-
PDF
- Split View
-
Views
-
Cite
Cite
Nikola Holtkamp, Nicolas Ziegenhagen, Elke Malzer, Christian Hartmann, Alf Giese, Andreas von Deimling, Characterization of the amplicon on chromosomal segment 4q12 in glioblastoma multiforme, Neuro-Oncology, Volume 9, Issue 3, July 2007, Pages 291–297, https://doi.org/10.1215/15228517-2007-009
Close - Share Icon Share
Abstract
A subset of glioblastomas (GBMs) carry gene amplifications on chromosomal segment 4q12. To characterize this amplicon in detail, we analyzed a set of 100 samples consisting of 65 GBMs, 10 WHO grade III astrocytomas, 12 oligodendrogliomas, and 13 glioma cell cultures. We applied multiplex ligation-dependent probe amplification to determine the gene dosage of PDGFRA, KIT, and KDR and the flanking genes USP46, RASL11B, LNX1, CHIC2, SEC3L1, and IGFBP7. The amplicon was highly variable in size and copy number and extended over a region of up to 5 Mb. Amplifications on 4q12 were observed in 15% of GBMs and 23% of GBM cell cultures but not in 22 other gliomas. We analyzed transcription and translation of some genes within this amplicon. Gene amplification generally correlated with high transcript levels but did not necessarily result in increased protein levels. However, we detected frequent expression of proteins encoded by PDGFRA, KIT, and KDR in GBMs and GBM cell cultures independent of the amplification status. Future treatment of GBM patients may include drugs targeting multiple kinases that are encoded by genes on chromosomal segment 4q12.
Median survival of patients with glioblastoma (GBM) is poor despite aggressive treatment with radio- and chemotherapy. Therefore, novel strategies for treatment need to be developed. A promising approach is the analysis of mechanisms responsible for tumor genesis or progression and the development of drugs targeting these mechanisms. Interesting candidates for therapy include genes with tumor-specific activation or deactivation. Gene amplification is a very effective mechanism for up-regulation of protein expression. Several genes are frequently amplified in GBM, such as EGFR1 and PDGFRA.2,3 An amplicon on 12q13-q15 containing several genes, such as MDM2, SAS, and CDK4, has been described.4 We and others recently detected an amplicon on 4q12 in malignant peripheral nerve sheath tumors (MPNSTs)5 and GBMs6 containing PDGFRA, KIT, and KDR clustering next to each other. These three genes encode platelet-derived growth factor (PDGF) receptor alpha, KIT, and kinase insert domain receptor, also known as vascular endothelial growth factor receptor (VEGFR) 2 or Flk-1. All three receptors belong to the type III subfamily of receptor tyrosine kinases and play an important role in cell survival, proliferation, and angiogenesis.
In order to analyze frequency and expansion of this amplicon, we applied multiplex ligation-dependent probe amplification (MLPA). MLPA allows simultaneous analysis of gene dosage in multiple loci.7 Studies comparing MLPA with fluorescence in situ hybridization (FISH) demonstrated a good congruity.8,9
Materials and Methods
Tumor Tissue, DNA and RNA Extraction, and Cell Culture
All samples were collected at the Charité-Universitätsmedizin Berlin (Germany). The study contained 100 tumor samples: 62 primary and 3 secondary GBMs, 6 GBM cell lines, 6 low-passage GBM cultures and one oligoastrocytoma culture (< 8 passages), 10 WHO grade III astrocytomas, and 12 oligodendrogliomas. The GBM cell lines U-373 MG, DBTRG-05 MG, SNB-19, T98G, LN229, and U-87 MG and low-passage cultures were maintained in Dulbecco's modified Eagle's medium Glutamax-I containing 10% fetal calf serum and gentamicin (50 μg/ml) from Invitrogen (Karlsruhe, Germany) with 10% CO2. Prior to extracting nucleic acids and protein from frozen tumor sections, each sample was examined histologically. Only samples that contained predominantly tumor cells and lacked necrotic areas were included. RNA was extracted using TRIzol (Invitrogen). DNA extraction was carried out with QIAamp DNA Mini Kit from Qiagen (Hilden, Germany). Lymphocyte DNA from healthy volunteers served as reference for MLPA. cDNA of temporal lobe from two patients with pharmacoresistant epilepsy served as reference for transcription studies. The investigations were carried out with the informed consent of the patients.
Detection of Expression by Western Blot and Real-Time PCR
Western blots were performed as described previously.5 Lysates (50 μg) of 10 GBMs, cell lines U-373 MG, DBTRG-05 MG, and SNB-19, and five low-passage GBM cultures were separated by electrophoresis and blotted. The PDGFRα antibody (C-20, 1:200 dilution) and VEGFR-2 antibody (A-3, 1:200 dilution) were obtained from Santa Cruz Biotechnology (Heidelberg, Germany). The KIT antibody (A4502, 1:600 dilution) was from DakoCytomation (Hamburg, Germany). The membranes were incubated for 1 h with biotin-conjugated secondary antibodies, washed, and incubated 1 h with ExtrAvidin (1:2,000 dilution) from Sigma (Munich, Germany). Visualization was performed with enhanced chemoluminescence (Amersham Biosciences, Freiburg, Germany). The β-actin antibody AC-15 (1:3,000 dilution) from Sigma and a second horseradish-conjugated antibody were employed to demonstrate equal loading. Ponceau S staining (Sigma) of the membranes was also performed. GBM 21830 was loaded on all gels as an internal standard to make blots comparable.
Reverse transcription of 2 μg DNAse-treated RNA was achieved with the SuperScript First-Strand Synthesis System (Invitrogen). Subsequent PCR reactions were performed in a volume of 25 μl containing cDNA equivalents of 15 ng of RNA and 0.625 units of Platinum Taq DNA polymerase (Invitrogen). PCRs were performed in triplicate with SYBR green I (1:5,000 dilution, Invitrogen) using the ABI Prism 7700 Sequence Detection System (Applied Biosystems, Weiterstadt, Germany). Data were accepted as valid if Ct values (threshold cycles) of triplicate reactions were within the same cycle. Primer sequences and amplification conditions are available on request. PCR products of the target genes PDGFRA and CHIC2 (product size, 148 bp and 153 bp) were compared to the reference gene RPS3 (product size, 205 bp). RPS3 was shown to be expressed with similar levels in benign and malignant nerve sheath tumors.5 PCR efficiency determined by serial dilution of cDNA demonstrated similar results for target and reference genes. ΔCt was defined as Ct (target) - Ct (reference). The value for n-fold amplification of targets in relation to the reference was calculated by n = 2(ΔCt). cDNAs from temporal lobes of two epilepsy patients were used to determine expression in nontumorous brain tissue. Expression of GBM samples was plotted relative to temporal lobe 23194, which was set to 1.
