Abstract
RNAi therapeutics provide an opportunity to correct faulty genes, and several RNAi have entered clinical evaluation. The existing quantification methods typically use radioactivity- or fluorescence-labeled RNAi, require large blood volumes, and/or have a limited dynamic detection range. We established a quantitative reverse transcriptase real-time polymerase chain reaction (RT-qPCR) assay to measure RNAi; the model analyte was survivin siRNA (siSurvivin). A second siRNA was used as the internal standard. The three major steps were (a) extraction of the two siRNAs from blood or water, (b) synthesis of their cDNA by poly-A extension, and (c) qPCR of cDNA. Standard curves were established. Utility of the assay was demonstrated in a pharmacokinetic study where all 12 samples for the blood concentration-time profile were obtained from a single mouse given an intravenous dose of 1 nmole siSurvivin (prepared as lipoplex with pegylated cationic liposomes). The RT-qPCR assay was sensitive (lower detection limit of 100 fM) and had a 5 × 107-fold dynamic range and low sample volume requirement (10 μL). The 16-point standard curves constructed using whole blood samples were linear (R 2 > 0.98). The intraday and interday variations for the slopes were ≤6%, although the variations for accuracy and precision at individual concentrations were substantially higher (58–145%). Standard curves prepared with water in place of blood showed similar results (<6% difference), indicating water may be used when blood is not available. The current RT-qPCR assay enabled the measurement of nonlabeled siRNA in small volume of blood samples.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
INTRODUCTION
RNA interference (RNAi) is an endogenous cellular process whereby siRNA of ∼20–30 nucleotides causes the degradation of complementary mRNA and consequently the knockdown of the target protein (1). This specificity makes siRNA a potentially useful tool to correct faulty genes, and several siRNA therapeutics have entered clinical evaluation (2).
Most of the pharmacokinetic (PK) studies of siRNA in animals and humans use radioisotope- or fluorescence-labeled siRNA, or biologic assays that measure the RNA levels (northern blot, ELISA) (3–8). Another method is to quantify the fluorescence-labeled siRNA carriers (9). These assays typically require >500 μL of blood, and therefore, in the case of small animals such as mice, require one animal per data point. A more recent study describes a quantitative real-time polymerase chain reaction (RT-qPCR) method to measure siRNA in plasma (10). Its utility in whole blood, e.g., in situations where the target genes are located within blood cells, has not been established.
The present study describes the development and validation of a RT-qPCR method to measure the target siRNA concentrations in blood. A second siRNA against metadherin (siMTDH) was used as the internal standard to correct for variations in the three major steps (extracting siRNA from blood, reverse transcription, PCR). The high sensitivity and low sample volume requirement (10 μL per time point) enabled the measurement of the entire blood PK profile in a single mouse.
MATERIALS AND METHODS
Chemicals and Reagents
Because naked siRNA is rapidly degraded, we used liposomes as the siRNA carrier. siRNA was formulated as a lipoplex in pegylated cationic liposomes (PCat).
Lipids (1,2-dioleoyl-3-trimethylammoniumpropane or DOTAP, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine or DOPE, 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy(polyethylene glycol)-2000] or DSPE-PEG, cholesterol) were purchased from Avanti Polar Lipids, Inc. (Alabaster, AL). 2-Mercaptoethanol and ethylenediaminetetraacetic acid (EDTA) were purchased from Sigma-Aldrich (St. Louis, MO), and heparin from Greiner Bio-One (Monroe, NC). siRNA against survivin (siSurvivin) and metadherin (siMTDH) were purchased from Integrated DNA Technologies (Coralville, IA). The sequences of the siRNA used were: siSurvivin, 5′-GGACCACCGCAUCUCUACATT-3′ (sense), 5′-UGUAGAGAUGCGGUGGUCCTT-3′ (antisense); siMTDH, 5′-UCACGUUUACGCUGCUGUCGUUUCUCU-3′ (sense), 5′-AGAAACGACAGCAGCGUAAACGUGA-3′ (antisense); Alexafluor 647 MTDH: 5′-UCACGUUUACGCUGCUGUCGUUUCUCU-3′ (sense), 5′-UCACGUUUACGCUGCUGUCGUUUCUCU/3AlexaF647N/-3′ (antisense). siRNA solutions were diluted to 10 μM, aliquoted and stored at −20°C. All reagents were used as received.
