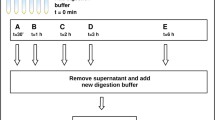
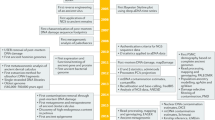
Abstract
The emergence and development of massive parallel sequencing (new generation sequencing) methods have opened up new prospects in the study of ancient organisms, including extinct ones. Numerous skeletal remains from archaeological and museum collections are often the only source of information on ancient species and populations. In this review, we discuss the features of human bone tissue and the advantages and disadvantages of bone material as a source of DNA for genomic analysis of ancient people. Here we present new methodological approaches to DNA extraction from ancient human skeletal remains and its preparation for large-scale parallel sequencing are presented, as well as prospects and directions for further research in a new interdisciplinary field, paleogenomics.




Similar content being viewed by others
REFERENCES
Hagelberg, E., Sykes, B., and Hedges, R., Ancient bone DNA amplified, Nature, 1989, vol. 342, no. 6249, p. 485. https://doi.org/10.1038/342485a0
Lindahl, T., Instability and decay of the primary structure of DNA, Nature, 1993, vol. 362, no. 6422, pp. 709—715. https://doi.org/10.1038/362709a0
Pääbo, S. and Wilson, A.C., Miocene DNA sequences—a dream come true?, Curr. Biol., 1991, vol. 1, no. 1, pp. 45—46. https://doi.org/10.1016/0960-9822(91)90125-G
Allentoft, M.E., Collins, M., Harker, D., et al., The half-life of DNA in bone: measuring decay kinetics in 158 dated fossils, Proc. R. Soc. London, Ser. B, 2012, vol. 279, no. 1748, pp. 4724—4733. https://doi.org/10.1098/rspb.2012.1745
van der Valk, T., Pečnerová, P., Díez-del-Molino, D., et al., Million-year-old DNA sheds light on the genomic history of mammoths, Nature, 2021, vol. 591, no. 7849, pp. 265—269. https://doi.org/10.1038/s41586-021-03224-9
Van der Plicht, J., Bronk Ramsey, C., Heaton, T.J., et al., Recent developments in calibration for archaeological and environmental samples, Radiocarbon, 2020, vol. 62, no. 4, pp. 1095—1117. https://doi.org/10.1017/RDC.2020.22
Horai, S., Hayasaka, K., Murayama, K., et al., DNA amplification from ancient human skeletal remains and their sequence analysis, Proc. Jpn. Acad., Ser. B Phys. Biol. Sci., 1989, vol. 65, no. 10, pp. 229—233. https://doi.org/10.2183/pjab.65.229
Briggs, A.W., Stenzel, U., Johnson, P.L.F., et al., Patterns of damage in genomic DNA sequences from a Neandertal, Proc. Natl. Acad. Sci. U.S.A., 2007, vol. 104, no. 37, pp. 14616—14621. https://doi.org/10.1073/pnas.0704665104
Frederico, L.A., Shaw, B.R., and Kunkel, T.A., A sensitive genetic assay for the detection of cytosine deamination: determination of rate constants and the activation energy, Biochemistry, 1990, vol. 29, no. 10, pp. 2532—2537. https://doi.org/10.1021/bi00462a015
Dabney, J., Meyer, M., and Pääbo, S., Ancient DNA damage, Cold Spring Harb. Perspect. Biol., 2013, vol. 5, no. 7. https://doi.org/10.1101/cshperspect.a012567
Heyn, P., Stenzel, U., Briggs, A.W., et al., Road blocks on paleogenomes-polymerase extension profiling reveals the frequency of blocking lesions in ancient DNA, Nucleic Acids Res., 2010, vol. 38, no. 16. https://doi.org/10.1093/nar/gkq572
Trueman, C.N. and Martill, D.M., The long-term survival of bone: the role of bioerosion, Archaeometry, 2002, vol. 44, no. 3, pp. 371—382. https://doi.org/10.1111/1475-4754.t01-1-00070
Bell, L.S., Skinner, M.F., and Jones, S.J., The speed of post mortem change to the human skeleton and its taphonomic significance, Forensic Sci. Int., 1996, vol. 82, no. 2, pp. 129—140. https://doi.org/10.1016/0379-0738(96)01984-6