MLPA Probe Selection, Design, and Analysis
Design of the synthetic probes was performed according to the recommendations of MRC-Holland (http://www.mrc-holland.com/pages/support_desing_synthetic_probespag.html). The probes were synthesized by Metabion International AG (Martinsried, Germany). All 5′ half-probes started with 5′-GGGTTCCCTAAGGGTTGGA-3′, and the 3′ half-probes were phosphorylated at the 5′ end and carried the primer binding tag 5′-TCTAGATTGGATCTTGCTGGCGC-3′ at the 3′ end. Gene-specific sequences of the probes are listed in Table 1. We designed a set of 36 half-probes in order to examine nine genes on 4q12 and four control genes mapping to other regions: 8q12-13 (IL7), 3q28 (FXR1), 2q35 (DES), 4q13 (EPHA5). On 4q12 PDGFRA, KIT, and KDR were chosen because of their biological and therapeutic importance. PDGFRA and KIT had been reported as amplified in tumors before.5,6 Further genes up- and downstream of the tyrosine kinase cluster were selected in steps of approximately 0.5 Mb. Fourteen probe pairs were designed to bind to genes on the chromosomal segment 4q12. The genes KIT, PDGFRA, KDR, IGFBP7, and CHIC2 were each recognized by two different probe pairs. All MLPA reagents were obtained from MRC-Holland (Amsterdam, Netherlands), and reactions were performed according to the manufacturer's protocol. We used 2.25 fmol of each synthetic half-probe for overnight hybridization to bind to 150 μg of tumor or lymphocyte DNA. PCR reactions were performed in a 25 μl volume for 30 cycles. The PCR products were analyzed on a semiautomated sequencer (ABI377, Applied Biosystems). One microliter of the PCR product was mixed with 4 μl of loading buffer and heat denatured. One-half microliter was loaded on acryl amid gels. TAMRA 500 (Applied Biosystems) was used as molecular weight marker.
Sequences of the MLPA half-probes
| Probe Number . | MLPA Probe . | 5′ Half-Probe . | 3′ Half-Probe . | Product Size (bp) . |
|---|---|---|---|---|
| 1 | IGFBP7ex4 | CTGTCCTTGGGAATTGGATGCA | TGGCACTCATATTCTCCAGCAT | 86 |
| 2 | II7ex6 | TCACATCCTTCATGCTCTCTCAGC | CTCATCACCATCTAGTTATGTGCT | 90 |
| 3 | KDRex30 | GCAGGGAGTCTGTGGCATCTGAAGG | CTCAAACCAGACAAGCGGCTACCAGTC | 94 |
| 4 | PDGFRAex21 | CTCTTGAGTTCTGTCCCCACA | GCTACGAGATCATGGTGAAATGCTGGAACAGTGAGC | 99 |
| 5 | CHIC2ex3 | GTGGCTGCCTTTGTTGCTGCTGCAC | ATTAGGTTGCAGTATGTGGCCAGTTATTTGCCTCAGT | 104 |
| 6 | FXR1intr16 | GCTGAGTCTCAGAGCAGACAAAGAAACCTCCCAAGGGA | AACTTTGGCTAAAAACAAGAAAGAAATGG | 109 |
| 7 | KITex10 | CAAATCCATCCCCACACCCTGTTCACTCCTTT | GCTGATTGGTTTCGTAATCGTAGCTGGCATGATGTGCATT | 114 |
| 8 | USP46ex7 | TGGTTTCTTAGGATGAGGGTAAAAAAGCTGCCCATG | ATCTTGGCCCTGCACCTAAAGCGGTTCAAGTACATGGAGC | 118 |
| 9 | DESex4 | TGGACATGTCTAAGCCAGACCTCACTGCCGCCCT | CAGGGACATCCGGGCTCAGTATGAGACCATCGCGGCTAAGAACATT | 122 |
| 10 | PDGFRAex22 | TCACCTGGACTTCCTGAAGAGTGACCATCCTGCTGTGGCACGCAT | GCGTGTGGACTCAGACAATGCATACATTGGTGTCACCTAC | 127 |
| 11 | KDRex29 | CAACCAGACGGACAGTGGTATGGTTCTTGCCTCAGAAGA | GCTGAAAACTTTGGAAGACAGAACCAAATTATCTCCATCTTTTGGGTAAGAC | 133 |
| 12 | CHIC2ex4 | TGGGAAAACAATAGGTTATACCACAAGGTGAGTACCACAGGGAGCA | GCTTCGTACTGTTGGGGTCTGAATTGCACAGTATGGAAACAGATGCTTTT | 138 |