Primers for qPCR (with proprietary sequences) were purchased from Quanta Biosciences (Gaithersburg, MD). The specificity of the primers was confirmed by the lack of off-target amplification products derived from naturally occurring miRNAs in blank blood samples, with up to 35 PCR cycle threshold (Ct). Amplification products that require above 35 Ct are generally accepted to be unreliable and are usually caused by amplification of primer dimers or reaction contaminants.
Preparation of PCat and Lipoplex
PCat was prepared using the dry film and extrusion method as described previously (11). Briefly, the four lipids in PCat (DOTAP:Cholesterol:DOPE:DSPE-PEG at a ratio of 50:30:19:1) were combined and dissolved in chloroform. Evaporation of the organic phase under vacuum in a rotovap (IKA RV06-ML, Wilmington, NC) yielded a thin lipid film, followed by drying under vacuum in a desiccator and subsequent hydration in RNase-free water (1 ml per 10 mg of lipids) at 60°C for 2 h with gentle rotation. The resulting liposomal suspension was passed through a liposome extruder with a 100-nm membrane to obtain PCat. siRNA lipoplex was formed by gently mixing PCat with aqueous siRNA solution (10 μM) at room temperature, in a 1:4 siRNA-to-DOTAP charge ratio. Particle size distribution was measured using the light scattering method with a Zetasizer Nano ZS90 (Malvern, Westborough, MA).
Animal Protocols
Animal experiments were performed according to a protocol approved by the Institutional Animal Care and Use Committee. Five-week-old female athymic nude mice (Charles River, Wilmington, MA) were used as blood donors for the assay development and for a pilot PK study. For blood donors, whole blood (∼1 mL) was collected via cardiac puncture in mice anesthetized with isoflurane (2% in 100% O2 gas), and added to a 15-mL conical tube containing anti-coagulant (EDTA or heparin). For the PK study, a mouse was given an intravenous injection of PCat-siSurvivin lipoplex (200 μL total volume, 1 nmole siRNA dose) at the anterior end of the left lateral tail vein. At predefined time points, 10 μL of whole blood was withdrawn from the right lateral tail vein with a 20-μL pipette and processed (see below). Blood samples were stored on ice or in a refrigerator until use.
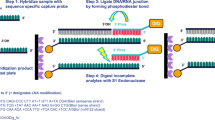
siRNA Extraction and Quantification
siRNA was extracted (from blood or water) and then quantified using RT-qPCR. Extraction used a commercial kit (mirVana™ PARIS™ RNA and Native Protein Purification Kit, Thermo Fisher Scientific, Waltham, MA). This kit, which gives the highest yield of total and small RNAs among similar commercial kits (12), extracts small RNAs in two steps. The first step is to separate small RNAs (i.e., siRNA and miRNA) from large RNA (i.e., mRNA). Briefly, siRNA-containing samples (10 μL) were mixed within 1 min, by up-and-down pipetting, with an ice-cold mixture of the internal standard (siMTDH, 10 μL of 100 pM solution), Cell Disruption Buffer (180 μL to dissociate siRNA from PCat), and 2X Denaturation Buffer containing beta-mercaptoethanol (200 μL, to inactive RNAse). The resulting mixture was vortexed with 400 μL phenolic chloroform. Following centrifugation at 20,000×g for 5 min at 4°C, an aliquot of the supernatant (350 μL) was transferred to a new Eppendorf tube and mixed with 100% ethanol (1/3 the sample volume) to precipitate the larger RNA molecules. The mixture was vortexed and applied to the column containing a glass fiber filter that immobilizes the large RNA precipitates. The column was centrifuged at 10,000×g for 30 s, yielding flow-through containing siRNA (about 450 μL). The second step was to extract siRNA from the flow-through. Briefly, the flow-through was mixed with 100% ethanol (at 2/3 the sample volume to precipitate siRNA) and applied to a second column with glass fiber filter, followed by one wash with Wash Buffer 1 and two washes with Wash Buffer 2. The siRNA was then eluted with warm Elution Buffer (90°C), yielding an eluate enriched with siRNA.
