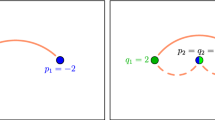
Abstract
We demonstrate that cell resampling can eliminate the bulk of negative event weights in large event samples of high multiplicity processes without discernible loss of accuracy in the predicted observables. The application of cell resampling to much larger data sets and higher multiplicity processes such as vector boson production with up to five jets has been made possible by improvements in the method paired with drastic enhancement of the computational efficiency of the implementation.
Similar content being viewed by others
1 Introduction
One of the greatest challenges in theoretical high-energy physics is to meet the demand for increasingly precise predictions. Even leaving aside conceptual issues, reducing theoretical uncertainties typically requires ever more complex calculations, which incur steeply rising computing costs. Already now Monte Carlo event generation for the LHC constitutes a notable fraction of the experimental computing budgets. Even with this large computing power investment event sample sizes have to be limited to a size where the resulting uncertainty can be non-negligible [1]. What is more, event generation is only the first step in the full simulation chain, and the already substantial computing costs of this step are often dwarfed by the subsequent simulation of the detector response. All these problems are expected to become even more severe in the future, in particular with the advent of the HL-LHC. This development is obviously at odds with general sustainability goals. Improving the efficiency of event simulation is one of the foremost tasks in particle physics phenomenology.
For a high-accuracy simulation a Monte Carlo generator needs to combine real and virtual corrections, match resummations in different limits and combine processes of different multiplicities while avoiding double counting. Many different prescriptions exist for each of these combination steps [2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24]. While they differ substantially in the details, a common theme is the introduction of a varying number of auxiliary events that subtract from the accumulated cross section instead of adding to it. If the number of such negative-weight events becomes large enough, they can severely impair the statistical convergence since large amounts of events are required for a sufficient precision to allow for accurate cancellation of the contributions with opposite signs. In fact, for a fractional negative-weight contribution \(r_-\) the number \(N(r_-)\) of required unweighted Monte Carlo events to reach a given statistics goal is (see e.g. [25])
Requiring a larger number of events not only increases the computational cost in the generation stage, but especially also in the subsequent detector simulation. A further problem is the increased disk space usage, inducing both short- and long-term storage costs. It is therefore highly desirable to keep the negative-weight contribution small, \(r_- \ll \frac{1}{2}\).
One avenue in this direction is to reduce the number of negative-weight events during event generation, see e.g. [25,26,27,28,29]. A second approach is to eliminate negative weights in the generated sample, before detector simulation [30,31,32,33]. In the following, we focus on the cell resampling approach proposed in [33], which in turn was inspired by positive resampling [30]. Cell resampling is independent of both the scattering process under consideration and any later stages of the event simulation chain. It only redistributes event weights, exactly preserving event kinematics. The effective range over which event weights are smeared decreases with an increasing event density. This implies that the smearing uncertainty decreases systematically with increasing statistics without the need to change the method. As demonstrated for the example of the production of a W boson with two jets at next-to-leading-order (NLO), a large fraction of negative weights can be eliminated without discernible distortion of any predicted observable. A limitation of the original formulation is the computational cost, which rises steeply with both the event sample size and the number of final-state particles.
In Sect. 2, we briefly review the method and describe a number of algorithmic improvements which allow us to overcome the original limitations through a speed-up by several orders of magnitude. In Sect. 3, we then apply cell resampling to high-multiplicity samples with up to several billions of events for the production of a W or Z boson in association with up to five jets at NLO. We conclude in Sect. 4.
2 Algorithmic improvements
In the following, we briefly review cell resampling and describe the main improvements that allow us to apply the method also to large high-multiplicity Monte Carlo samples. For a detailed motivation and description of the original algorithm see [33].
2.1 Cell resampling
At its heart, cell resampling consists of repeatedly selecting a subset of events – referred to as cells – and redistributing event weights within the selected set. The steps are as follows.
-
1.
Select an event with negative weight as the first event (the “seed”) of a new cell \({{\mathcal {C}}}\).
-
2.
Out of all events outside the cell, add the one with the smallest distance from the seed to the cell. Repeat for as long as the accumulated weight of all events within the cell is negative.
-
3.
Redistribute the weights of the events inside the cell such that the accumulated weight is preserved and none of the final weights is negative.Footnote 1
-
4.
Start anew with the next cell, i.e. with step 1.
Note that an event can be part of several cells, but will only be chosen as a cell seed at most once. In practice, we usually want to limit the maximum cell size and abort step 2 once the distance between the cell seed and its nearest neighbour outside the cell becomes too large. We denote the maximum cell radius, i.e. the maximum allowed distance between the cell seed and any other event within the cell, by \(d_\text {max}\). It is a parameter of the cell resampling algorithm. If we limit the cell size in this way, we can only achieve a partial cancellation between negative and positive event weights in the following step 3. For practical applications this is often sufficient, since the small remaining contribution from negative weights has a much reduced impact on the statistical convergence, cf. Eq. (1).