| 13 | KITex13 | CGGGAAGCCCTCATGTCTGAACTCAAAGTCCTGACTTACCTTGGTAATCA | CATGAATATTGTGAATCTACTTGGAGCCTGCACCATTGGAGGTAAAGCCCGT | 143 |
| 14 | LNX1ex2 | CAGAGCCAAAGCTCTTCTTCACCTCCCCGCTGGTTCTGTTACAGCTGTAAAGG | TGCCTCCCACTACGGCCTGACCAAAGATAGGAAGAGGCGCTCACAAGATGGCT | 148 |
| 15 | SEC3L1ex3 | CCGATAAGGGAGATTTCTACAAAAGGCAGATTGCATGGGCCCTTCGAGATCTT | GCTGTGGTAGATGCCAAAGATGCTATCAAAGTAGGTTTTTCTCAGTTCTTTGACCTGT | 153 |
| 16 | IGFBP7ex3 | CCAGAAAAGCATGAAGTAACTGGCTGGGTGCTGGTGAGTACCAGTACCAAG | GCTGGCAAGAGTTTCATTCTTAGCATTAGACATAAGCAGACCTGGCAAAGGTGCCTGATGTACC | 158 |
| 17 | EPHA5ex4 | CCAGAGCTGCGGCAAATGTCCACCTCACAGTTATACCCATGAGGAAGCTTCAACCTCTTGTGTCT | GTGAAAAGGATTATTTCAGGAGAGAGTCTGATCCACCCACAATGGCATGCACAAGT | 163 |
| 18 | RASL11Bex3 | CCTTCATAGGTAATCTCTATACTAGACAAGTTCAGATAGAAGGTGAAACCCTGGCTCTTCAGGT | TCAAGACACTCCAGGTATTCAGGTGAGAAGCTCTGAGACTCTGGGTGAAAGGGGAACCTACT | 168 |
| Probe Number . | MLPA Probe . | 5′ Half-Probe . | 3′ Half-Probe . | Product Size (bp) . |
|---|---|---|---|---|
| 1 | IGFBP7ex4 | CTGTCCTTGGGAATTGGATGCA | TGGCACTCATATTCTCCAGCAT | 86 |
| 2 | II7ex6 | TCACATCCTTCATGCTCTCTCAGC | CTCATCACCATCTAGTTATGTGCT | 90 |
| 3 | KDRex30 | GCAGGGAGTCTGTGGCATCTGAAGG | CTCAAACCAGACAAGCGGCTACCAGTC | 94 |
| 4 | PDGFRAex21 | CTCTTGAGTTCTGTCCCCACA | GCTACGAGATCATGGTGAAATGCTGGAACAGTGAGC | 99 |
| 5 | CHIC2ex3 | GTGGCTGCCTTTGTTGCTGCTGCAC | ATTAGGTTGCAGTATGTGGCCAGTTATTTGCCTCAGT | 104 |
| 6 | FXR1intr16 | GCTGAGTCTCAGAGCAGACAAAGAAACCTCCCAAGGGA | AACTTTGGCTAAAAACAAGAAAGAAATGG | 109 |
| 7 | KITex10 | CAAATCCATCCCCACACCCTGTTCACTCCTTT | GCTGATTGGTTTCGTAATCGTAGCTGGCATGATGTGCATT | 114 |
| 8 | USP46ex7 | TGGTTTCTTAGGATGAGGGTAAAAAAGCTGCCCATG | ATCTTGGCCCTGCACCTAAAGCGGTTCAAGTACATGGAGC | 118 |
| 9 | DESex4 | TGGACATGTCTAAGCCAGACCTCACTGCCGCCCT | CAGGGACATCCGGGCTCAGTATGAGACCATCGCGGCTAAGAACATT | 122 |
| 10 | PDGFRAex22 | TCACCTGGACTTCCTGAAGAGTGACCATCCTGCTGTGGCACGCAT | GCGTGTGGACTCAGACAATGCATACATTGGTGTCACCTAC | 127 |
| 11 | KDRex29 | CAACCAGACGGACAGTGGTATGGTTCTTGCCTCAGAAGA | GCTGAAAACTTTGGAAGACAGAACCAAATTATCTCCATCTTTTGGGTAAGAC | 133 |
| 12 | CHIC2ex4 | TGGGAAAACAATAGGTTATACCACAAGGTGAGTACCACAGGGAGCA | GCTTCGTACTGTTGGGGTCTGAATTGCACAGTATGGAAACAGATGCTTTT | 138 |
| 13 | KITex13 | CGGGAAGCCCTCATGTCTGAACTCAAAGTCCTGACTTACCTTGGTAATCA | CATGAATATTGTGAATCTACTTGGAGCCTGCACCATTGGAGGTAAAGCCCGT | 143 |
| 14 | LNX1ex2 | CAGAGCCAAAGCTCTTCTTCACCTCCCCGCTGGTTCTGTTACAGCTGTAAAGG | TGCCTCCCACTACGGCCTGACCAAAGATAGGAAGAGGCGCTCACAAGATGGCT | 148 |
| 15 | SEC3L1ex3 | CCGATAAGGGAGATTTCTACAAAAGGCAGATTGCATGGGCCCTTCGAGATCTT | GCTGTGGTAGATGCCAAAGATGCTATCAAAGTAGGTTTTTCTCAGTTCTTTGACCTGT | 153 |
| 16 | IGFBP7ex3 | CCAGAAAAGCATGAAGTAACTGGCTGGGTGCTGGTGAGTACCAGTACCAAG | GCTGGCAAGAGTTTCATTCTTAGCATTAGACATAAGCAGACCTGGCAAAGGTGCCTGATGTACC | 158 |
| 17 | EPHA5ex4 | CCAGAGCTGCGGCAAATGTCCACCTCACAGTTATACCCATGAGGAAGCTTCAACCTCTTGTGTCT | GTGAAAAGGATTATTTCAGGAGAGAGTCTGATCCACCCACAATGGCATGCACAAGT | 163 |
| 18 | RASL11Bex3 | CCTTCATAGGTAATCTCTATACTAGACAAGTTCAGATAGAAGGTGAAACCCTGGCTCTTCAGGT | TCAAGACACTCCAGGTATTCAGGTGAGAAGCTCTGAGACTCTGGGTGAAAGGGGAACCTACT | 168 |
Probe sequences are given without the primer binding tags (see Materials and Methods). Product size corresponds to PCR fragments generated after successful ligation of the half-probes.
