Abstract
Background
Despite a greater understanding of the molecular heterogeneity of breast cancer, current therapeutic strategies still cannot overcome the relatively poor prognosis of triple-negative breast cancer (TNBC). Deregulation of fibroblast growth factor (FGF) signaling has been found in breast cancer, and blocking this pathway has been suggested as a potential therapeutic target. We therefore evaluated the expression and copy number changes of FGF family members in TNBC.
Methods
We retrospectively evaluated 148 primary TNBC in 2009 for FGFR1, FGFR2, and FGF2 expression by immunohistochemistry. FGFR1 and FGFR2 gene copy numbers were analyzed by fluorescence in situ hybridization. The Cancer Genome Atlas (TCGA) data was used to study correlations between gene expression and amplification or methylation of FGFR1, FGFR2, and FGF2 in basal-like TNBC.
Results
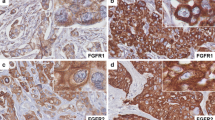
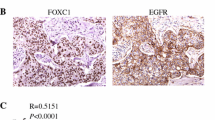
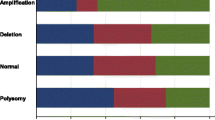
FGFR1, FGFR2, and FGF2 expression were found in 16.2 % (24 of 148), 12.8 % (19 of 148), and 12.8 % (19 of 148) of TNBCs, respectively. FGFR1 gene amplification was observed in 4.1 % (6 of 145), and FGFR1 high polysomy was detected in 6.9 % (10 of 145) of the cases examined. FGFR2 gene amplification and high polysomy were identified in 4.7 % (6 of 129 cases) and 0.8 % (1 of 129 cases), respectively. FGF2 expression was found to be associated with basal-like TNBC. The expression of FGF family members and FGFR1 or FGFR2 gene amplification did not affect patient survival. TCGA data revealed that promoter methylation of the 3 genes was significantly associated with mRNA expression.
Conclusions
Even though the implications for patient outcomes are not significant, subsets of TNBCs harbor FGFR1 or FGFR2 gene amplification and FGFR1, FGFR2, or FGF2 protein overexpression.

Similar content being viewed by others
References
Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121:2750–67.
Ossovskaya V, Wang Y, Budoff A, Xu Q, Lituev A, Potapova O, et al. Exploring molecular pathways of triple-negative breast cancer. Genes Cancer. 2011;2:870–9.
Turner N, Lambros MB, Horlings HM, Pearson A, Sharpe R, Natrajan R, et al. Integrative molecular profiling of triple negative breast cancers identifies amplicon drivers and potential therapeutic targets. Oncogene. 2010;29:2013–23.
Brooks AN, Kilgour E, Smith PD. Molecular pathways: fibroblast growth factor signaling: a new therapeutic opportunity in cancer. Clin Cancer Res. 2012;18:1855–62.
Ahmad I, Iwata T, Leung HY. Mechanisms of FGFR-mediated carcinogenesis. Biochim Biophys Acta. 2012;1823:850–60.
Elbauomy Elsheikh S, Green AR, Lambros MB, Turner NC, Grainge MJ, Powe D, et al. FGFR1 amplification in breast carcinomas: a chromogenic in situ hybridisation analysis. Breast Cancer Res. 2007;9:R23.
Hammond ME, Hayes DF, Dowsett M, Allred DC, Hagerty KL, Badve S, et al. American Society of Clinical Oncology/College Of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J Clin Oncol. 2010;28:2784–95.
Koh YW, Lee HJ, Lee JW, Kang J, Gong G. Dual-color silver-enhanced in situ hybridization for assessing HER2 gene amplification in breast cancer. Mod Pathol. 2011;24:794–800.
Jang MH, Kim EJ, Choi Y, Lee HE, Kim YJ, Kim JH, et al. FGFR1 is amplified during the progression of in situ to invasive breast carcinoma. Breast Cancer Res. 2012;14:R115.
Cappuzzo F, Hirsch FR, Rossi E, et al. Epidermal growth factor receptor gene and protein and gefitinib sensitivity in non-small-cell lung cancer. J Natl Cancer Inst. 2005;97:643-55.
Stephens PJ, Tarpey PS, Davies H, Van Loo P, Greenman C, Wedge DC, et al. The landscape of cancer genes and mutational processes in breast cancer. Nature. 2012;486:400–4.
Nielsen TO, Hsu FD, Jensen K, Cheang M, Karaca G, Hu Z, et al. Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin Cancer Res. 2004;10:5367–74.
Cheang MC, Voduc D, Bajdik C, Leung S, McKinney S, Chia SK, et al. Basal-like breast cancer defined by five biomarkers has superior prognostic value than triple-negative phenotype. Clin Cancer Res. 2008;14:1368–76.
Beroukhim R, Getz G, Nghiemphu L, Barretina J, Hsueh T, Linhart D, et al. Assessing the significance of chromosomal aberrations in cancer: methodology and application to glioma. Proc Natl Acad Sci USA. 2007;104:20007–12.
Massabeau C, Sigal-Zafrani B, Belin L, Savignoni A, Richardson M, Kirova YM, et al. The fibroblast growth factor receptor 1 (FGFR1), a marker of response to chemoradiotherapy in breast cancer? Breast Cancer Res Treat. 2012;134:259–66.
Sun S, Jiang Y, Zhang G, Song H, Zhang X, Zhang Y, et al. Increased expression of fibroblastic growth factor receptor 2 is correlated with poor prognosis in patients with breast cancer. J Surg Oncol. 2012;105:773–9.
Gelsi-Boyer V, Orsetti B, Cervera N, Finetti P, Sircoulomb F, Rougé C, et al. Comprehensive profiling of 8p11-12 amplification in breast cancer. Mol Cancer Res. 2005;3:655–67.
Turner N, Pearson A, Sharpe R, Lambros M, Geyer F, Lopez-Garcia MA, et al. FGFR1 amplification drives endocrine therapy resistance and is a therapeutic target in breast cancer. Cancer Res. 2010;70:2085–94.
Sharpe R, Pearson A, Herrera-Abreu MT, Johnson D, Mackay A, Welti JC, et al. FGFR signaling promotes the growth of triple-negative and basal-like breast cancer cell lines both in vitro and in vivo. Clin Cancer Res. 2011;17:5275–86.
Jain VK, Turner NC. Challenges and opportunities in the targeting of fibroblast growth factor receptors in breast cancer. Breast Cancer Res. 2012;14:208.
Michor F, Polyak K. The origins and implications of intratumor heterogeneity. Cancer Prev Res Phila. 2010;3:1361–4.
Author information
Authors and Affiliations
Corresponding author
Additional information
Hee Jin Lee and An Na Seo are contributed equally to this work.
Rights and permissions
About this article
Cite this article
Lee, H.J., Seo, A.N., Park, S.Y. et al. Low Prognostic Implication of Fibroblast Growth Factor Family Activation in Triple-negative Breast Cancer Subsets. Ann Surg Oncol 21, 1561–1568 (2014). https://doi.org/10.1245/s10434-013-3456-x
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1245/s10434-013-3456-x