The computational cost of cell resampling tends to be completely dominated by the nearest-neighbour search in step 2. In a naive approach, one has to calculate the distances between the cell seed and each other event in the sample. Since the number of cells is proportional to the sample size N, the total computational complexity is \({{\mathcal {O}}}(N^2)\). This renders the naive approach unfeasible for samples with more than a few million events. For this reason, an alternative approximate nearest-neighbour search based on locality-sensitive hashing (LSH) [34, 35] was considered in [33]. While this lead to an improved scaling behaviour, the quality of the approximate search was also found to deteriorate with an increasing sample size. An improved version of this algorithm, discussed in Appendix A, still appears to suffer from the same problem. In Sect. 2.2, we introduce an exact search algorithm that is orders of magnitude faster than the naive search.
The problem of costly distance calculations is further exacerbated by the fact that a direct implementation of the originally proposed distance function suffers from poor scaling for high multiplicities. To compute the distance between two events e and \(e'\), we first cluster the outgoing particles into infrared-safe physics objects, e.g. jets. We collect objects of the same type t into sets \(s_t\) for e and \(s_t'\) for s. The distance between the two events is then
where \(d(s_t, s_t')\) is the distance between the two sets \(s_t, s_t'\). It is given by
where \(p_1,\dots , p_P\) are the momenta of the objects in \(s_t\) and \(q_1,\dots , q_P\) the momentaFootnote 2 in \(s_t'\). A naive minimisation procedure considers all permutations \(\sigma \) in the symmetric group \(S_P\), i.e. P! possibilities. For large multiplicities P a direct calculation quickly becomes prohibitively expensive. In [33], it was therefore suggested to use an approximate scheme in this case. In Sect. 2.3 we discuss how the set-to-set distance can be calculated both exactly and efficiently.
2.2 Nearest-neighbour search
Our improved nearest-neighbour search is based on vantage-point trees [36, 37]. To construct a vantage-point tree, we choose a single event as the first vantage point. We then compute the distance to the vantage point for each event. The closer half of the events lie within a hypersphere with radius given by the median distance to the vantage point. We call the populated part of this hypersphere the inside region and its complement the outside region. We then recursively construct vantage-point trees inside each of the two regions. The construction terminates in regions that only contain a single point.
To find the nearest neighbour for any event e, we start at the root of the tree, namely the first chosen vantage point. We calculate the distance D between this vantage point and e. If D is less than the radius R of the hypersphere defining the inside region, we first continue the search in the inside subtree, otherwise we choose the outside subtree first. Let us first consider the case that the inside region is the preferred one. It will contain a nearest-neighbour candidate with a distance d to the initial event e. By the triangle inequality we deduce that the actual nearest neighbour can have a distance of at most \(D+d\) to the current vantage point. Therefore, if \(D+d < R\), the actual nearest neighbour cannot be in the outside region. Conversely, if we started our search in the outside region and found a nearest-neighbour candidate with \(D-d > R\), then the actual nearest neighbour cannot lie in the inside region. In summary, if \(d < |R - D|\) only the preferred region has to be considered.
Vantage-point tree search is indeed very well suited for cell resampling. The construction is completely agnostic to the chosen distance function. In particular, unlike the LSH-based methods considered in [33] and Appendix A, it does not require a Euclidean metric. For an event sample of size N, the tree construction requires \({{\mathcal {O}}}(N \log N)\) steps and can be easily parallelised. In the ideal case where only the preferred regions are probed, each nearest-neighbour search requires \(\log _2 N\) comparisons, which again results in an overall asymptotic complexity of \({{\mathcal {O}}}(N \log N)\). While this means that for sufficiently large event samples cell resampling will eventually require more computing time than the \({{\mathcal {O}}}(N)\) event generation, we find that this is not the case for samples with up to several billion events. Timings for a practical application are given in Sect. 3.3.
We further optimise the nearest-neighbour search in several aspects. Most importantly, if we limit the maximum cell size to \(d_{\text {max}}\), we can dramatically increase the probability that only the preferred regions have to be considered. In fact, if \(|R - D| > d_{\text {max}}\) then any suitable nearest neighbours have to lie inside the preferred region. We can further enhance the probability through a judicious choice of the vantage points. Since input events near the boundary between inside and outside regions require checking both regions for nearest neighbours, the general goal is to minimise this surface. To this end, we choose our first vantage point at the boundary of the populated phase space. We select a random event, calculate the distance to all other events, and choose the event with the largest distance as the vantage point. Then, when constructing the subtrees for the inside and outside regions, we choose as vantage points those events that have the largest distance to the parent vantage point.