Sequences of the MLPA half-probes
| Probe Number . | MLPA Probe . | 5′ Half-Probe . | 3′ Half-Probe . | Product Size (bp) . |
|---|---|---|---|---|
| 1 | IGFBP7ex4 | CTGTCCTTGGGAATTGGATGCA | TGGCACTCATATTCTCCAGCAT | 86 |
| 2 | II7ex6 | TCACATCCTTCATGCTCTCTCAGC | CTCATCACCATCTAGTTATGTGCT | 90 |
| 3 | KDRex30 | GCAGGGAGTCTGTGGCATCTGAAGG | CTCAAACCAGACAAGCGGCTACCAGTC | 94 |
| 4 | PDGFRAex21 | CTCTTGAGTTCTGTCCCCACA | GCTACGAGATCATGGTGAAATGCTGGAACAGTGAGC | 99 |
| 5 | CHIC2ex3 | GTGGCTGCCTTTGTTGCTGCTGCAC | ATTAGGTTGCAGTATGTGGCCAGTTATTTGCCTCAGT | 104 |
| 6 | FXR1intr16 | GCTGAGTCTCAGAGCAGACAAAGAAACCTCCCAAGGGA | AACTTTGGCTAAAAACAAGAAAGAAATGG | 109 |
| 7 | KITex10 | CAAATCCATCCCCACACCCTGTTCACTCCTTT | GCTGATTGGTTTCGTAATCGTAGCTGGCATGATGTGCATT | 114 |
| 8 | USP46ex7 | TGGTTTCTTAGGATGAGGGTAAAAAAGCTGCCCATG | ATCTTGGCCCTGCACCTAAAGCGGTTCAAGTACATGGAGC | 118 |
| 9 | DESex4 | TGGACATGTCTAAGCCAGACCTCACTGCCGCCCT | CAGGGACATCCGGGCTCAGTATGAGACCATCGCGGCTAAGAACATT | 122 |
| 10 | PDGFRAex22 | TCACCTGGACTTCCTGAAGAGTGACCATCCTGCTGTGGCACGCAT | GCGTGTGGACTCAGACAATGCATACATTGGTGTCACCTAC | 127 |
| 11 | KDRex29 | CAACCAGACGGACAGTGGTATGGTTCTTGCCTCAGAAGA | GCTGAAAACTTTGGAAGACAGAACCAAATTATCTCCATCTTTTGGGTAAGAC | 133 |
| 12 | CHIC2ex4 | TGGGAAAACAATAGGTTATACCACAAGGTGAGTACCACAGGGAGCA | GCTTCGTACTGTTGGGGTCTGAATTGCACAGTATGGAAACAGATGCTTTT | 138 |
| 13 | KITex13 | CGGGAAGCCCTCATGTCTGAACTCAAAGTCCTGACTTACCTTGGTAATCA | CATGAATATTGTGAATCTACTTGGAGCCTGCACCATTGGAGGTAAAGCCCGT | 143 |
| 14 | LNX1ex2 | CAGAGCCAAAGCTCTTCTTCACCTCCCCGCTGGTTCTGTTACAGCTGTAAAGG | TGCCTCCCACTACGGCCTGACCAAAGATAGGAAGAGGCGCTCACAAGATGGCT | 148 |
| 15 | SEC3L1ex3 | CCGATAAGGGAGATTTCTACAAAAGGCAGATTGCATGGGCCCTTCGAGATCTT | GCTGTGGTAGATGCCAAAGATGCTATCAAAGTAGGTTTTTCTCAGTTCTTTGACCTGT | 153 |
| 16 | IGFBP7ex3 | CCAGAAAAGCATGAAGTAACTGGCTGGGTGCTGGTGAGTACCAGTACCAAG | GCTGGCAAGAGTTTCATTCTTAGCATTAGACATAAGCAGACCTGGCAAAGGTGCCTGATGTACC | 158 |
| 17 | EPHA5ex4 | CCAGAGCTGCGGCAAATGTCCACCTCACAGTTATACCCATGAGGAAGCTTCAACCTCTTGTGTCT | GTGAAAAGGATTATTTCAGGAGAGAGTCTGATCCACCCACAATGGCATGCACAAGT | 163 |
| 18 | RASL11Bex3 | CCTTCATAGGTAATCTCTATACTAGACAAGTTCAGATAGAAGGTGAAACCCTGGCTCTTCAGGT | TCAAGACACTCCAGGTATTCAGGTGAGAAGCTCTGAGACTCTGGGTGAAAGGGGAACCTACT | 168 |
| Probe Number . | MLPA Probe . | 5′ Half-Probe . | 3′ Half-Probe . | Product Size (bp) . |
|---|---|---|---|---|
| 1 | IGFBP7ex4 | CTGTCCTTGGGAATTGGATGCA | TGGCACTCATATTCTCCAGCAT | 86 |
| 2 | II7ex6 | TCACATCCTTCATGCTCTCTCAGC | CTCATCACCATCTAGTTATGTGCT | 90 |
| 3 | KDRex30 | GCAGGGAGTCTGTGGCATCTGAAGG | CTCAAACCAGACAAGCGGCTACCAGTC | 94 |
| 4 | PDGFRAex21 | CTCTTGAGTTCTGTCCCCACA | GCTACGAGATCATGGTGAAATGCTGGAACAGTGAGC | 99 |
| 5 | CHIC2ex3 | GTGGCTGCCTTTGTTGCTGCTGCAC | ATTAGGTTGCAGTATGTGGCCAGTTATTTGCCTCAGT | 104 |
| 6 | FXR1intr16 | GCTGAGTCTCAGAGCAGACAAAGAAACCTCCCAAGGGA | AACTTTGGCTAAAAACAAGAAAGAAATGG | 109 |
| 7 | KITex10 | CAAATCCATCCCCACACCCTGTTCACTCCTTT | GCTGATTGGTTTCGTAATCGTAGCTGGCATGATGTGCATT | 114 |
| 8 | USP46ex7 | TGGTTTCTTAGGATGAGGGTAAAAAAGCTGCCCATG | ATCTTGGCCCTGCACCTAAAGCGGTTCAAGTACATGGAGC | 118 |
| 9 | DESex4 | TGGACATGTCTAAGCCAGACCTCACTGCCGCCCT | CAGGGACATCCGGGCTCAGTATGAGACCATCGCGGCTAAGAACATT | 122 |
| 10 | PDGFRAex22 | TCACCTGGACTTCCTGAAGAGTGACCATCCTGCTGTGGCACGCAT | GCGTGTGGACTCAGACAATGCATACATTGGTGTCACCTAC | 127 |
| 11 | KDRex29 | CAACCAGACGGACAGTGGTATGGTTCTTGCCTCAGAAGA | GCTGAAAACTTTGGAAGACAGAACCAAATTATCTCCATCTTTTGGGTAAGAC | 133 |
| 12 | CHIC2ex4 | TGGGAAAACAATAGGTTATACCACAAGGTGAGTACCACAGGGAGCA | GCTTCGTACTGTTGGGGTCTGAATTGCACAGTATGGAAACAGATGCTTTT | 138 |
| 13 | KITex13 | CGGGAAGCCCTCATGTCTGAACTCAAAGTCCTGACTTACCTTGGTAATCA | CATGAATATTGTGAATCTACTTGGAGCCTGCACCATTGGAGGTAAAGCCCGT | 143 |
| 14 | LNX1ex2 | CAGAGCCAAAGCTCTTCTTCACCTCCCCGCTGGTTCTGTTACAGCTGTAAAGG | TGCCTCCCACTACGGCCTGACCAAAGATAGGAAGAGGCGCTCACAAGATGGCT | 148 |
| 15 | SEC3L1ex3 | CCGATAAGGGAGATTTCTACAAAAGGCAGATTGCATGGGCCCTTCGAGATCTT | GCTGTGGTAGATGCCAAAGATGCTATCAAAGTAGGTTTTTCTCAGTTCTTTGACCTGT | 153 |
| 16 | IGFBP7ex3 | CCAGAAAAGCATGAAGTAACTGGCTGGGTGCTGGTGAGTACCAGTACCAAG | GCTGGCAAGAGTTTCATTCTTAGCATTAGACATAAGCAGACCTGGCAAAGGTGCCTGATGTACC | 158 |
| 17 | EPHA5ex4 | CCAGAGCTGCGGCAAATGTCCACCTCACAGTTATACCCATGAGGAAGCTTCAACCTCTTGTGTCT | GTGAAAAGGATTATTTCAGGAGAGAGTCTGATCCACCCACAATGGCATGCACAAGT | 163 |
| 18 | RASL11Bex3 | CCTTCATAGGTAATCTCTATACTAGACAAGTTCAGATAGAAGGTGAAACCCTGGCTCTTCAGGT | TCAAGACACTCCAGGTATTCAGGTGAGAAGCTCTGAGACTCTGGGTGAAAGGGGAACCTACT | 168 |
Probe sequences are given without the primer binding tags (see Materials and Methods). Product size corresponds to PCR fragments generated after successful ligation of the half-probes.
