Abstract
Background
Pancreatic ductal adenocarcinoma (PDA) is a fatal cancer for which even unfavorable clinicopathological factors occasionally fail to preclude long-term survival. We sought to establish a scoring system that utilizes measurable pre-intervention factors for predicting survival following surgical resection.
Methods
We retrospectively analyzed 34 patients who died from short-term recurrences and 32 long-term survivors among 310 consecutively resected patients with PDA. A logistic regression model was used to define factors related to clinical parameters, molecular profiles of 18 pancreatic cancer-associated genes, and aberrant expression of major tumor suppressors.
Results
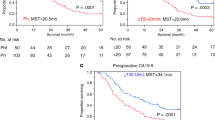
Carbohydrate antigen 19-9 (CA19-9) had the best ability to classify patients with short-term recurrence and long-term survivors [odds ratio 21.04, 95% confidence interval (CI) 4.612–96.019], followed by SMAD4 and TP53 mutation scoring (odds ratio 41.322, 95% CI 3.156–541.035). Missense TP53 mutations were strongly associated with the nuclear expression of p53, whereas truncating mutations were associated with the absence of nuclear p53. The former subset was associated with a worse prognosis. The combination of aberrant SMAD4 and mutation types of TP53 exhibited a better resolution for distinguishing patients with short-term recurrences from long-term survivors (compared with the assessment of the number of mutated KRAS, CDKN2A, TP53, and SMAD4 genes). Calibration of mutation scores combined with CA19-9 in a logistic regression model setting demonstrated a practical effect in classifying long survivors and patients with early recurrence (c-statistic = 0.876).
Conclusions
Genetic information, i.e., TP53 mutation types and SMAD4 abnormalities, combined with CA19-9, will be a valuable tool for improving surgical strategies for pancreatic cancer.


Similar content being viewed by others
References
Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74(11):2913–21.
Majumder S, Philip NA, Singh Nagpal SJ, et al. High-grade dysplasia in resected main-duct intraductal papillary mucinous neoplasm (MD-IPMN) is associated with an increased risk of subsequent pancreatic cancer. Am J Gastroenterol. 2019;114(3):524–9.
McGuigan A, Kelly P, Turkington RC, Jones C, Coleman HG, McCain RS. Pancreatic cancer: a review of clinical diagnosis, epidemiology, treatment and outcomes. World J Gastroenterol. 2018;24(43):4846–61.
Mizrahi JD, Surana R, Valle JW, Shroff RT. Pancreatic cancer. Lancet. 2020;395(10242):2008–20.
Jamieson NB, Denley SM, Logue J, et al. A prospective comparison of the prognostic value of tumor- and patient-related factors in patients undergoing potentially curative surgery for pancreatic ductal adenocarcinoma. Ann Surg Oncol. 2011;18(8):2318–28.
Imaoka H, Mizuno N, Hara K, et al. Evaluation of modified Glasgow prognostic score for pancreatic cancer: a retrospective cohort study. Pancreas. 2016;45(2):211–7.
Kim J, Bamlet WR, Oberg AL, et al. Detection of early pancreatic ductal adenocarcinoma with thrombospondin-2 and CA19-9 blood markers. Sci Transl Med. 2017;9(398):eaah5583.
Zhou Y, Wei Q, Fan J, Cheng S, Ding W, Hua Z. Prognostic role of the neutrophil-to-lymphocyte ratio in pancreatic cancer: a meta-analysis containing 8252 patients. Clin Chim Acta. 2018;479:181–9.
Melo SA, Luecke LB, Kahlert C, et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature. 2015;523(7559):177–82.
Krantz BA, O’Reilly EM. Biomarker-based therapy in pancreatic ductal adenocarcinoma: an emerging reality? Clin Cancer Res. 2018;24(10):2241–50.
Yachida S, White CM, Naito Y, et al. Clinical significance of the genetic landscape of pancreatic cancer and implications for identification of potential long-term survivors. Clin Cancer Res. 2012;18(22):6339–47.