Turner-Walker, G., Nielsen-Marsh, C.M., Syversen, U., et al., Sub-micron spongiform porosity is the major ultra-structural alteration occurring in archaeological bone, Int. J. Osteoarchaeol., 2002, vol. 12, no. 6, pp. 407—414. https://doi.org/10.1002/oa.642
Romanowski, G., Lorenz, M.G., and Wackernagel, W., Adsorption of plasmid DNA to mineral surfaces and protection against DNase I, Appl. Environ. Microbiol., 1991, vol. 57, no. 4, pp. 1057—1061. https://doi.org/10.1128/aem.57.4.1057-1061.1991
Demanèche, S., Jocteur-Monrozier, L., Quiquampoix, H., and Simonet, P., Evaluation of biological and physical protection against nuclease degradation of clay-bound plasmid DNA, Appl. Environ. Microbiol., 2001, vol. 67, no. 1, pp. 293—299. https://doi.org/10.1128/AEM.67.1.293-299.2001
Brundin, M., Figdor, D., Sundqvist, G., and Sjögren, U., DNA binding to hydroxyapatite: a potential mechanism for preservation of microbial DNA, J. Endod., 2013, vol. 39, no. 2, pp. 211—216. https://doi.org/10.1016/j.joen.2012.09.013
Sosa, C., Vispe, E., Núñez, C., et al., Association between ancient bone preservation and DNA yield: a multidisciplinary approach, Am. J. Phys. Anthropol., 2013, vol. 151, no. 1, pp. 102—109. https://doi.org/10.1002/ajpa.22262
Rogaev, E.I., Moliaka, Y.K., Malyarchuk, B.A., et al., Complete mitochondrial genome and phylogeny of pleistocene mammoth Mammuthus primigenius, PLoS Biol., 2006, vol. 4, no. 3, pp. 0403—0410. https://doi.org/10.1371/journal.pbio.0040073
Götherström, A., Collins, M.J., Angerbjörn, A., and Lidén, K., Bone preservation and DNA amplification, Archaeometry, 2002, vol. 44, no. 3, pp. 395—404. https://doi.org/10.1111/1475-4754.00072
Schwarz, C., Debruyne, R., Kuch, M., et al., New insights from old bones: DNA preservation and degradation in permafrost preserved mammoth remains, Nucleic Acids Res., 2009, vol. 37, no. 10, pp. 3215—3229. https://doi.org/10.1093/nar/gkp159
Poinar, H.N. and Stankiewicz, B.A., Protein preservation and DNA retrieval from ancient tissues, Proc. Natl. Acad. Sci. U.S.A., 1999, vol. 96, no. 15, pp. 8426—8431. https://doi.org/10.1073/pnas.96.15.8426
Colson, I.B., Bailey, J.F., Vercauteren, M., et al., The preservation of ancient DNA and bone diagenesis, Anc. Biomol., 1997, vol. 1, no. 2, pp. 109–117.
Smith, C.I., Chamberlain, A.T., Riley, M.S., et al., The thermal history of human fossils and the likelihood of successful DNA amplification, J. Hum. Evol., 2003, vol. 45, no. 3, pp. 203—217. https://doi.org/10.1016/S0047-2484(03)00106-4
Pääbo, S., Molecular cloning of ancient Egyptian mummy DNA, Nature, 1985, vol. 314, no. 6012, pp. 644—645. https://doi.org/10.1038/314644a0
Schuenemann, V.J., Peltzer, A., Welte, B., et al., Ancient Egyptian mummy genomes suggest an increase of Sub-Saharan African ancestry in post-Roman periods, Nat. Commun., 2017, vol. 8. https://doi.org/10.1038/ncomms15694
Gad, Y.Z., Abu-Mandil Hassan, N., Mousa, D.M., et al., Insights from ancient DNA analysis of Egyptian human mummies: clues to disease and kinship, Hum. Mol. Genet., 2021, vol. 30, no. 2, pp. R24—R28. https://doi.org/10.1093/hmg/ddaa223
Rogaev, E.I., Grigorenko, A.P., Moliaka, Y.K., et al., Genomic identification in the historical case of the Nicholas II royal family, Proc. Natl. Acad. Sci. U.S.A., 2009, vol. 106, no. 13, pp. 5258—5263. https://doi.org/10.1073/pnas.0811190106