Normalization of the samples was performed by dividing each peak area by the combined peak areas of all peaks in a lane. This procedure generates proportions of individual peaks of the total peak area (relative peak value). The mean value of the four internal control probes was calculated, and all relative peak values were divided by the mean value of the controls. Tumor values were then divided by the values obtained from lymphocyte DNA to calculate a ratio. Ratios of > 1.5 were scored as gene amplification because lymphocyte DNAs never yielded values higher than 1.3. For genes recognized by two pairs of half-probes, mean values were calculated.
Gene Amplification Analysis by Real-Time PCR
Quantitative real-time PCR was performed with SYBR green I (1:5,000 dilution; Molecular Probes, Leiden, Netherlands) using the ABI Prism 7700 Sequence Detection System (Applied Biosystems). PCR products of the target genes CHIC2 (exon 3; PCR product, 153 bp), KIT (exon 17; PCR product, 185 bp), PDGFRA (exon 21; PCR product, 148 bp), and KDR (exon 30; PCR product, 163 bp) were compared to the reference gene DES (exon 8; PCR product, 180 bp) on chromosomal segment 2q35 encoding desmin. PCR efficiency, determined by serial dilution of DNA, proved to be comparable for target genes and DES. All samples were analyzed in triplicate in 25 μl reaction mixes containing 1.25 units of Platinum Taq DNA polymerase (Invitrogen). Data were accepted as valid if Ct values of triplicate reactions were within the same cycle. Primer sequences and amplification conditions are available from the author on request. Evaluation of data was performed using the ΔΔCt method: ΔΔCt = ΔCt tumor DNA - ΔCt blood DNA. ΔCt is the Ct value of the reference gene minus the Ct of the target gene. Fold increase of the target gene was calculated by 2(ΔΔCt). A reference:target ratio of 1.7 or more was defined as gene amplification.
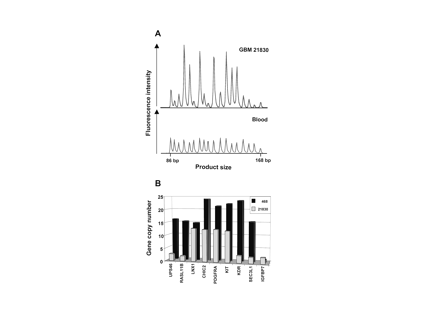
Electropherogram and gene amplification pattern of glioblastomas (GBMs). (A) Electropherogram of multiplex ligation-dependent probe amplification (MLPA) products of GBM 21830 and lymphocyte DNA. Product size is shown on the x-axis and ranges from 86 to 168 base pairs. Peak areas corresponding to amplified genes are enhanced relative to unaffected control genes. (B) The diagram shows gene amplification pattern of GBMs 468 and 21830 with high copy numbers.
Results
The region analyzed spanned 5 Mb from USP46 close to the centromere to the distal IGFBP7 and encodes 29 genes. Our probe set detected nine genes in this region (Table 1). The tyrosine kinase cluster containing the genes PDGFRA, KIT, and KDR is situated in the center of the region analyzed. Electropherograms from MLPA with our probe set invariably yielded unambiguous peaks (Fig. 1A). Our results show that synthetic probes of up to 88 bp in length yield consistent results.
Thirteen of 65 GBMs carried amplifications in at least one gene (Table 2). None of the three secondary GBMs carried a gene amplification. Amplification patterns of GBMs 468 and 21830 with high copy numbers (>10) are shown in Fig. 1B. GBMs 116, 238, 410, and 468 were previously known to harbor PDGFRA amplifications.3 The frequency of gene amplification on 4q12 in the nonselected fraction of GBMs in this study was 15% (9 of 61 cases). The EPHA5 gene located about 8 Mb distal from IGFBP7 on 4q13 yielded normal values in the tumors, indicating amplicon restriction to 4q12 and absence of polysomy. Six GBMs had amplifications close to the cut-off (between 1.5 and 2) in either LNX1 or RASL11B or in RASL11B, LNX1, and CHIC2. Non-integer copy numbers may be caused by heterogeneous cell populations within the tumor, such as nontumorous cells or tumor subclones. Cell cultures derived from GBMs carried amplified genes in 3 of 13 (23%) samples. A remarkable amplification pattern was detected in U-373 MG cells (Table 2). PDGFRA appears as the center of the amplicon with 19-fold increase, framed by CHIC2 and KIT with only 4-fold amplification. A copy number increase was seen in the flanking LNX1 and KDR with values of 10 and 7. No amplification was detected in tumors other than GBMs (10 WHO grade III astrocytomas, 12 oligodendrogliomas).