When constructing a cell, we have to find nearest neighbours until either the accumulated weight becomes non-negative or the distance exceeds the maximum cell radius. This corresponds to a so-called k nearest neighbour search, where in this case the required number k of nearest neighbours is a priori unknown. To speed up successive searches, we cache the results of distance calculations, i.e. all values of D for a given input event.
Finally, we note that the vantage-point tree can also be employed for approximate nearest-neighbour search if one only searches the preferred region in each step. We exploit this property by first partitioning the input events into the inside and outside regions of a shallow vantage point tree, aborting the construction already after the first few steps. We then apply cell resampling to each partition independently. This approach allows efficient parallelisation, while yielding much better results than the independent cell resampling of randomly chosen partial samples. The price to pay is that the quality of the nearest-neighbour search and therefore also of the overall resampling deteriorates to some degree. In practice this effect appears to be minor, see also Sect. 3.2.
2.3 Set-to-set distance at high multiplicities
The distance between two events as defined in [33] is the sum of distances between sets of infrared-safe physics objects, see Eq. (2). To define the distance between two such sets \(s_t, s_t'\), we aim to find the optimal pairing between the momenta \(p_1,\dots , p_P\) of the objects in \(s_t\) and the momenta \(q_1,\dots , q_P\) of the objects in \(s_t'\). The naive approach of considering all possible pairings, cf. Eq. (3), scales very poorly with the number of objects. However, the task of finding an optimal pairing is an instance of the well-studied assignment problem.
Let us introduce the matrix \({{\mathbb {D}}}\) of distances with
An efficient method for minimising \(\sum _{i=1}^{P} {{\mathbb {D}}}_{i\sigma (i)}\) was first found by Jacobi [38, 39]. It was later rediscovered independently and popularised under the name “Hungarian method” [40,41,42,43,44]. The algorithm mutates the entries of \({{\mathbb {D}}}_{ij}\) in such a way that the optimal pairing is preserved during each step. After each step, one marks a minimum number of rows and columns such that each vanishing entry is part of a marked row or column. The algorithm terminates as soon as all rows (or columns) have to be marked. The mutating steps are as follows:
-
1.
Replace each \({{\mathbb {D}}}_{ij}\) by \({{\mathbb {D}}}_{ij} - \min _{k} {{\mathbb {D}}}_{ik}\).
-
2.
Replace each \({{\mathbb {D}}}_{ij}\) by \({{\mathbb {D}}}_{ij} - \min _{k} {{\mathbb {D}}}_{kj}\).
-
3.
Find the smallest non-vanishing entry. Subtract it from all unmarked rows and add it to all marked columns.
Step 3 is repeated until the termination criterion is fulfilled. In our code, we use the implementation in the pathfinding [45] package. Like the remainder of our implementation of cell resampling it is written in the Rust programming language.
Using the Hungarian algorithm instead of a brute-force search improves the scaling behaviour for sets with P momenta from \({{\mathcal {O}}}(P!)\) to \({{\mathcal {O}}}(P^3)\). In practice, we find it superior for \(P > 3\). The FlowAssign algorithm proposed by Ramshaw and Tarjan [46] would scale even better, with a time complexity of \({{\mathcal {O}}}\big (P^{5 / 2}\log (D P)\big )\). The caveat is that the scaling also depends logarithmically on the range D of distances encountered. Since the maximum multiplicity reached in current NLO computations is limited, an auction [47] (or equivalently push-relabel [48, 49]) algorithm may still perform better in practice despite formally inferior scaling behaviour. We leave a detailed comparison to future work.
3 Negative weight elimination in vector boson plus jets production at NLO
We are now in a position to apply cell resampling to large high-multiplicity event samples. We consider the production of a vector boson in association with jets at NLO, using ROOT [50] ntuple [51, 52] event files generated with BlackHat [53] and Sherpa 2.1 [54]. Jets are defined according to the anti-\(k_t\) algorithm [55] with \(R = 0.4\) and a minimum transverse momentum of \(25\,\)GeV. More details on the event generation are given in [52, 56]. The various samples with their most salient properties are listed in Table 1.
We apply cell resampling to each of the samples, defining infrared-safe physics objects according to the above jet definition. We use the distance function defined in [30], which follows from Eqs. (2), (3), and the momentum distance
Here, we set \(\tau = 0\) and limit the maximum cell radius to 10 GeV for samples Z1, Z2, and W5, and to 2 GeV for sample Z3. To examine the impact of these choices for the maximum radius we additionally compare to a resampling run with a maximum cell size of 100 GeV for sample W5.
For better parallelisation and general performance we pre-partition each input sample into several regions according to one of the upper levels of a vantage-point tree, as explained in Sect. 2.2. Here, we use the seventh level, corresponding to 128 regions.