The extracted siRNA served as the template to synthesize cDNA using the poly-A tail method (qScript™ microRNA cDNA Synthesis Kit, Quanta Biosciences). This method uses two steps. The first step uses poly A polymerase to add a poly-A tail on to the 3′-end of either strand of RNAs, such as siRNA and miRNA, that do not have a poly-A tail. The second step uses an Oligo-dT adapter primer with a universal PCR primer sequence to reverse transcribe the resulting siRNA-poly-A tail sequence. The resulting cDNA template was diluted in RNAse-free water. As discussed in “Results”, the cDNA synthesis step was nonlinear and samples containing high siRNA concentrations were diluted 100- or 1000-fold before cDNA synthesis. Fifteen microliters of a master-mix containing PerfeCTa® SYBR® Green SuperMix (Quanta Biosciences), PerfeCTa® Universal PCR Primer, and primers for survivin or MTDH, was added to 5 μL of cDNA template, and qPCR was performed using the following program on a CFX96 Real-Time PCR Detection System (Bio-Rad Laboratories, Hercules, CA): 3 min at 95°C, 40 cycles of 15 s at 95°C and 1 min at 61°C. We selected 200 relative fluorescence units (RFU), which was within the log-linear range of the resulting amplification curves, as the threshold. The differences in the resulting Ct for the target and internal standard siRNA (siSurvivin Ct minus siMTDH Ct) were used to calculate the siRNA concentrations based on standard curves established using similarly processed samples containing known concentrations of PCat-siSurvivin lipoplex.
We found the RNA concentrations for the highest standard curves sample were at or below the lower limit of detection (2 ng/μL) for the instrument being used (Nanodrop 2000, ThermoFisher), likely due to the small sample volumes and the fact that siRNA is ∼100-fold shorter than the average mammalian mRNA molecule. Accordingly, quantitation of siRNA concentration prior to cDNA synthesis was not possible.
Verification of siRNA Release from PCat
The completeness of extracting siRNA from PCat-siRNA lipoplex by Cell Disruption Buffer was studied using two methods. Confocal microscopy was used to visualize the dissociation of fluorescent AF-647 siMTDH from the lipoplex. An aliquot of 50 μL of the lipoplex was placed on a Lab-Tek® Chambered Coverglass slide (Electron Microscopy Sciences, Hatfield, PA) and visualized using a Leica SP8 spectral confocal microscope (Wetzlar, Germany) equipped with an environmental chamber and 405, 488, 552, and 638 nm solid-state lasers. Time-lapse images were taken every 0.2 s at 1400 Hz (256 × 256 resolution) using the 638 laser line, before and after adding Cell Disruption Buffer (150 μL), for a total of 2 min. The second method monitored the size change of the PCat-siRNA lipoplex nanoparticles upon mixing with Cell Disruption Buffer (150 μL). Particle size was measured using the light scattering method with Zetasizer Nano ZS90, and intensity values were expressed as kilo counts per second (kcps). The control samples for both methods used water instead of Cell Disruption Buffer.
Effect of Anti-Coagulant on RT-qPCR siRNA Analysis
It has been reported that heparin interferes with PCR reactions and RNA stability (13,14), whereas another anti-coagulant, EDTA, does not (15). We used the internal standard siMTDH to evaluate the effect of these two anticoagulants on RT-qPCR assay results. Briefly, 10 μL of whole blood stabilized with heparin (15 U heparin/mL blood) or EDTA (1.8 mg/mL of blood) was mixed with siMTDH (10 μL of 100 pM solution) and analyzed using the methods described above.