Amplified genes on chromosomal segment 4q12 in GBMs and GBM cell cultures
| Tumor ID . | USP46 . | RASL11B . | LNX1 . | CHIC2 . | PDGFRA . | KIT . | KDR . | SEC3L1 . | IGFBP7 . | EPHA5 . |
|---|---|---|---|---|---|---|---|---|---|---|
| 468 | 16 | 15 | 15 | 24 (11) | 21 (20) | 22 (12) | 23 (29) | 15 | 1 | 1 |
| 21830 | 3 | 2 | 13 | 12 (12) | 13 (13) | 12 (13) | 3 (3) | 2 | 2 | 1 |
| 116 | 8 | 10 | 8 | 11 (8) | 11 (7) | 12 (12) | 11 (5) | 7 | 7 | 1 |
| 238 | 4 | 4 | 4 | 4 (3) | 4 (3) | 4 (4) | 4 (4) | 4 | 4 | 1 |
| 410 | 5 | 5 | 6 | 5 (8) | 5 (3) | 5 (5) | 5 (7) | 6 | 9 | 1 |
| 25508 | 1 | 2 | 2 | 3 (5) | 3 (7) | 1 (3) | 1 (2) | 2 | 1 | 1 |
| 26640 | 1 | 2 | 2 | 2 | 1 | 2 | 1 | 1 | 1 | 1 |
| 21828 | 1 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 |
| 24538 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 25520 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 22616 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 22548 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 23480 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Cell culture | ||||||||||
| U-373 MG | 5 | 6 | 10 | 4 | 19 | 4 | 7 | 4 | 6 | 1 |
| 30620 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 1 | 1 |
| SNB-19 | 1 | 1 | 2 | 3 | 3 | 2 | 2 | 2 | 2 | 1 |
| Tumor ID . | USP46 . | RASL11B . | LNX1 . | CHIC2 . | PDGFRA . | KIT . | KDR . | SEC3L1 . | IGFBP7 . | EPHA5 . |
|---|---|---|---|---|---|---|---|---|---|---|
| 468 | 16 | 15 | 15 | 24 (11) | 21 (20) | 22 (12) | 23 (29) | 15 | 1 | 1 |
| 21830 | 3 | 2 | 13 | 12 (12) | 13 (13) | 12 (13) | 3 (3) | 2 | 2 | 1 |
| 116 | 8 | 10 | 8 | 11 (8) | 11 (7) | 12 (12) | 11 (5) | 7 | 7 | 1 |
| 238 | 4 | 4 | 4 | 4 (3) | 4 (3) | 4 (4) | 4 (4) | 4 | 4 | 1 |
| 410 | 5 | 5 | 6 | 5 (8) | 5 (3) | 5 (5) | 5 (7) | 6 | 9 | 1 |
| 25508 | 1 | 2 | 2 | 3 (5) | 3 (7) | 1 (3) | 1 (2) | 2 | 1 | 1 |
| 26640 | 1 | 2 | 2 | 2 | 1 | 2 | 1 | 1 | 1 | 1 |
| 21828 | 1 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 |
| 24538 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 25520 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 22616 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 22548 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 23480 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Cell culture | ||||||||||
| U-373 MG | 5 | 6 | 10 | 4 | 19 | 4 | 7 | 4 | 6 | 1 |
| 30620 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 1 | 1 |
| SNB-19 | 1 | 1 | 2 | 3 | 3 | 2 | 2 | 2 | 2 | 1 |
Genes are listed in their physical order. With the exception of EPHA5, which localizes to chromosomal segment 4q13, all other genes localize to 4q12. Numbers indicate n-fold increase of gene dosage relative to the normal gene dosage. Amplification levels that were obtained by real-time PCR are given in parentheses.
Amplified genes on chromosomal segment 4q12 in GBMs and GBM cell cultures
| Tumor ID . | USP46 . | RASL11B . | LNX1 . | CHIC2 . | PDGFRA . | KIT . | KDR . | SEC3L1 . | IGFBP7 . | EPHA5 . |
|---|---|---|---|---|---|---|---|---|---|---|
| 468 | 16 | 15 | 15 | 24 (11) | 21 (20) | 22 (12) | 23 (29) | 15 | 1 | 1 |
| 21830 | 3 | 2 | 13 | 12 (12) | 13 (13) | 12 (13) | 3 (3) | 2 | 2 | 1 |
| 116 | 8 | 10 | 8 | 11 (8) | 11 (7) | 12 (12) | 11 (5) | 7 | 7 | 1 |
| 238 | 4 | 4 | 4 | 4 (3) | 4 (3) | 4 (4) | 4 (4) | 4 | 4 | 1 |
| 410 | 5 | 5 | 6 | 5 (8) | 5 (3) | 5 (5) | 5 (7) | 6 | 9 | 1 |
| 25508 | 1 | 2 | 2 | 3 (5) | 3 (7) | 1 (3) | 1 (2) | 2 | 1 | 1 |
| 26640 | 1 | 2 | 2 | 2 | 1 | 2 | 1 | 1 | 1 | 1 |
| 21828 | 1 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 |
| 24538 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 25520 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 22616 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 22548 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 23480 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Cell culture | ||||||||||
| U-373 MG | 5 | 6 | 10 | 4 | 19 | 4 | 7 | 4 | 6 | 1 |
| 30620 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 1 | 1 |
| SNB-19 | 1 | 1 | 2 | 3 | 3 | 2 | 2 | 2 | 2 | 1 |
| Tumor ID . | USP46 . | RASL11B . | LNX1 . | CHIC2 . | PDGFRA . | KIT . | KDR . | SEC3L1 . | IGFBP7 . | EPHA5 . |
|---|---|---|---|---|---|---|---|---|---|---|
| 468 | 16 | 15 | 15 | 24 (11) | 21 (20) | 22 (12) | 23 (29) | 15 | 1 | 1 |
| 21830 | 3 | 2 | 13 | 12 (12) | 13 (13) | 12 (13) | 3 (3) | 2 | 2 | 1 |
| 116 | 8 | 10 | 8 | 11 (8) | 11 (7) | 12 (12) | 11 (5) | 7 | 7 | 1 |
| 238 | 4 | 4 | 4 | 4 (3) | 4 (3) | 4 (4) | 4 (4) | 4 | 4 | 1 |
| 410 | 5 | 5 | 6 | 5 (8) | 5 (3) | 5 (5) | 5 (7) | 6 | 9 | 1 |
| 25508 | 1 | 2 | 2 | 3 (5) | 3 (7) | 1 (3) | 1 (2) | 2 | 1 | 1 |
| 26640 | 1 | 2 | 2 | 2 | 1 | 2 | 1 | 1 | 1 | 1 |
| 21828 | 1 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 |
| 24538 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 25520 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 22616 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 22548 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 23480 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Cell culture | ||||||||||
| U-373 MG | 5 | 6 | 10 | 4 | 19 | 4 | 7 | 4 | 6 | 1 |
| 30620 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 1 | 1 |
| SNB-19 | 1 | 1 | 2 | 3 | 3 | 2 | 2 | 2 | 2 | 1 |
Genes are listed in their physical order. With the exception of EPHA5, which localizes to chromosomal segment 4q13, all other genes localize to 4q12. Numbers indicate n-fold increase of gene dosage relative to the normal gene dosage. Amplification levels that were obtained by real-time PCR are given in parentheses.