To interpret our results we use standard Rivet [57] analyses. We verify that the event count and total cross section of each sample is preserved using the MC_XS analysis. Furthermore, we employ this analysis to assess the degree to which negative weights are eliminated. For the sample W5 we additionally use the MC_WINC and MC_WJETS analysis, and their counterparts MC_ZINC and MC_ZJETS for the remaining samples involving a Z boson. We investigate the impact of additional cuts applied after cell resampling using the ATLAS analysis ATLAS_2017_I1514251 [58] for inclusive Z boson production.
3.1 Comparison of predictions
We first assert that predictions remain equivalent by comparing a number of distributions before and after cell resampling. Figure 1 shows a variety of distributions for sample W5. In Fig. 2 we show selected distributions for the Z1, Z2, and Z3 samples. In all cases, we find that the differences between original and resampled predictions are comparable to or even smaller than the statistical bin-to-bin fluctuations in the original. A more indirect way to estimate the bias introduced by cell resampling is to consider the characteristic cell radii and the spread of measured observables within the cells. This is discussed in Appendix 1.
Comparison of distributions before and after cell resampling for sample W5 in Table 1. The blue lines indicate cell resampling with a maximum cell radius of 10 GeV, the green lines result from a radius limit of 100 GeV. Distributions are normalised according to the total cross section for sample W5
Comparison of distributions before and after cell resampling for samples Z1, Z2, and Z3 in Table 1. a–d show jet transverse momentum and rapidity distributions taken from the ATLAS_2017_I1514251 Rivet analysis. e is a jet rapidity distribution taken from MC_ZJETS
3.2 Improvement in sample quality
In order to assess the improvement achieved through cell resampling, we first consider the reduction in the negative weight contribution. To this end, we determine how much larger the original and the resampled event samples have to be to reach the same statistical power as an event sample without negative weights. In other words, we compute \(N(r_-)\) as defined in Eq. (1), where the fractional negative-weight contribution \(r_- = \sigma _-/(\sigma _+ + \sigma _-)\) is obtained from the contribution \(\sigma _+\) of positive-weight events to the total cross section \(\sigma \) and the absolute value of the negative-weight cross section contribution \(\sigma _- = \sigma _+ - \sigma \).
As demonstrated in Fig. 3, cell resampling leads to a drastic improvement by roughly two to three orders of magnitude. Increasing the maximum cell radius leads to an even stronger reduction, at the cost of increased computing time and potentially larger systematic errors introduced by the procedure. To assess the impact of pre-partioning the event samples, we alternatively resample Z1 without prior partitioning. This leads to a slight reduction of \(N(r_-)/N(0)\) from 18.4 with pre-partitioning to 17.1 without pre-partitioning.
Required number of events relative to an ideal event sample without negative weights before and after resampling. Event samples are labeled as listed in Table 1
Cell resampling not only reduces the amount of negative weights, but as a by-product also results in a narrower weight distribution, enhancing the unweighting efficiency. Indeed, after standard unweighting we retain only 320 out of the \(8.21\times 10^8\) events in the Z1 sample. If we apply resampling beforehand, unweighting yields 11,574 events. The resulting unweighted sample is not only larger, but also contains a lower fraction of negative-weight events. We show the gain in statistical power by selecting a subset of 320 randomly chosen events and compare to the unweighted sample based on the original events. Selected distributions are shown in Fig. 4.
Comparison between unweighted samples before and after cell resampling. Lines labeled “original” show the reference prediction from the original weighted event sample Z1. After standard unweighting, the lines with the label “unweighted” are obtained. Applying cell resampling followed by standard unweighting to the sample Z1 yields a sample represented by the “resampled + unweighted” lines. Out of this sample, we randomly select a subset matching the size of the original “unweighted” sample. This leads to the “resampled + unweighted (small sample)” lines. Data points are taken from [58]
3.3 Runtime requirements
Cell resampling with the improvements presented in Sect. 2 and a maximum cell size of 10 GeV typically takes a few hours of wall-clock time for samples with about a billion events. As an example, let us consider the resampling for the W5 sample listed in Table 1. The combined size of the original compressed event files is approximately 150 GB. The resampling program requires about 450 GB of memory and the total runtime is about 9 h on a machine with 24 Intel Xeon E5-2643 processors. The memory usage could of course be reduced significantly at the cost of computing time by not keeping all events in memory, but we have not explored this option in our current implementation. Reading in the events and converting them to a space-efficient internal format that only retains the information needed for resampling takes about 2 h. This is followed by approximately 30 min spent for the pre-partitioning of the event sample and less than 3 h for resampling itself, including the construction of the search trees, cf. Sect. 2.2. Since the event information in the internal format is incomplete, we finally read in the original events again and write them to disk after updating their weights. This final step takes roughly 4 h. While input and output do not benefit from parallelisation, the pre-partitioning and the resampling are performed in parallel and the total CPU time spent is 55 h.