Hayashi H, Kohno T, Ueno H, et al. Utility of assessing the number of mutated KRAS, CDKN2A, TP53, and SMAD4 genes using a targeted deep sequencing assay as a prognostic biomarker for pancreatic cancer. Pancreas. 2017;46(3):335–40.
Qian ZR, Rubinson DA, Nowak JA, et al. Association of alterations in main driver genes with outcomes of patients with resected pancreatic ductal adenocarcinoma. JAMA Oncol. 2018;4(3):e173420.
Dan A, Fujii D, Soda S, Machimura T, Ike M. Removal of phenol, bisphenol A, and 4-tert-butylphenol from synthetic landfill leachate by vertical flow constructed wetlands. Sci Total Environ. 2017;578:566–76.
Witkiewicz AK, McMillan EA, Balaji U, et al. Whole-exome sequencing of pancreatic cancer defines genetic diversity and therapeutic targets. Nat Commun. 2015;6:6744.
Qian Y, Gong Y, Fan Z, et al. Molecular alterations and targeted therapy in pancreatic ductal adenocarcinoma. J Hematol Oncol. 2020;13(1):130.
Isaji S, Murata Y, Kishiwada M. New Japanese classification of pancreatic cancer. In: JP Neoptolemos, R Urrutia, JL Abbruzzese, MW Büchler, editors. Pancreatic cancer. New York, NY: Springer, New York; 2018. p. 1021–37.
Sobin LH, Gospodarowicz MK, Wittekind C. TNM classification of malignant tumours. NJ: John Wiley & Sons; 2011.
Omori Y, Ono Y, Tanino M, et al. Pathways of progression from intraductal papillary mucinous neoplasm to pancreatic ductal adenocarcinoma based on molecular features. Gastroenterology. 2019;156(3):647–661 e642.
Liu J, Lichtenberg T, Hoadley KA, et al. An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell. 2018;173(2):400–416 e411.
Nakamura T, Asano T, Okamura K, et al. A preoperative prognostic scoring system to predict prognosis for resectable pancreatic cancer: who will benefit from upfront surgery? J Gastrointest Surg. 2019;23(5):990–6.
Bournet B, Buscail C, Muscari F, Cordelier P, Buscail L. Targeting KRAS for diagnosis, prognosis, and treatment of pancreatic cancer: hopes and realities. Eur J Cancer. 2016;54:75–83.
McIntyre CA, Lawrence SA, Richards AL, et al. Alterations in driver genes are predictive of survival in patients with resected pancreatic ductal adenocarcinoma. Cancer. 2020;126(17):3939–49.
Bournet B, Muscari F, Buscail C, et al. KRAS G12D Mutation subtype is a prognostic factor for advanced pancreatic adenocarcinoma. Clin Transl Gastroenterol. 2016;7:e157.
Maddalena M, Mallel G, Nataraj NB, et al. TP53 missense mutations in PDAC are associated with enhanced fibrosis and an immunosuppressive microenvironment. Proc Natl Acad Sci USA. 2021;118(23)
Kang H, Kim SS, Sung MJ, et al. Radiographic portal or superior mesenteric vein invasion is an independent prognostic factor in non-metastatic pancreatic ductal adenocarcinoma: a missing block of clinical T staging? Pancreatology. 2020;20(5):952–9.
Elshaer M, Gravante G, Kosmin M, Riaz A, Al-Bahrani A. A systematic review of the prognostic value of lymph node ratio, number of positive nodes and total nodes examined in pancreatic ductal adenocarcinoma. Ann R Coll Surg Engl. 2017;99(2):101–6.
Puli SR, Bechtold ML, Buxbaum JL, Eloubeidi MA. How good is endoscopic ultrasound-guided fine-needle aspiration in diagnosing the correct etiology for a solid pancreatic mass?: A meta-analysis and systematic review. Pancreas. 2013;42(1):20–6.
Wang W, Shpaner A, Krishna SG, et al. Use of EUS-FNA in diagnosing pancreatic neoplasm without a definitive mass on CT. Gastrointest Endosc. 2013;78(1):73–80.