To verify MLPA results with an independent method, real-time PCR was performed. A subset of GBMs with amplifications according to MLPA was analyzed for CHIC2, PDGFRA, KIT, and KDR amplification status by PCR (Table 2). All cases with amplification according to MLPA were confirmed by PCR. In nearly all cases, copy numbers obtained by MLPA and PCR matched well. However, GBM 468 exhibited lower amplification levels for CHIC2 and KIT if tested by real-time PCR than if tested by MLPA.
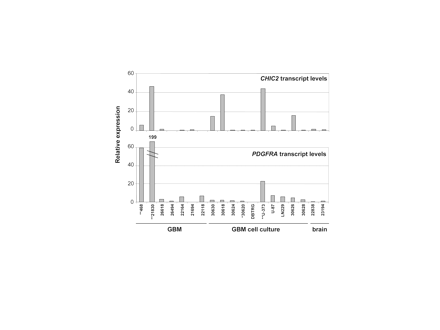
Next, we examined gene expression levels. CHIC2 expression was determined since amplification levels were similar to those of the receptor tyrosine kinase genes and antibodies are not available. PDGFRA expression was analyzed to study the relations of RNA and protein levels. Increased CHIC2 expression relative to brain tissue was seen in 41% of the tumors (Fig. 2). Three of these cases (GBMs 468 and 21830, U-373 MG) carried several CHIC2 copies. Increased CHIC2 levels without underlying gene amplification were observed in cell cultures only and may have been induced by culture conditions. PDGFRA expression was increased in 10 of 17 (59%) of the GBM samples (at least two times higher than in brain tissue). GBMs 468 and 21830 and cell line U-373 MG with high PDGFRA copy numbers (>10) demonstrated extraordinarily high PDGFRA transcript levels, reaching a 199-fold increase over brain tissue (Fig. 2). In GBM 468 and cell line U-373 MG, the transcript levels were efficiently translated, leading to very high amounts of PDGFRA, whereas protein levels in GBM 21830 were low (Fig. 3).
Quantification of CHIC2 and PDGFRA transcript levels in GBMs, GBM cell cultures, and temporal lobe (brain) by real-time PCR. Bars correspond to the mean value of triplicates. *Samples with low-grade amplification (<10); **samples with high-grade amplification (>10).
We determined relative protein expression levels by Western blotting in 18 GBM and glioma cultures. PDGFRA was detected in 83% of the samples, with considerable variation in protein quantities (Fig. 3). All cell cultures were PDGFRA positive. PDGFRA occurred in three different isoforms ranging from 140 to 180 kDa. KIT was detected in 67% of the samples. High amounts were observed in GBM 21830 harboring a 12-fold KIT amplification. KIT delineated as a single band of 140 kDa. KDR expression was found in 83% of the samples. All but one of the cell cultures were positive. KDR appeared in three isoforms with a major band at 140 kDa and two faint bands up to 220 kDa. Cytochemistry on glioma cell lines demonstrated a cytoplasmic and/or membranous localization of the receptors (data not shown).
Discussion
We provide evidence for amplification of multiple genes on the chromosomal segment 4q12 in a fraction of GBM and GBM cell cultures. The amplicon size and degree of amplification in individual tumors varied considerably. PDGFRA, KIT, and KDR in the center of the amplicon encode growth factor receptors of therapeutic interest. Although activating mutations in these genes have not been described in brain tumors,10,11 the encoded receptors are still likely to play an important role in tumor biology due to frequent expression of receptors and their ligands. Imatinib mesylate that targets PDGF receptors and KIT was recently shown to mediate durable anti-tumor activity in combination with hydroxyurea in patients with recurrent GBM.12
Gene amplifications of KIT and PDGFRA have been detected in GBM, MPNST, and seminoma.3,5,6,11,13 However, in seminomas, KIT was generally amplified alone and not together with the flanking genes PDGFRA and KDR, as in GBM. A study that screened 43 GBMs by FISH for PDGFRA, KIT, and KDR amplification detected increased copy numbers in 29%, 47%, and 39% of the tumors.6 Our data support the finding of an amplicon that encompasses PDGFRA, KIT, and KDR. We observed a lower amplification frequency than in the previous study. The discrepancy may be due to different tumor sets in the studies or due to differences in the detection system. However, our data match well with previous reports on PDGFRA amplification in GBM, ranging from 8% to 16%.2,3
Protein levels of PDGFRA, KIT, and KDR in GBMs and GBM cell cultures determined by Western blot. Amplification status of the genes is depicted under the lanes. Samples that were also analyzed by real-time PCR are gray. Note that Ponceau S staining showed that in the U-373 MG sample total protein was comparable to the other samples despite the weaker β-actin band. *Samples with low-grade amplification (<10); **samples with high-grade amplification (>10).
PDGFRA expression is well documented in gliomas. In contrast, limited studies have determined KIT expression. A recent study demonstrated KIT in only 4% of 101 GBMs.14 Another study analyzed KIT expression in small specimen numbers of astrocytomas, medulloblastomas, meningiomas, and oligodendrogliomas and detected KIT in one of eight GBMs and one of three medulloblastomas.11 Both immunopositive tumors carried high KIT copy numbers. Joensuu et al.6 confirmed that KIT expression is rare in GBM and is restricted to tumors with high copy numbers of KIT. In contrast, we detected KIT expression in the majority of GBMs. Possibly, application of different techniques is responsible for variable KIT expression frequencies. In immunohistochemistry, weak homogeneous protein expression may be difficult to differentiate from background staining, complicating interpretation. Western blot analysis unambiguously detects even low expression levels, if present, in the majority of the tumor cells.