One important optimisation discussed in Sect. 2.2 is trimming the nearest-neighbour search according to the maximum cell radius. In fact, when increasing the allowed radius from 10 to 100 GeV the wall clock time needed for resampling rises to several weeks, with a corresponding increase in total CPU time. Extrapolating from smaller sample sizes, the expected total required CPU time without any of the new optimisations would be of the order of 1600 years for the much simpler process of W boson production with two jets considered in [33].
4 Conclusions
We have demonstrated that the fraction of negative event weights in existing large high-multiplicity samples can be reduced by more than an order of magnitude, whilst preserving predictions for observables within statistical uncertainties. Concretely, we have employed the cell resampling method proposed in [33] with NLO event samples for Z boson production with up to three jets and W boson production with five jets produced with Sherpa and BlackHat.
For the first time, cell resampling has been applied to samples with up to several billions of events. This was made possible by algorithmic improvements leading to a speed-up by several orders of magnitude. Our updated implementation can be retrieved from https://cres.hepforge.org/.
The advances in the development of the cell resampling method presented in this work pave the way for future applications to processes with high-multiplicities, in particular including parton showered predictions. It will be necessary to quantify the uncertainty introduced by the weight smearing. Variations in the maximum cell size parameter and different prescriptions for weight redistribution within a cell can serve as handles to assess this uncertainty. Another promising avenue for further exploration is the analysis of the information on weight distribution within phase space collected during cell resampling. Regions with insufficient Monte Carlo statistics could be identified by their accumulated negative weight, thereby guiding the event generation. We leave the investigation of these questions to future work.
References
HSF Physics Event Generator WG collaboration, S. Amoroso et al., Challenges in Monte Carlo event generator software for high-luminosity LHC. Comput. Softw. Big Sci. 5, 12 (2021). https://doi.org/10.1007/s41781-021-00055-1. arXiv:2004.13687
S. Frixione, Z. Kunszt, A. Signer, Three jet cross-sections to next-to-leading order. Nucl. Phys. B 467, 399–442 (1996). https://doi.org/10.1016/0550-3213(96)00110-1. arXiv:hep-ph/9512328
S. Catani, M.H. Seymour, A general algorithm for calculating jet cross-sections in NLO QCD. Nucl. Phys. B 485, 291–419 (1997). https://doi.org/10.1016/S0550-3213(96)00589-5. arXiv:hep-ph/9605323
S. Catani, S. Dittmaier, M.H. Seymour, Z. Trocsanyi, The dipole formalism for next-to-leading order QCD calculations with massive partons. Nucl. Phys. B 627, 189–265 (2002). https://doi.org/10.1016/S0550-3213(02)00098-6. arXiv:hep-ph/0201036
Z. Nagy, D.E. Soper, General subtraction method for numerical calculation of one loop QCD matrix elements. JHEP 09, 055 (2003). https://doi.org/10.1088/1126-6708/2003/09/055. arXiv:hep-ph/0308127
A. GehrmannDeRidder, T. Gehrmann, E.W.N. Glover, Antenna subtraction at NNLO. JHEP 09, 056 (2005). https://doi.org/10.1088/1126-6708/2005/09/056. arXiv:hep-ph/0505111
M. Czakon, A novel subtraction scheme for double-real radiation at NNLO. Phys. Lett. B 693, 259–268 (2010). https://doi.org/10.1016/j.physletb.2010.08.036. arXiv:1005.0274
G. Somogyi, Z. Trocsanyi, V. Del Duca, Matching of singly- and doubly-unresolved limits of tree-level QCD squared matrix elements. JHEP 06, 024 (2005). https://doi.org/10.1088/1126-6708/2005/06/024. arXiv:hep-ph/0502226
G. Somogyi, Z. Trocsanyi, V. Del Duca, A subtraction scheme for computing QCD jet cross sections at NNLO: regularization of doubly-real emissions. JHEP 01, 070 (2007). https://doi.org/10.1088/1126-6708/2007/01/070. arXiv:hep-ph/0609042
J. Gaunt, M. Stahlhofen, F.J. Tackmann, J.R. Walsh, N-jettiness subtractions for NNLO QCD calculations. JHEP 09, 058 (2015). https://doi.org/10.1007/JHEP09(2015)058. arXiv:1505.04794
M. Cacciari, F.A. Dreyer, A. Karlberg, G.P. Salam, G. Zanderighi, Fully differential vector-boson-fusion Higgs production at next-to-next-to-leading order. Phys. Rev. Lett. 115, 082002 (2015). https://doi.org/10.1103/PhysRevLett.115.082002. arXiv:1506.02660