Assay Variability
Assay variability was measured in two ways; both used standard curve samples containing 100 fM–5 μM of siSurvivin and a constant concentration of the internal standard siMTDH (50 pM). First, in order to evaluate the relative contribution of each step to assay variations, we used variances (Ct2, squared value of standard deviation (SD) due to its additive property). The overall assay variance is the sum of all three steps (siRNA extraction, cDNA synthesis, and qPCR) and was determined by extracting three sets of standard curve samples (16 concentrations). The variance for the second plus last steps (cDNA synthesis and cDNA amplification) was determined using one set of the post-extraction samples in triplicates, and the variance for the last step (qPCR) was determined using one set of cDNA template in triplicates. We calculated, for each concentration, the variance among three measurements and expressed the results as mean ± SD and coefficient of variation (CV). Second, the intraday and interday assay accuracy and precision were determined at low, medium, and high concentrations (1 pM, 1 nM, and 1 μM), using three standard curves (13 concentrations between 100 fM and 5 μM) prepared on the same day or one set per day for three consecutive days. Accuracy was the (average of values quantified using three separate standard curves) divided by (known value)*100%, and precision was the variation among the triplicate concentrations. Intraday and interday variations of the slopes of the three standard curves were also determined.
Data Analysis
All experiments were performed in triplicates. Statistical analysis was performed and standard curves were established using log-linear regression using GraphPad Prism Software ver. 6.0 (La Jolla, CA). Analysis of the blood concentration-time profile was performed using Phoenix WinNonlin (Pharsight, Princeton, NJ).
RESULTS
Verification of siRNA Release from PCat
Figure 1a shows the representative confocal microscopy images. The time-lapse microscopy results are available in Supplementary Materials. The undiluted stock solution of PCat-siMTDH lipoplex (10 μM) showed punctated fluorescence signals, indicating the lipoplex was present as discrete particles. Addition of three volumes of water diluted the signals but did not alter the punctated appearance. In contrast, replacing water with Cell Disruption Buffer caused the signals to become diffuse, indicating siRNA was no longer associated with PCat. Figure 1b shows the particle size analysis results. The PCat-siMTDH lipoplex showed a single peak with an average diameter of 190 nm. In contrast, the mixture of lipoplex and Cell Disruption Buffer showed no identifiable peaks and showed intensity readings comparable to the background readings of Disruption Buffer (3.3 kcps). These data confirmed that treatment with Cell Disruption Buffer caused the complex dissociation of siRNA from the lipoplex.
Effect of Anticoagulants on RT-qPCR Quantification of siRNA in Whole Blood
Figure 2 shows the RT-qPCR results of samples containing the internal standard siMTDH (10 μL of 100 pM solution). The RNAse-free water control samples required 17.5 Ct to produce the threshold amplification product (200 RFU) of siMTDH. In comparison, the freshly obtained blood without coagulants required 18.5 Ct to yield the same PCR product, which was increased to 19.5 Ct for the EDTA-treated blood and 28 Ct for the heparin-treated blood. Because the amount of PCR products doubled with each Ct, the corresponding reduction in detection limit, relative to fresh blood, was 21 or 2-fold by EDTA and 210 or 1000-fold by heparin. Subsequent studies used EDTA as the anticoagulant.
Effect of anti-coagulant on siRNA analysis by RT-qPCR. RT-qPCR amplification data of the internal standard siMTDH: the number of Ct required to produce 200 relative fluorescence units (RFU) increased from 17.5 in water to 18.5 in fresh blood, 19.5 in EDTA-treated blood, and 28 in heparin-stabilized blood. (n = 3)
Linearity, sensitivity, and variability of RT-qPCR siRNA assay
Figure 3a shows the standard curve constructed with blood samples containing known amounts of siSurvivin; the relationship shows deviations from linearity at concentrations above 1 nM. We next determined the source of nonlinearity.
