt signaling pathway (Figure 7B). Therefore, miR-1182 and miR-1246 and their target genes might play crucial roles in CRC tumorigenesis.
Although CRC treatment has been revolutionized in recent years, the current therapeutic strategies, including surgery, chemotherapy and molecular targeted therapy, still do not satisfy the ever-rising demand for early diagnostic and prognostic improvements for patients. Under this circumstance, it is pivotal to elucidate the mechanisms underlying CRC initiation and progression and to identify useful biomarkers for the early diagnosis of CRC. In this study, we first revealed that circ_0000375 and circ_0011536 are potential biomarkers for CRC diagnosis and then explored their function as tumor suppressors in CRC progression.
Compelling evidence indicates that circRNAs play crucial roles in diverse pathological processes in various cancer types. In our study, we compared the expression profiles in GSE126095 datasets and selected circ_0000375 and circ_0011536 as biomarker candidates. The expression levels of these 2 circRNAs were downregulated in CRC cell lines compared with normal colonic mucosa cells. Furthermore, we observed low circ_0000375 and circ_0011536 expression in CRC tissues and serum samples, with high diagnostic value, respectively, or combined. Interestingly, the serum expression levels of these 2 circRNAs were elevated in CRC patients 30 d postsurgery compared with preoperative patients, further supporting our discovery. Considering that serum exosomal circRNAs are abundant, stable and easy to test, circRNAs in the serum might have substantial advantages as promising noninvasive biomarkers for accurately diagnosing CRC, assessing disease stage and predicting prognosis. Previously, several circRNAs have been shown to be diagnostic biomarkers of CRC, including hsa_circ_0004771[11] and circ-PNN[21] in the serum. Metastasis and progression biomarkers of CRC have been revealed, including circPTK2[10] and circ-133[22], which might be new therapeutic targets. Four circRNAs, hsa_circ_0122319, hsa_circ_0087391, hsa_circ_0079480, and hsa_circ_0008039, were identified as reliable prognostic tools for postoperative recurrence of stage Ⅱ/Ⅲ CRC[23]. Furthermore, in the present study, we found that CRC patients with lymph node metastasis or advanced TNM stage tended to exhibit high circ_0011536 expression compared with patients in the early stage, suggesting that circ_0011536 is a more sensitive indicator for early CRC and that a complicated regulation system might impact the expression level of circ_0011536 in CRC tissues during CRC progression and metastasis. The expression levels of circ_0000375 in tissues and serum and circ_0011536 in serum samples from CRC patients were relatively stable and not associated with clinical features. Overall, we provide the first evidence of the potential of circ_0000375 and circ_0011536 as tumor biomarkers in tissues and serum samples from CRC patients, but the relationship of circRNA expression and clinicopathological features still needs to be confirmed by further investigation in a large cohort.
In our study, tumorigenic and metastatic properties, such as cell proliferation, migration and invasion, were suppressed by circ_0000375 and circ_0011536 overexpression in CRC cells. Next, miR-1182 and miR-1246 were predicted as target miRNAs of circ_0000375 and circ_0011536, respectively, through bioinformatics analysis, and these findings were validated by luciferase reporter assays. Furthermore, the expression levels of miR-1182 and miR-1246 in CRC cells were significantly decreased after transfection with circ_0000375 and circ_0011536 overexpression plasmids, further supporting our discovery. To date, an increasing number of studies have reported circRNAs as tumor regulators through direct binding to different miRNA molecules in CRC. For instance, CRC cell proliferation, metastasis and apoptosis can be regulated by circPACRGL via the miR-142-3p/miR-506-3p-TGF-β1 axis[24], and by circHIPK3 sponging of miR-7[25]. Moreover, circRNAs can play crucial roles in the therapeutic response of CRC as miRNA sponges. Hsa_circ_0005963 was reported to induce chemoresistance and modulate glycolysis in CRC through the miR-122-PKM2 axis[26]. Additionally, Liu et al[27] found that synthetic circRNA targeting miR-21 could efficiently inhibit cancer cell proliferation and achieve loss-of-function, revealing the enormous promise of circRNA sponges as innovative tools for molecular therapy in the future. In summary, our results described the tumor-suppressive role of circ_0000375 and circ_0011536 and identified the specific binding sites between circ_0000375 and miR-1182 and between circ_0011536 and miR-1246 for the first time, providing exploitable targets for CRC therapeutic strategies.