R. Bonciani, S. Catani, M. Grazzini, H. Sargsyan, A. Torre, The \(q_T\) subtraction method for top quark production at hadron colliders. Eur. Phys. J. C 75, 581 (2015). https://doi.org/10.1140/epjc/s10052-015-3793-y. arXiv:1508.03585
L. Magnea, E. Maina, G. Pelliccioli, C. Signorile-Signorile, P. Torrielli, S. Uccirati, Local analytic sector subtraction at NNLO. JHEP 12, 107 (2018). https://doi.org/10.1007/JHEP12(2018)107. arXiv:1806.09570
S. Frixione, B.R. Webber, Matching NLO QCD computations and parton shower simulations. JHEP 06, 029 (2002). https://doi.org/10.1088/1126-6708/2002/06/029. arXiv:hep-ph/0204244
S. Frixione, P. Nason, C. Oleari, Matching NLO QCD computations with Parton Shower simulations: the POWHEG method. JHEP 11, 070 (2007). https://doi.org/10.1088/1126-6708/2007/11/070. arXiv:0709.2092
K. Hamilton, P. Nason, G. Zanderighi, MINLO: multi-scale improved NLO. JHEP 10, 155 (2012). https://doi.org/10.1007/JHEP10(2012)155. arXiv:1206.3572
S. Höche, Y. Li, S. Prestel, Drell–Yan lepton pair production at NNLO QCD with parton showers. Phys. Rev. D 91, 074015 (2015). https://doi.org/10.1103/PhysRevD.91.074015. arXiv:1405.3607
S. Jadach, W. Płaczek, S. Sapeta, A. Siódmok, M. Skrzypek, Matching NLO QCD with parton shower in Monte Carlo scheme-the KrkNLO method. JHEP 10, 052 (2015). https://doi.org/10.1007/JHEP10(2015)052. arXiv:1503.06849
P.F. Monni, P. Nason, E. Re, M. Wiesemann, G. Zanderighi, MiNNLO\(_{PS}\): a new method to match NNLO QCD to parton showers. JHEP 05, 143 (2020). https://doi.org/10.1007/JHEP05(2020)143. arXiv:1908.06987
S. Prestel, Matching N3LO QCD calculations to parton showers. JHEP 11, 041 (2021). https://doi.org/10.1007/JHEP11(2021)041. arXiv:2106.03206
S. Catani, F. Krauss, R. Kuhn, B.R. Webber, QCD matrix elements + parton showers. JHEP 11, 063 (2001). https://doi.org/10.1088/1126-6708/2001/11/063. arXiv:hep-ph/0109231
L. Lonnblad, Correcting the color dipole cascade model with fixed order matrix elements. JHEP 05, 046 (2002). https://doi.org/10.1088/1126-6708/2002/05/046. arXiv:hep-ph/0112284
R. Frederix, S. Frixione, Merging meets matching in MC@NLO. JHEP 12, 061 (2012). https://doi.org/10.1007/JHEP12(2012)061. arXiv:1209.6215
L. Lönnblad, S. Prestel, Merging multi-leg NLO matrix elements with parton showers. JHEP 03, 166 (2013). https://doi.org/10.1007/JHEP03(2013)166. arXiv:1211.7278
R. Frederix, S. Frixione, S. Prestel, P. Torrielli, On the reduction of negative weights in MC@NLO-type matching procedures. arXiv:2002.12716
C. Gao, J. Isaacson, C. Krause, i-flow: high-dimensional integration and sampling with normalizing flows. Mach. Learn. Sci. Technol. 1, 045023 (2020). https://doi.org/10.1088/2632-2153/abab62. arXiv:2001.05486
E. Bothmann, T. Janßen, M. Knobbe, T. Schmale, S. Schumann, Exploring phase space with neural importance sampling. SciPost Phys. 8, 069 (2020). https://doi.org/10.21468/SciPostPhys.8.4.069. arXiv:2001.05478
C. Gao, S. Höche, J. Isaacson, C. Krause, H. Schulz, Event generation with normalizing flows. Phys. Rev. D 101, 076002 (2020). https://doi.org/10.1103/PhysRevD.101.076002. arXiv:2001.10028
K. Danziger, S. Höch, F. Siegert, Reducing negative weights in Monte Carlo event generation with Sherpa. arXiv:2110.15211
J.R. Andersen, C. Gütschow, A. Maier, S. Prestel, A positive resampler for Monte Carlo events with negative weights. Eur. Phys. J. C 80, 1007 (2020). https://doi.org/10.1140/epjc/s10052-020-08548-w. arXiv:2005.09375
B. Nachman, J. Thaler, Neural resampler for Monte Carlo reweighting with preserved uncertainties. Phys. Rev. D 102, 076004 (2020). https://doi.org/10.1103/PhysRevD.102.076004. arXiv:2007.11586
B. Stienen, R. Verheyen, Phase space sampling and inference from weighted events with autoregressive flows. arXiv:2011.13445
J.R. Andersen, A. Maier, Unbiased elimination of negative weights in Monte Carlo samples. Eur. Phys. J. C 82, 433 (2022). https://doi.org/10.1140/epjc/s10052-022-10372-3. arXiv:2109.07851
P. Indyk, R. Motwani, Approximate nearest neighbors: Towards removing the curse of dimensionality, in Proceedings of the 30th ACM Symposium on Theory of Computing (1998), p. 604–613