Wang LH, Wu CF, Rajasekaran N, Shin YK. Loss of tumor suppressor gene function in human cancer: an overview. Cell Physiol Biochem. 2018;51(6):2647–93.
Hingorani SR, Wang L, Multani AS, et al. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell. 2005;7(5):469–83.
Bargonetti J, Prives C. Gain-of-function mutant p53: history and speculation. J Mol Cell Biol. 2019;11(7):605–9.
Escobar-Hoyos LF, Penson A, Kannan R, et al. Altered RNA splicing by mutant p53 activates oncogenic RAS signaling in pancreatic cancer. Cancer Cell. 2020;38(2):198–211 e198.
Kobel M, Piskorz AM, Lee S, et al. Optimized p53 immunohistochemistry is an accurate predictor of TP53 mutation in ovarian carcinoma. J Pathol Clin Res. 2016;2(4):247–58.
Ryan DP, Hong TS, Bardeesy N. Pancreatic adenocarcinoma. N Engl J Med. 2014;371(11):1039–49.
Singhi AD, George B, Greenbowe JR, et al. Real-time targeted genome profile analysis of pancreatic ductal adenocarcinomas identifies genetic alterations that might be targeted with existing drugs or used as biomarkers. Gastroenterology. 2019;156(8):2242–2253 e2244.
Heining C, Horak P, Uhrig S, et al. NRG1 fusions in KRAS wild-type pancreatic cancer. Cancer Discov. 2018;8(9):1087–95.
Luo G, Liu C, Guo M, et al. Potential biomarkers in Lewis negative patients with pancreatic cancer. Ann Surg. 2017;265(4):800–5.
Kusama K, Okamoto Y, Saito K, et al. Reevaluation of Pholiota squarrosa lectin-reactive haptoglobin as a pancreatic cancer biomarker using an improved ELISA system. Glycoconj J. 2017;34(4):537–44.
Dal Molin M, Zhang M, de Wilde RF, et al. Very long-term survival following resection for pancreatic cancer is not explained by commonly mutated genes: results of whole-exome sequencing analysis. Clin Cancer Res. 2015;21(8):1944–50.
Okada T, Mizukami Y, Ono Y, et al. Digital PCR-based plasma cell-free DNA mutation analysis for early-stage pancreatic tumor diagnosis and surveillance. J Gastroenterol. 2020;55(12):1183–93.
Lee B, Lipton L, Cohen J, et al. Circulating tumor DNA as a potential marker of adjuvant chemotherapy benefit following surgery for localized pancreatic cancer. Ann Oncol. 2019;30(9):1472–8.
Acknowledgements
We would like to thank Atsuko Sumi and Munehiko Ogata (Sapporo Higashi Tokushukai Hospital) for technical support regarding genetic analyses. We thank other members of Gastroenterological Surgery II at Hokkaido Medical University and the laboratory staff of the Institute of Biomedical Research at Sapporo Higashi Tokushukai Hospital for helpful suggestions throughout the course of this project and critical reading of the manuscript. We are also very grateful to Osamu Takahashi (Thermo-Fisher Scientific) for technical support regarding the sequencing analyses. This work was supported by JSPS KAKENHI grant number 20K07671 (to Y. Ono), 21K08724 (to T.N.), and 20H03655 (to Y.M.).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Disclosure
Yusuke Ono and Yusuke Mizukami received funding from the Hitachi High-Tech Corporation. The other authors declare that they have no known competing financial interests or personal relationships that could have influenced the work reported in this paper.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ono, M., Ono, Y., Nakamura, T. et al. Predictors of Long-Term Survival in Pancreatic Ductal Adenocarcinoma after Pancreatectomy: TP53 and SMAD4 Mutation Scoring in Combination with CA19-9. Ann Surg Oncol 29, 5007–5019 (2022). https://doi.org/10.1245/s10434-022-11630-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1245/s10434-022-11630-0