We next discovered significantly higher miR-1182 and miR-1246 expression levels in CRC cell lines than in normal colonic mucosa cells. Consistently, the expression levels of these 2 miRNAs were also upregulated in CRC tissues in three GEO datasets, GSE41655, GSE49246 and GSE115513. The critical roles of miR-1182 and miR-1246 have been investigated in some cancer types. For instance, miR-1182 was reported to attenuate proliferation and metastasis by targeting telomerase reverse transcriptase in gastric cancer[28]. Moreover, circCSPP1 in liver cancer[29], circ_0067934 in non-small cell lung cancer[30], circWHSC1 in ovarian cancer[31] and circFMN2 in CRC[32] can modulate the malignant behavior of tumors by sponging miR-1182. Moreover, miR-1246 plays an oncogenic role in metastasis by upregulating the methyltransferase METT3 in CRC[33]. Recent advances have indicated that infection with the CRC-associated bacterium Fusobacterium nucleatum might stimulate the generation of miR-1246/92b-3p/27a-3p-rich exosomes from tumor cells to promote the prometastatic ability of uninfected cells[34]. Mutant P53 might reprogram macrophages into tumor-supporting macrophages with anti-inflammatory immunosuppressive activity via exosomal miR-1246 in colon cancer cells, which could promote the progression and metastasis of CRC[35]. In summary, miR-1182 and miR-1246 might serve as biomarkers and function as oncogenic miRNAs in CRC carcinogenesis, but the diagnostic value of miR-1182 and miR-1246 in CRC tissues and serum samples still requires further investigation in the future.
In the present study, we also predicted the downstream genes of miR-1182 and miR-1246 as well as the potential signaling pathways involved in the regulation of these 2 miRNAs. The Wnt/beta-catenin signaling pathway is central to CRC tumorigenesis and might have promising value as a therapeutic target[36,37]. Zhao et al[30] found that miR-1182 might regulate the Wnt/beta-catenin pathway in lung cancer. In liver cancer, miR-1246 was reported to activate the Wnt/beta-catenin pathway in liver cancer stem cells[38] and regulate the RORalpha-Wnt/beta-catenin axis to promote liver cancer progression[39]. Moreover, miR-1246 was found to promote lung cancer metastasis and invasion through the Wnt/beta-catenin pathway by targeting GSK3B[40] and to induce apoptosis and overcome drug resistance by targeting AXIN2 and GSK3B[41]. DYRK1A was also reported to be a potential target of miR-1246 because overexpression of miR-1246 induced by P53 could downregulate the DYRK1A level[42]. In a nutshell, the targets of miR-1182 and miR-1246 were preliminarily identified in this study, and the Wnt/beta-catenin pathway might be involved in the regulation of these two miRNAs in CRC, which needs further exploration in the future.
In summary, we identified circ_0000375 and circ_0011536 as promising diagnostic biomarkers and tumor suppressors in CRC. To our knowledge, this is the first study to reveal the dysregulation of circ_0000375 and circ_0011536 in CRC tissues and serum samples and uncover their function in CRC pathogenesis. This is also the first study to investigate the interaction between circ_0000375 and miR-1182 and between circ_0011536 and miR-1246. However, some limitations also exist in our study. First, the robustness of these circRNAs and miRNAs as diagnostic and prognostic biomarkers should be confirmed with large-scale clinical samples. In addition, whether the tumor-suppressive effects of circ_0000375 and circ_0011536 can be counteracted by miR-1182 or miR-1246 remains unclear and should be validated through further comprehensive exploration. Finally, we tried to predict molecular mechanisms in this study, but the specific downstream genes and signaling pathways should be further elucidated later by performing experiments in vivo and in vitro.
To sum up, our findings demonstrated that circ_0000375 and circ_0011536 are downregulated in CRC cell lines, tissues and serum samples and exhibit potential diagnostic performances. Furthermore, these 2 circRNAs significantly suppress CRC cell proliferation, migration and invasion. As potential targets of circ_0000375 and circ_0011536, miR-1182 and miR-1246, respectively, are overexpressed in CRC cells and tissues. Our study not only further elucidates the mechanism of circRNA regulation in CRC cell growth and progression but also provides novel diagnostic biomarkers and promising therapeutic targets for CRC.