KDR and VEGF have been detected by Western blot in 100% of astrocytic neoplasms (n = 37).15 We confirm that KDR expression is common to GBM. It is thought that VEGF secreted by glioma cells acts on endothelial cells and promotes angiogenesis. However, recent observations provide evidence for an autocrine VEGF loop in different tumor entities, including GBM.16-18 Our observation of KDR expression in GBM cell cultures confirmed these findings. Most likely, up-regulation of KDR together with ligand expression provides an advantage for tumor cells. This observation makes therapeutic approaches targeting KDR particularly attractive because several survival strategies of the tumor (induction of angiogenesis, proliferation, and invasion) would be targeted via the same molecule.
Only high copy numbers of PDGFRA and KIT had a pronounced effect on protein levels. GBMs 468 and 21830 with more than 10 copies of PDGFRA and KIT displayed very high levels of either PDGFRA or KIT, possibly indicating that just one of these receptors is required to provide a survival advantage. This scenario resembles the alternate oncogenic mechanism in gastrointestinal stromal tumor that contains either mutant PDGFRA or mutant KIT. GBM 21830 demonstrated strong transcription of PDGFRA but only minute amounts of the corresponding protein. This points toward translational regulation as a mechanism in PDGFRA expression control. PDGFRA, KIT, and KDR were frequently expressed in GBMs and GBM cell cultures, independent of the gene amplification status. The expansion of the amplicon over a region of 5 Mb indicates that, besides the receptor tyrosine kinases, coamplified genes might contribute to the malignant phenotype of GBM. Functional relevance of genes coamplified together with ERBB2 at 17q12 in breast cancer cell lines was recently shown.19 Although not much is known about the genes coamplified together with PDGFRA, KIT, and KDR, the gene RASL11B, a member of the RAS-like family, and CHIC2 with increased transcript levels might be interesting candidates. Notably, cases of acute myeloid leukemia with a CHIC2-ETV6 fusion gene have been reported, indicating a possible role of CHIC2 (previously BTL) in cancer.20 The CHIC2 protein localizes to vesicular structures and the plasma membrane, but the function remains unknown. Further analyses are required to clarify functional contribution of individual genes included in amplicon 4q12.
For the time being, it appears rational to evaluate, in addition to imatinib, other drugs approved by the U.S. Food and Drug Administration, such as sunitinib or sorafenib, for their therapeutic effect in targeting PDGF receptors, KIT, and KDR. It will be of interest to learn whether GBMs with amplification of these genes respond better than those with normal copy numbers.
We thank Jana Mucha for her excellent technical assistance and Dr. M. Baier for helpful discussion. This study was supported by Deutsche Krebshilfe (Nationales Verbundprojekt Gliome).
References
Liu L, Backlund LM, Nilsson BR, et al. Clinical significance of EGFR amplification and the aberrant EGFRvIII transcript in conventionally treated astrocytic gliomas.
Fleming T, Saxena A, Clark C, et al. Amplification and/or overexpression of platelet-derived growth factor receptors and epidermal growth factor receptor in human glial tumors.
Hermanson M, Funa K, Koopmann J, et al. Association of loss of heterozygosity on chromosome 17p with high platelet-derived growth factor alpha receptor expression in human malignant gliomas.
Reifenberger G, Ichimura K, Reifenberger J, Elkahloun AG, Meltzer PS, Collins VP. Refined mapping of 12q13-q15 amplicons in human malignant gliomas suggests CDK4/SAS and MDM2 as independent amplification targets.
Holtkamp N, Okuducu AF, Mucha J, et al. Mutation and expression of PDGFRA and KIT in malignant peripheral nerve sheath tumors, and its implications for imatinib sensitivity.
Joensuu H, Puputti M, Sihto H, Tynninen O, Nupponen NN. Amplification of genes encoding KIT, PDGFRalpha and VEGFR2 receptor tyrosine kinases is frequent in glioblastoma multiforme.
Schouten JP, McElgunn CJ, Waaijer R, Zwijnenburg D, Diepvens F, Pals G. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification.
Palomares M, Delicado A, Lapunzina P, et al. MLPA vs multiprobe FISH: comparison of two methods for the screening of subtelomeric rearrangements in 50 patients with idiopathic mental retardation.
Slater H, Bruno D, Ren H, et al. Improved testing for CMT1A and HNPP using multiplex ligation-dependent probe amplification (MLPA) with rapid DNA preparations: comparison with the interphase FISH method.
Hartmann C, Xu X, Bartels G, et al. Pdgfr-alpha in 1p/19q LOH oligodendrogliomas.
Sihto H, Sarlomo-Rikala M, Tynninen O, et al. KIT and platelet-derived growth factor receptor alpha tyrosine kinase gene mutations and KIT amplifications in human solid tumors.
Reardon DA, Egorin MJ, Quinn JA, et al. Phase II study of imatinib mesylate plus hydroxyurea in adults with recurrent glioblastoma multiforme.
McIntyre A, Summersgill B, Grygalewicz B, et al. Amplification and overexpression of the KIT gene is associated with progression in the seminoma subtype of testicular germ cell tumors of adolescents and adults.
Haberler C, Gelpi E, Marosi C, et al. Immunohistochemical analysis of platelet-derived growth factor receptor-alpha, -beta, c-kit, c-abl, and arg proteins in glioblastoma: possible implications for patient selection for imatinib mesylate therapy.
Carroll RS, Zhang J, Bello L, Melnick MB, Maruyama T, Black PM. KDR activation in astrocytic neoplasms.
Lacal PM, Ruffini F, Pagani E, D'Atri S. An autocrine loop directed by the vascular endothelial growth factor promotes invasiveness of human melanoma cells.
Steiner HH, Karcher S, Mueller MM, Nalbantis E, Kunze S, Herold-Mende C. Autocrine pathways of the vascular endothelial growth factor (VEGF) in glioblastoma multiforme: clinical relevance of radiation-induced increase of VEGF levels.
Vieira JM, Santos SC, Espadinha C, et al. Expression of vascular endothelial growth factor (VEGF) and its receptors in thyroid carcinomas of follicular origin: a potential autocrine loop.
Kao J, Pollack JR. RNA interference-based functional dissection of the 17q12 amplicon in breast cancer reveals contribution of coamplified genes.