Assay nonlinearity. a Nonlinear standard curve at higher siSurvivin concentrations. b Source of nonlinearity. Expected Ct changes at different sample dilutions vs. observed values. Solid line: no nonlinearity. Solid squares: cDNA samples for qPCR. Open squares: siRNA extracts for cDNA synthesis. c Dilution improved linearity. Samples containing 1–30 and 100–5000 nM siSurvivin diluted 1:100 and 1:1000-fold, respectively, after siRNA extraction. (n = 3 for all experiments)
The three main steps in the siRNA quantitation assay are column extraction of siRNA from whole blood, synthesis of cDNA from the extracted siRNA, and qPCR of the cDNA template. Regarding siRNA extraction, we calculated the column capacity to be 3 × 10−4 g using an assumption of 30 pg RNA/cell (16,17). Because this value is about 400-times the maximum siRNA amount of 7 × 10−7 g used in the standard curve samples, we ruled out column saturation as a potential source of nonlinearity. To determine if cDNA synthesis from the extracted siRNA and/or PCR amplification of the cDNA template caused the nonlinearity, we compared the respective expected Ct changes to reach the RFU with the observed Ct changes, at different dilutions of siRNA extract or cDNA template (diluted 1:20, 1:100 and 1:1000 folds). The results are shown in Fig. 3b; the cDNA amplification of samples at all four dilutions show nearly identical Ct changes indicating linearity for this step. In contrast, the cDNA synthesis step showed identical observed and expected Ct changes only for the more diluted samples (1:100 and 1:1000 dilution), whereas the more concentrated samples show substantial deviations. These results indicate saturable cDNA synthesis as the primary source of assay nonlinearity. This was subsequently confirmed by the finding that dilution of siRNA extracts prior to cDNA synthesis by 1:100 fold (1–30 nM concentration) or 1:1000-fold (>30 nM) yielded a linear standard curve (Fig. 3c). Note that the internal standard siMTDH was added prior to extraction and hence was diluted together with the analyte siSurvivin. Subsequent standard curves used 1:100 and 1:1000 dilution, respectively, for samples containing >1 and >30 nM siRNA.
Figure 4 shows a representative standard curve of PCat-siSurvivin in whole blood; the relationship was log-linear between 100 fM and 5 μM siRNA (5 × 107-fold range) with a coefficient of determination (R 2) of >0.98. The blank blood samples showed no detectable amplification products at the upper limit of 35 Ct. In comparison, amplification products were readily detected at ∼30 Ct for samples containing 100 fM siRNA, or at least 25 or 32-fold above the value in blank samples. Accordingly, 100 fM was used as the lower detection limit. Note the 1:100- and 1:1000-fold diluted samples yielded nearly identical linear relationship as the undiluted samples. A similarly prepared standard curve using water in place of whole blood yielded a nearly superimposable line.
PCat-siSurvivin standard curves. Samples were diluted as in Fig. 3c. Lines: best-fitted linearly regressed lines. Comparable standard curves in water (circles, solid line; y = 2.84–3.2*log(x), R 2 = 0.98) and whole blood (square, broken line; y = 2.30–2.8*log(x), R 2 = 0.98). Inset: PCat-siSurvivin pharmacokinetics in a mouse. Symbols: siRNA concentrations determined using the two standard curves. Lines: best-fit curves using a two-compartment model
We further determined the extents of assay variability due to siRNA extraction, cDNA synthesis and cDNA amplification (Table Ia), using the property of additivity of variances of independent variables. The results of standard curve samples were used to compare the overall assay variance with the variances of the cDNA synthesis and cDNA amplification steps. The mean overall variance was 0.125 Ct2. The variance due to the sum of cDNA synthesis and cDNA amplification was 0.015 Ct2 and the variance due to qPCR was 0.006 Ct2. Hence, the net variance due to cDNA synthesis was 0.009 Ct2. The respective variances for cDNA synthesis and cDNA amplification, therefore, corresponded to 7.3% and 4.8% of the overall variance, indicating the majority of assay variation was attributable to siRNA extraction (88% of overall variance).
Table Ib summarizes the intraday and interday assay accuracy and precision at low, medium, and high siSurvivin concentrations (1 pM, 1 nM, and 1 μM). The variations in assay precision (about 60% for both) were lower compared to the variations in assay accuracy (113% for intraday and 145% for interday). For the latter, the variation at 1 pM was generally lower than at higher concentrations (e.g., ∼60% vs. between 150–220%). The intraday and interday variations in the standard curve slopes were substantially lower, at ≤6% for both.