Colorectal cancer (CRC) is a common tumor worldwide. Circular ribonucleic acids (circRNAs) have been reported to regulate CRC as microRNA (miRNA) sponges. However, the aberrant expression and biological functions of circRNAs in CRC remain to be further explored.
CircRNAs might serve as novel biomarker candidates for CRC diagnosis. Further exploration of useful biomarkers to improve CRC diagnosis and treatment is crucial.
This study aimed to identify circRNAs that could be potential biomarkers of CRC and explore their functions in CRC carcinogenesis.
Cir_0000375 and circ_0011536 were selected as CRC biomarker candidates using the Gene Expression Omnibus (GEO) database. Quantitative real-time polymerase chain reaction was utilized to evaluate the expression levels of these 2 circRNAs in CRC tissues, serum samples and cell lines. Functional experiments, including cell counting kit-8 (CCK-8), wound healing and Transwell invasion assays, were carried out after overexpression of cir_0000375 and circ_0011536 in CRC cell lines. Furthermore, the expression levels of target miRNAs in CRC tissues were explored in GEO datasets and CRC cell lines. Then, the interactions of circRNAs and miRNAs and enrichment analysis of the miRNAs and target genes were further performed.
Our findings demonstrated that circ_0000375 and circ_0011536 were downregulated in CRC cell lines, tissues and serum samples and showed good diagnostic performance. Furthermore, these 2 circRNAs significantly inhibited CRC cell proliferation, migration and invasion. As potential targets of circ_0000375 and circ_0011536, miR-1182 and miR-1246, respectively, were overexpressed in CRC cells and tissues.
Circ_0000375 and circ_0011536 may function as tumor suppressors and serve as novel biomarkers for CRC diagnosis and are promising candidates for therapeutic exploration.
Our findings uncover the mechanisms by which 2 circRNAs, circ_0000375 and circ_0011536, suppress CRC cell growth and progression and provide novel diagnostic biomarkers and promising therapeutic targets for CRC.
We thank the Gene Expression Omnibus project for providing invaluable datasets for bioinformatic analysis.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Janvilisri T, Kołat D S-Editor: Guo XR L-Editor: A P-Editor: Yu HG
| 1. | Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-249. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 50630] [Cited by in F6Publishing: 43376] [Article Influence: 14458.7] [Reference Citation Analysis (47)] |
| 2. | Siegel RL, Miller KD, Goding Sauer A, Fedewa SA, Butterly LF, Anderson JC, Cercek A, Smith RA, Jemal A. Colorectal cancer statistics, 2020. CA Cancer J Clin. 2020;70:145-164. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2268] [Cited by in F6Publishing: 2755] [Article Influence: 688.8] [Reference Citation Analysis (2)] |
| 3. | Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet. 2014;383:1490-1502. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1965] [Cited by in F6Publishing: 2135] [Article Influence: 213.5] [Reference Citation Analysis (1)] |
| 4. | Gao Y, Wang J, Zhou Y, Sheng S, Qian SY, Huo X. Evaluation of Serum CEA, CA19-9, CA72-4, CA125 and Ferritin as Diagnostic Markers and Factors of Clinical Parameters for Colorectal Cancer. Sci Rep. 2018;8:2732. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 94] [Cited by in F6Publishing: 156] [Article Influence: 26.0] [Reference Citation Analysis (0)] |
| 5. | Su M, Xiao Y, Ma J, Tang Y, Tian B, Zhang Y, Li X, Wu Z, Yang D, Zhou Y, Wang H, Liao Q, Wang W. Circular RNAs in Cancer: emerging functions in hallmarks, stemness, resistance and roles as potential biomarkers. Mol Cancer. 2019;18:90. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 188] [Cited by in F6Publishing: 260] [Article Influence: 52.0] [Reference Citation Analysis (0)] |
| 6. | Chen B, Huang S. Circular RNA: An emerging non-coding RNA as a regulator and biomarker in cancer. Cancer Lett. 2018;418:41-50. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 167] [Cited by in F6Publishing: 219] [Article Influence: 36.5] [Reference Citation Analysis (0)] |