J. Leskovec, A. Rajaraman, J. Ullman, Mining of Massive Datasets (Cambridge University Press, Cambridge, 2020)
J.K. Uhlmann, Satisfying general proximity/similarity queries with metric trees. Inf. Process. Lett. 40, 175–179 (1991). https://doi.org/10.1016/0020-0190(91)90074-R
P.N. Yianilos, Data structures and algorithms for nearest neighbor search in general metric spaces, in Proceedings of the Fourth Annual ACM-SIAM Symposium on Discrete Algorithms, SODA ’93, (USA), Society for Industrial and Applied Mathematics, (1993), p. 311–321
C.G.J. Jacobi, De investigando ordine systematis aequationum differentialum vulgarium cujuscunque, in C.G.J. Jacobi’s gesammelte Werke, fünfter Band ed. by K. Weierstrass (Verlag Georg Reimer, 1890), p. 193–216
C.G.J. Jacobi, De aequationum differentialum systemate non normali ad formam normalem revocando, in C.G.J. Jacobi’s gesammelte Werke, fünfter Band ed. by K. Weierstrass (Verlag Georg Reimer, 1890), p. 485–513
H.W. Kuhn, The Hungarian method for the assignment problem. Nav. Res. Logist. Q. 2, 83–97 (1955)
H.W. Kuhn, Variants of the Hungarian method for assignment problems. Nav. Res. Logist. Q. 3, 253–258 (1956)
J. Munkres, Algorithms for the assignment and transportation problems. J. Soc. Ind. Appl. Math. 5, 32–38 (1957)
J. Edmonds, R.M. Karp, Theoretical improvements in algorithmic efficiency for network flow problems. J. ACM 19, 248–264 (1972). https://doi.org/10.1145/321694.321699
N. Tomizawa, On some techniques useful for solution of transportation network problems. Networks 1, 173–194 (1971). https://doi.org/10.1002/net.3230010206
S. Tardieu, “pathfinding.” https://crates.io/crates/pathfinding, (2022)
L. Ramshaw, R.E. Tarjan, A weight-scaling algorithm for min-cost imperfect matchings in bipartite graphs 581–590 (2012). https://doi.org/10.1109/FOCS.2012.9
D.P. Bertsekas, Auction algorithms for network flow problems: a tutorial introduction. Comput. Optim. Appl. 1, 7–66 (1992)
A. Goldberg, R. Kennedy, An efficient cost scaling algorithm for the assignment problem. Math. Program. 71, 153–177 (1995). https://doi.org/10.1007/BF01585996
C. Alfaro, S.P. Perez, C. Valencia, M.V. Magañ, The assignment problem revisited. Optim. Lett. (2022). https://doi.org/10.1007/s11590-021-01791-4
R. Brun, F. Rademakers, ROOT: an object oriented data analysis framework. Nucl. Instrum. Methods A 389, 81–86 (1997). https://doi.org/10.1016/S0168-9002(97)00048-X
Z. Bern, L.J. Dixon, F. Febres Cordero, S. Höche, H. Ita, D.A. Kosower et al., Ntuples for NLO events at hadron colliders. Comput. Phys. Commun. 185, 1443–1460 (2014). https://doi.org/10.1016/j.cpc.2014.01.011. arXiv:1310.7439
F.R. Anger, F. FebresCordero, S. Höche, D. Maître, Weak vector boson production with many jets at the LHC \(\sqrt{s}= 13\) TeV. Phys. Rev. D 97, 096010 (2018). https://doi.org/10.1103/PhysRevD.97.096010. arXiv:1712.08621
C.F. Berger, Z. Bern, L.J. Dixon, F. FebresCordero, D. Forde, H. Ita et al., An automated implementation of on-shell methods for one-loop amplitudes. Phys. Rev. D 78, 036003 (2008). https://doi.org/10.1103/PhysRevD.78.036003. arXiv:0803.4180
T. Gleisberg, S. Hoeche, F. Krauss, M. Schonherr, S. Schumann, F. Siegert et al., Event generation with SHERPA 1.1. JHEP 02, 007 (2009). https://doi.org/10.1088/1126-6708/2009/02/007. arXiv:0811.4622
M. Cacciari, G.P. Salam, G. Soyez, The anti-\(k_t\) jet clustering algorithm. JHEP 04, 063 (2008). https://doi.org/10.1088/1126-6708/2008/04/063. arXiv:0802.1189
Z. Bern, L.J. Dixon, F. FebresCordero, S. Höche, H. Ita, D.A. Kosower et al., Next-to-leading order \(W + 5\)-jet production at the LHC. Phys. Rev. D 88, 014025 (2013). https://doi.org/10.1103/PhysRevD.88.014025. arXiv:1304.1253
C. Bierlich et al., Robust independent validation of experiment and theory: Rivet version. SciPost Phys. 8, 026 (2020). https://doi.org/10.21468/SciPostPhys.8.2.026. arXiv:1912.05451
ATLAS collaboration, M. Aaboud et al., Measurements of the production cross section of a \(Z\) boson in association with jets in pp collisions at \(\sqrt{s} = 13\) TeV with the ATLAS detector. Eur. Phys. J. C 77, 361 (2017). https://doi.org/10.1140/epjc/s10052-017-4900-z. arXiv:1702.05725
Acknowledgements
AM thanks Zahari Kassabov for encouragement to reconsider the use of nearest neighbour search trees. The work of JRA and DM is supported by the STFC under Grant ST/P001246/1.