Application of RT-qPCR Assay to Study siRNA Pharmacokinetics in an Individual Mouse
The inset in Fig. 4 shows the PK profile in an individual mouse given an intravenous dose of PCat-siSurvivin (1 nmole). A total of 12 samples, 10 μL each, were obtained from 0 min to 24 h post-injection. The blood concentration-time profile, established using a standard curve constructed with whole blood as the matrix, showed biphasic disposition, with a rapid initial half-life of 72 min followed by a slow terminal phase. In comparison, the PK profile obtained using a standard curve constructed with water showed similar results; the individual data points and the areas under the two profiles deviated by less than 6% (e.g., 10.8 vs. 11.5 μmol/L*min). Over 90% and >99% of the initial blood concentration was eliminated by 5 and 8 h, respectively.
DISCUSSION
Two published studies used RT-qPCR to quantify siRNA. A 2010 study measured the siRNA amount associated to the Ago2 enzyme in the RNA-induced silencing complex, in mouse liver tissue following intravenous injection of siRNA-containing liposomes (17). In this study, the siRNA-Ago2 complex was isolated from cell/tissue extracts using immunoprecipitation, followed by extraction of Ago2-associated siRNA and endogenous miRNA. The amount of siRNA was quantified by stem-loop RT-qPCR and the results were expressed relative to that of miRNA. Hence, this method is useful for quantifying the amount of siRNA delivered to its target site, but, because the siRNA in blood is not associated with the intracellular Ago2, cannot be used in PK studies. A 2014 study used RT-qPCR and radioactivity to measure siRNA in plasma and tissues (10); the RT-qPCR results show detection of siRNA over a 105-fold dynamic range (10 pM to 2 μM). The major differences between the current study and the 2014 study are that the current assay (a) extracted the total siRNA from whole blood, and therefore included the lipoplex in blood cells, and (b) used an internal standard to correct for experimental variations during the multistep assay.
Table II compares the present method and the methods described in the literature for siRNA quantification (4,6–8,10). The previously used methods have one or more of the following limitations: (a) require the use of plasma instead of whole blood and therefore cannot account for lipoplexes associated with blood cells, (b) require the use of radioactivity, (c) require the use of fluorescence labels, which may alter the structure and interfere with distribution, and (d) have a limited dynamic range or sensitivity. The current method offers several advantages. First, its dynamic linear range is orders of magnitude higher compared to ELISA (4) and radioactivity detection (7). Second, it has 50- and 600- times greater sensitivity compared to the previous assays (5 pM detection limit by ELISA and 60 pM by RT-qPCR of plasma). This feature enables the detection of siRNA at lower levels and hence extends the duration of detecting siRNA in biological matrices. For example, Fig. 4 shows appreciable siRNA levels for at least 24 h after dosing and identified a slow terminal disposition phase. In comparison, PK studies performed using ELISA, fluorescence, and radioactivity assays were limited to <2 h post-administration and omitted the later slow phase (4,6,8). Another important advantage derived from the high sensitivity is the low sample volume requirement, i.e., the assay requires only 10 μL of blood. Because this low volume can be readily and repeatedly withdrawn from a tail vein in rodents, it is possible to obtain from the same animal (e.g., a mouse) multiple blood samples, such that a complete PK profile in individual animals can be defined. This approach is cost-effective and eliminates the inter-animal variability compared to situations that require using one animal per time point (e.g., assays that require much larger sample volumes).