| 7. | Han B, Chao J, Yao H. Circular RNA and its mechanisms in disease: From the bench to the clinic. Pharmacol Ther. 2018;187:31-44. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 403] [Cited by in F6Publishing: 540] [Article Influence: 90.0] [Reference Citation Analysis (0)] |
| 8. | Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384-388. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4631] [Cited by in F6Publishing: 5500] [Article Influence: 500.0] [Reference Citation Analysis (0)] |
| 9. | Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20:675-691. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1947] [Cited by in F6Publishing: 2523] [Article Influence: 504.6] [Reference Citation Analysis (0)] |
| 10. | Yang H, Li X, Meng Q, Sun H, Wu S, Hu W, Liu G, Yang Y, Chen R. CircPTK2 (hsa_circ_0005273) as a novel therapeutic target for metastatic colorectal cancer. Mol Cancer. 2020;19:13. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 138] [Cited by in F6Publishing: 140] [Article Influence: 35.0] [Reference Citation Analysis (0)] |
| 11. | Pan B, Qin J, Liu X, He B, Wang X, Pan Y, Sun H, Xu T, Xu M, Chen X, Xu X, Zeng K, Sun L, Wang S. Identification of Serum Exosomal hsa-circ-0004771 as a Novel Diagnostic Biomarker of Colorectal Cancer. Front Genet. 2019;10:1096. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 108] [Cited by in F6Publishing: 146] [Article Influence: 29.2] [Reference Citation Analysis (0)] |
| 12. | Ding B, Yao M, Fan W, Lou W. Whole-transcriptome analysis reveals a potential hsa_circ_0001955/hsa_circ_0000977-mediated miRNA-mRNA regulatory sub-network in colorectal cancer. Aging (Albany NY). 2020;12:5259-5279. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 55] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 13. | Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175-205. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2105] [Cited by in F6Publishing: 2246] [Article Influence: 132.1] [Reference Citation Analysis (0)] |
| 14. | Lu TX, Rothenberg ME. MicroRNA. J Allergy Clin Immunol. 2018;141:1202-1207. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 853] [Cited by in F6Publishing: 1429] [Article Influence: 204.1] [Reference Citation Analysis (0)] |
| 15. | Marcuello M, Vymetalkova V, Neves RPL, Duran-Sanchon S, Vedeld HM, Tham E, van Dalum G, Flügen G, Garcia-Barberan V, Fijneman RJ, Castells A, Vodicka P, Lind GE, Stoecklein NH, Heitzer E, Gironella M. Circulating biomarkers for early detection and clinical management of colorectal cancer. Mol Aspects Med. 2019;69:107-122. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 127] [Cited by in F6Publishing: 168] [Article Influence: 33.6] [Reference Citation Analysis (0)] |
| 16. | Long J, He Q, Yin Y, Lei X, Li Z, Zhu W. The effect of miRNA and autophagy on colorectal cancer. Cell Prolif. 2020;53:e12900. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 43] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 17. | Savardashtaki A, Shabaninejad Z, Movahedpour A, Sahebnasagh R, Mirzaei H, Hamblin MR. miRNAs derived from cancer-associated fibroblasts in colorectal cancer. Epigenomics. 2019;11:1627-1645. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 53] [Cited by in F6Publishing: 45] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 18. | Dong J, Tai JW, Lu LF. miRNA-Microbiota Interaction in Gut Homeostasis and Colorectal Cancer. Trends Cancer. 2019;5:666-669. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 25] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 19. | Ren A, Dong Y, Tsoi H, Yu J. Detection of miRNA as non-invasive biomarkers of colorectal cancer. Int J Mol Sci. 2015;16:2810-2823. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 57] [Cited by in F6Publishing: 60] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 20. | Yin TF, Zhao DY, Zhou YC, Wang QQ, Yao SK. Identification of the circRNA-miRNA-mRNA regulatory network and its prognostic effect in colorectal cancer. World J Clin Cases. 