Author information
Authors and Affiliations
Corresponding author
Appendices
Appendix A: Improved search based on locality-sensitive hashing
Locality-sensitive hashing (LSH) is a method for approximate nearest-neighbour search where points are inserted into a number of hash tables, with hashes that are calculated from the coordinates in such a way that nearby points end up inside the same hash table buckets with high probability. To search for a point’s nearest neighbour, one only checks points that share at least a given number of hash table buckets. An equivalent formulation in the language of particle physics is to consider a number of one-dimensional histograms, where the observables are chosen such that similar events end up in the same histogram bins. To find events that are nearby in phase space, one only checks those events that share a large number of histogram bins. A first LSH-based search algorithm for cell resampling was proposed in [33]. In the following, we discuss an improved version, where the histogram observables have a closer relation to the exact distance measure.
The first step in defining the locality-sensitive observables is the same as in the exact distance calculation: we cluster the outgoing particles in each event into infrared-safe physics objects and group them according to their types. As usual, we add objects with vanishing momentum to ensure that all events have the same number of objects for each type. For each object type t, we then choose a random axis \(a_t\) in three-dimensional Euclidean space. We choose a final axis A in a Euclidean space whose dimension is equal to the total number of infrared-safe physics objects in an event.
For a given event, we then calculate the observable as follows. For each object type t, we project the spatial momentum of each object onto the previously chosen axis \(a_t\) and sort the resulting coordinates. We concatenate all coordinates obtained in this way into a single vector. Finally, we obtain the observable by projecting this vector onto the axis A.
We find that the LSH-based search based on the present observables performs significantly better than the original version [33]. However, it still suffers from the same problem. For constant (or at most logarithmically growing) numbers of histograms and bin sizes we observe that the typical distance between an event and the approximate nearest neighbour fails to decrease with a growing sample size. Hence, we mainly focus on the exact tree-based search presented in Sect. 2.2.
Appendix B: Cell sizes
Larger cell sizes naturally lead to stronger smearing effects. Ideally, all cells should be small compared to the experimental resolution, which is limited both by the detector and by statistics.
In the upper pane of Fig. 5 we show the distribution of cell radii obtained for the Z + 1 jet sample Z1, cf. Table 1. We have omitted cells where aside from the seed no further event is found within the maximum cell radius of 10 GeV. The shape of the distribution is similar to the one found for W + 2 jets [33].
The cell diameter imposes an upper limit on the spread in any single direction. However, especially in a higher-dimensional phase space, the smearing range in one-dimensional distributions will be typically much smaller, as pointed out in our earlier work [33]. To illustrate this point, we compute the difference \(\Delta p_\perp (\text {jet})\) between the transverse momenta of the softest jet and the hardest jet among all events within a cell. The distribution is shown in the lower pane of Fig. 5. We observe a steep decline with a median of 0.4 GeV. There is a notable drop where \(\Delta p_\perp (\text {jet})\) reaches the maximum cell radius of 10 GeV. While the theoretical upper limit is given by the maximum cell diameter of 20 GeV, we find that the largest transverse momentum spread in the considered sample is approximately 15 GeV.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
Funded by SCOAP3. SCOAP3 supports the goals of the International Year of Basic Sciences for Sustainable Development.
About this article
Cite this article
Andersen, J.R., Maier, A. & Maître, D. Efficient negative-weight elimination in large high-multiplicity Monte Carlo event samples. Eur. Phys. J. C 83, 835 (2023). https://doi.org/10.1140/epjc/s10052-023-11905-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1140/epjc/s10052-023-11905-0