The results of the current study indicate several aspects that require attention. First, our results identified variations in siRNA extraction as the major cause of assay variability, similar to the observation for miRNA extraction (18). The confocal microscopy results and the size distribution results established that the PCat-siRNA lipoplex was fully solubilized after treatment with the commercial proprietary lysis buffer. However, the possibility that lipids in the samples affected the siRNA extraction cannot be ruled out and warrant additional investigations. Another error-prone step is that the separation of small RNAs from large RNAs is based on the use of different volumes of ethanol; it is conceivable that this step is subjected to substantial variability and that the assay performance may be improved when more specific separation methods become available. Loss of the siRNA-binding glass fibers from the siRNA extraction column during centrifugation, as reported previously (12), might also have contributed to the variability. The current assay uses an internal standard siRNA to control for the variations in siRNA extraction, cDNA synthesis, and PCR. We recommend using internal standards (a) with amplification efficiencies similar to the target siRNA, and (b) at concentrations that require between 18 and 30 Ct to yield sufficient amplification products, in order to achieve good reproducibility and linearity (e.g., 50 pM in the current study). Second, our results identified saturable cDNA synthesis of the extracted siRNA as the source of assay nonlinearity. The nonlinearity was likely caused by the limited amount of reverse transcriptase enzyme (we used the maximum recommended volume of 1 μL per 20 μL reaction as prescribed by the manufacturer). Theoretically, this problem can be overcome by increasing the amount of enzyme. However, because of its high costs, we took the alternative approach of diluting the sample prior to cDNA synthesis. We recommend using pilot studies to establish the linearity of this step and/or diluting the extracted siRNA prior to cDNA synthesis. We used 1:100 and 1:1000 dilution, respectively, for samples containing ≥1 and ≥100 nM siRNA in the present study. However, based on the nearly identical results of the two dilutions, we recommend using 1:1000 dilution for ≥1 nM. Third, while whole blood is the most suitable matrix for standard curves, our finding that the standard curve constructed using nuclease-free water yielded comparable PK results (∼6% difference) suggests water may be used as a substitute in the event that blank blood is not readily available. Fourth, the accuracy and precision of the assay were less robust compared to chemical assays. This was largely due to the nature of qPCR where quantitation is based on the number of amplification cycles; the increase of one Ct would result in a 200% change in concentration terms. This happens even though the CV of Ct values was low (∼15%). Other sources of variation include the small amount of RNA and the multistep sample preparation, which, at the current state-of-the-art, are unavoidable.
Finally, the specificity of RT-qPCR assays, either the stem-loop method used in previous studies (17) or the poly-A tail method used in the present study, depends on the specificity of primer for the RNAi-of-interest. The present study used a 20 nucleotide-long primer to detect the 21 nucleotide-long siRNA, in order to increase the specificity for the intact siRNA. Measurements of truncated siRNA would require the use of additional primers (e.g., shorter primers) or other methods to separate the siRNA metabolites prior to analysis.
CONCLUSION
In summary, the emerging importance of RNAi therapeutics requires the development of robust, quantitative assays to measure unmodified and modified RNAi. The present RT-qPCR assay provides a means for measuring the total siRNA blood concentration, is generally applicable to all siRNA for which there are specific primers, and can be applied to study PK in small animals. The accuracy and precision of RT-qPCR assays may be improved by using larger blood volumes although the benefits may be off-set by the fewer data points from the same animal, and by reducing the variations in siRNA enrichment and extraction.
Abbreviations
- Ct:
-
PCR cycle threshold
- DOPE:
-
1,2-dioleoyl-sn-glycero-3-phosphoethanolamine
- DOTAP:
-
1,2-dioleoyl-3-trimethylammoniumpropane
- kcps:
-
Kilo counts per second
- PK:
-
Pharmacokinetics
- RT-qPCR:
-
Quantitative real-time polymerase chain reaction
- siMTDH:
-
Metadherin siRNA
- siSurvivin:
-
Survivin siRNA
- R 2 :
-
Coefficient of determination
References
Rw C, Ej S. Origins and mechanisms of Mirnas and siRNAs. Cell. 2009;136:642–55.
Haussecker D. The business of Rnai therapeutics. Hum Gene Ther. 2008;19:451–62.
Iversen F, Yang C, Dagnaes-Hansen F, Schaffert D, Kjems J, Gao S. Optimized siRNA-peg conjugates for extended blood circulation and reduced urine excretion in mice. Theranostics. 2013;3:201–9.