2021;9:4520-4541. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 7] [Cited by in F6Publishing: 6] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 21. | Xie Y, Li J, Li P, Li N, Zhang Y, Binang H, Zhao Y, Duan W, Chen Y, Wang Y, Du L, Wang C. RNA-Seq Profiling of Serum Exosomal Circular RNAs Reveals Circ-PNN as a Potential Biomarker for Human Colorectal Cancer. Front Oncol. 2020;10:982. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 49] [Cited by in F6Publishing: 64] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 22. | Yang H, Zhang H, Yang Y, Wang X, Deng T, Liu R, Ning T, Bai M, Li H, Zhu K, Li J, Fan Q, Ying G, Ba Y. Hypoxia induced exosomal circRNA promotes metastasis of Colorectal Cancer via targeting GEF-H1/RhoA axis. Theranostics. 2020;10:8211-8226. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 67] [Cited by in F6Publishing: 122] [Article Influence: 30.5] [Reference Citation Analysis (0)] |
| 23. | Ju HQ, Zhao Q, Wang F, Lan P, Wang Z, Zuo ZX, Wu QN, Fan XJ, Mo HY, Chen L, Li T, Ren C, Wan XB, Chen G, Li YH, Jia WH, Xu RH. A circRNA signature predicts postoperative recurrence in stage II/III colon cancer. EMBO Mol Med. 2019;11:e10168. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 57] [Cited by in F6Publishing: 78] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 24. | Shang A, Gu C, Wang W, Wang X, Sun J, Zeng B, Chen C, Chang W, Ping Y, Ji P, Wu J, Quan W, Yao Y, Zhou Y, Sun Z, Li D. Exosomal circPACRGL promotes progression of colorectal cancer via the miR-142-3p/miR-506-3p- TGF-β1 axis. Mol Cancer. 2020;19:117. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 133] [Cited by in F6Publishing: 250] [Article Influence: 62.5] [Reference Citation Analysis (0)] |
| 25. | Zeng K, Chen X, Xu M, Liu X, Hu X, Xu T, Sun H, Pan Y, He B, Wang S. CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell Death Dis. 2018;9:417. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 363] [Cited by in F6Publishing: 463] [Article Influence: 77.2] [Reference Citation Analysis (0)] |
| 26. | Wang X, Zhang H, Yang H, Bai M, Ning T, Deng T, Liu R, Fan Q, Zhu K, Li J, Zhan Y, Ying G, Ba Y. Exosome-delivered circRNA promotes glycolysis to induce chemoresistance through the miR-122-PKM2 axis in colorectal cancer. Mol Oncol. 2020;14:539-555. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 294] [Cited by in F6Publishing: 317] [Article Influence: 79.3] [Reference Citation Analysis (0)] |
| 27. | Liu X, Abraham JM, Cheng Y, Wang Z, Zhang G, Ashktorab H, Smoot DT, Cole RN, Boronina TN, DeVine LR, Talbot CC Jr, Liu Z, Meltzer SJ. Synthetic Circular RNA Functions as a miR-21 Sponge to Suppress Gastric Carcinoma Cell Proliferation. Mol Ther Nucleic Acids. 2018;13:312-321. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 101] [Cited by in F6Publishing: 134] [Article Influence: 22.3] [Reference Citation Analysis (0)] |
| 28. | Zhang D, Xiao YF, Zhang JW, Xie R, Hu CJ, Tang B, Wang SM, Wu YY, Hao NB, Yang SM. miR-1182 attenuates gastric cancer proliferation and metastasis by targeting the open reading frame of hTERT. Cancer Lett. 2015;360:151-159. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 52] [Cited by in F6Publishing: 59] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 29. | Jia N, Song Z, Chen B, Cheng J, Zhou W. A Novel Circular RNA circCSPP1 Promotes Liver Cancer Progression by Sponging miR-1182. Onco Targets Ther. 2021;14:2829-2838. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 6] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 30. | Zhao M, Ma W, Ma C. Circ_0067934 promotes non-small cell lung cancer development by regulating miR-1182/KLF8 axis and activating Wnt/β-catenin pathway. Biomed Pharmacother. 2020;129:110461. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 32] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 31. | Zong ZH, Du YP, Guan X, Chen S, Zhao Y. CircWHSC1 promotes ovarian cancer progression by regulating MUC1 and hTERT through sponging miR-145 and miR-1182. J Exp Clin Cancer Res. 2019;38:437. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 69] [Cited by in F6Publishing: 92] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 32. | Li Y, Li C, Xu R, Wang Y, Li D, Zhang B. A novel circFMN2 promotes tumor proliferation in CRC by regulating the miR-1182/hTERT signaling pathways. Clin Sci (Lond). 2019;133:2463-2479. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 48] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 33. | Peng W, Li J, Chen R, Gu Q, Yang P, Qian W, Ji D, Wang Q, Zhang Z, Tang J, Sun Y. Upregulated METTL3 promotes metastasis of colorectal Cancer via miR-1246/SPRED2/MAPK signaling pathway. J Exp Clin Cancer Res. 2019;38:393. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 151] [Cited by in F6Publishing: 237] [Article Influence: 47.4] [Reference Citation Analysis (0)] |
| 34. | Guo S, Chen J, Chen F, Zeng Q, Liu WL, Zhang G. Exosomes derived from Fusobacterium nucleatum-infected colorectal cancer cells facilitate tumour metastasis by selectively carrying miR-1246/92b-3p/27a-3p and CXCL16. Gut. 2020;. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 95] [Cited by in F6Publishing: 88] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 35. | Cooks T, Pateras IS, Jenkins LM, Patel KM, Robles AI, Morris J, Forshew T, Appella E, Gorgoulis VG, Harris CC. Mutant p53 cancers reprogram macrophages to tumor supporting macrophages via exosomal miR-1246. Nat Commun. 2018;9:771. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 247] [Cited by in F6Publishing: 328] [Article Influence: 54.7] [Reference Citation Analysis (0)] |
| 36. | Bian J, Dannappel M, Wan C, Firestein R. Transcriptional Regulation of Wnt/β-Catenin Pathway in Colorectal Cancer. Cells. 2020;9. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 47] [Cited by in F6Publishing: 110] [Article Influence: 27.5] [Reference Citation Analysis (0)] |
| 37. | Cheng X, Xu X, Chen D, Zhao F, Wang W. Therapeutic potential of targeting the Wnt/β-catenin signaling pathway in colorectal cancer. Biomed Pharmacother. 2019;110:473-481. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 171] [Cited by in F6Publishing: 241] [Article Influence: 40.2] [Reference Citation Analysis (0)] |
| 38. | Chai S, Ng KY, Tong M, Lau EY, Lee TK, Chan KW, Yuan YF, Cheung TT, Cheung ST, Wang XQ, Wong N, Lo CM, Man K, Guan XY, Ma S. Octamer 4/microRNA-1246 signaling axis drives Wnt/β-catenin activation in liver cancer stem cells. Hepatology. 2016;64:2062-2076. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 115] [Cited by in F6Publishing: 142] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 39. | Huang JL, Fu YP, Gan W, Liu G, Zhou PY, Zhou C, Sun BY, Guan RY, Zhou J, Fan J, Yi Y, Qiu SJ. Hepatic stellate cells promote the progression of hepatocellular carcinoma through microRNA-1246-RORα-Wnt/β-Catenin axis. Cancer Lett. 2020;476:140-151. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 33] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 40. | Yang F, Xiong H, Duan L, Li Q, Li X, Zhou Y. MiR-1246 Promotes Metastasis and Invasion of A549 cells by Targeting GSK-3β‒Mediated Wnt/β-Catenin Pathway. Cancer Res Treat. 2019;51:1420-1429. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 61] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 41. | Xie B, Li L, Zhang Z, Zhao L, Cheng J, Zhou C, Yan J, Chen J, Yi J, Wang B, Jin S, Wei H. MicroRNA-1246 by Targeting AXIN2 and GSK-3β Overcomes Drug Resistance and Induces Apoptosis in Chemo-resistant Leukemia Cells. J Cancer. 2021;12:4196-4208. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 5] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 42. | Zhang Y, Liao JM, Zeng SX, Lu H. p53 downregulates Down syndrome-associated DYRK1A through miR-1246. EMBO Rep. 2011;12:811-817. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 106] [Cited by in F6Publishing: 108] [Article Influence: 8.3] [Reference Citation Analysis (0)] |