Ej K, Park Tg O, Yk SC. Assessment of siRNA pharmacokinetics using Elisa-based quantification. J Contrl Rel. 2010;143:80–7.
Kim S, Jeong J, Lee S, Sw K, Tg P. Local and systemic delivery of Vegf siRNA using polyelectrolyte complex micelles for effective treatment of cancer. J Contrl Rel. 2008;129:107–16.
Om M, Librizzi D, Pfestroff A, Schurrat T, Buyens K, Sanders N. Stability of siRNA polyplexes from poly (Ethylenimine) and poly (Ethylenimine)-G-Poly (Ethylene Glycol) under in vivo conditions: effects on pharmacokinetics and biodistribution measured by fluorescence fluctuation spectroscopy and single photon emission computed tomography (SPECT) imaging. J Contrl Rel. 2009;138:148–59.
Mui B, Tam YK, Jayaraman M, Ansell S, Du X, Tam YY. Influence of polyethylene glycol lipid desorption rates on pharmacokinetics and pharmacodynamics of siRNA lipid nanoparticles. Mol Ther Nucleic Acids. 2013;2:E139.
Bm G, Jr O, Quinlan A, Darcy R, Griffin BT, Cryan JF. Pegylated cyclodextrins as novel siRNA nanosystems: correlations between polyethylene glycol length and nanoparticle stability. Int J Pharmaceut. 2014;473:105–12.
Yk O, Tg P. siRNA delivery systems for cancer treatment. Adv Drug Deliv Rev. 2009;61:850–62.
Christensen J, Litherland K, Faller T, Van De Kerkhof E, Natt F, Hunziker J, et al. Biodistribution and metabolism studies of lipid nanoparticle-formulated internally [3h]-labeled siRNA in mice. Drug Metab Dispos. 2014;42:431–40.
Wang J, Lu Z, Yeung Bz, Wientjes Mg, Cole Dj, Au Jls. Tumor priming enhances siRNA delivery and transfection in intraperitoneal tumors. J Contrl Rel. 2014.
Kl B, Javaherian A, Bomprezzi R, Ghaffari L, Rhodes S, Courtright A. Identification of extracellular Mirna in human cerebrospinal fluid by next-generation sequencing. RNA. 2013;19:712–22.
Bai X, Fischer S, Keshavjee S, Liu M. Heparin interference with reverse transcriptase polymerase chain reaction of Rna extracted from lungs after ischemia-reperfusion. Transpl Int. 2000;13:146–50.
Jb M, Mohr S, Pfister M, Visvikis-Siest S. Collection and storage of human blood cells for Mrna expression profiling: a 15-month stability STUDY. Clin Chem. 2005;51:1250–2.
Me G, Jl B, Caballero J, Gargallo-Viola D. Anticoagulants interfere with Pcr used to diagnose invasive aspergillosis. J Clin Microbiol. 2002;40:1567–8.
Ps L, Schelter J, Burchard J, Kibukawa M, Martin MM, Bartz SR. Transcripts targeted by the microrna-16 family cooperatively regulate cell cycle progression. Mol Cell Biol. 2007;27:2240–52.
Pei Y, Hancock P, Zhang H, Bartz R, Cherrin C, Innocent N. Quantitative evaluation of siRNA delivery in vivo. RNA. 2010;16:2553–63.
Js M, Milosevic D, Reddi HV, Grebe SK, Algeciras-Schimnich A. Analysis of circulating microrna: preanalytical and analytical challenges. Clin Chem. 2011;57:833–40.
Acknowledgments
Supported in part by research grants, RO1CA158300 (GW, JA) and RO1CA163015 (GW, JA) from National Cancer Institute and National Institute of Biomedical Imaging and Bioengineering, NIH, DHHS.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Yeung, B.Z., Lu, Z., Wientjes, G.M. et al. High Sensitivity RT-qPCR Assay of Nonlabeled siRNA in Small Blood Volume for Pharmacokinetic Studies: Application to Survivin siRNA. AAPS J 17, 1475–1482 (2015). https://doi.org/10.1208/s12248-015-9812-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1208/s12248-015-9812-y