Grela, M., Jakubczak, A., Kowalczyk, M., et al., Effectiveness of various methods of DNA isolation from bones and teeth of animals exposed to high temperature, J. Forensic Leg. Med., 2021, vol. 78. https://doi.org/10.1016/j.jflm.2021.102131
Emery, M.V., Bolhofner, K., Winingear, S., et al., Reconstructing full and partial STR profiles from severely burned human remains using comparative ancient and forensic DNA extraction techniques, Forensic Sci. Int. Genet., 2020, vol. 46. https://doi.org/10.1016/j.fsigen.2020.102272
Ottoni, C., Koon, H.E.C., Collins, M.J., et al., Preservation of ancient DNA in thermally damaged archaeological bone, Naturwissenschaften, 2009, vol. 96, no. 2, pp. 267—278. https://doi.org/10.1007/s00114-008-0478-5
Latham, K.E. and Miller, J.J., DNA recovery and analysis from skeletal material in modern forensic contexts, Forensic Sci. Res., 2019, vol. 4, no. 1, pp. 51—59. https://doi.org/10.1080/20961790.2018.1515594
Prado, M., Franco, C.M., Fente, C.A., et al., Comparison of extraction methods for the recovery, amplification and species-specific analysis of DNA from bone and bone meals, Electrophoresis, 2002, vol. 23, nos. 7—8, pp. 1005—1012. https://doi.org/10.1002/1522-2683(200204)23:7/8<1005::AID-ELPS1005>3.0.CO;2-1
Pruvost, M., Schwarz, R., Correia, V.B., et al., Freshly excavated fossil bones are best for amplification of ancient DNA, Proc. Natl. Acad. Sci. U.S.A., 2007, vol. 104, no. 3, pp. 739—744. https://doi.org/10.1073/pnas.0610257104
Bowes, J.H. and Murray, M.M., The chemical composition of teeth, Biochem. J., 1935, vol. 29, no. 12, pp. 2721—2727. https://doi.org/10.1042/bj0292721
Beniash, E., Stifler, C.A., Sun, C.Y., et al., The hidden structure of human enamel, Nat. Commun., 2019, vol. 10, no. 1. https://doi.org/10.1038/s41467-019-12185-7
Malaver, P.C. and Yunis, J.J., Different dental tissues as source of DNA for human identification in forensic cases, Croat. Med. J., 2003, vol. 44, no. 3, pp. 306—309.
Trivedi, R., Chattopadhyay, P., and Kashyap, V.K., A new improved method for extraction of DNA from teeth for the analysis of hypervariable loci, Am. J. Forensic Med. Pathol., 2002, vol. 23, no. 2, pp. 191—196. https://doi.org/10.1097/00000433-200206000-00016
Pötsch, L., Meyer, U., Rothschild, S., et al., Application of DNA techniques for identification using human dental pulp as a source of DNA, Int. J. Legal Med., 1992, vol. 105, no. 3, pp. 139—143. https://doi.org/10.1007/BF01625165
Schuenemann, V.J., Singh, P., Mendum, T.A., et al., Genome-wide comparison of medieval and modern Mycobacterium leprae, Science, 2013, vol. 341, no. 6142, pp. 179—183. https://doi.org/10.1126/SCIENCE.1238286
Higgins, D. and Austin, J.J., Teeth as a source of DNA for forensic identification of human remains: a review, Sci. Justice, 2013, vol. 53, no. 4, pp. 433—441. https://doi.org/10.1016/j.scijus.2013.06.001
Dabney, J. and Meyer, M., Extraction of highly degraded DNA from ancient bones and teeth, Methods Mol. Biol., 2019, vol. 1963, pp. 25—29. https://doi.org/10.1007/978-1-4939-9176-1_4
Adler, C.J., Haak, W., Donlon, D., and Cooper, A., Survival and recovery of DNA from ancient teeth and bones, J. Archaeol. Sci., 2011, vol. 38, no. 5, pp. 956—964. https://doi.org/10.1016/j.jas.2010.11.010
Campos, P.F., Craig, O.E., Turner-Walker, G., et al., DNA in ancient bone—where is it located and how should we extract it?, Ann. Anat., 2012, vol. 194, no. 1, pp. 7—16. https://doi.org/10.1016/j.aanat.2011.07.003
Andreeva, T.V., Malyarchuk, A.B., Grigorenko, A.P., et al., Archaeogenetic analysis of an individual from a burial site at the ancient Yaroslavl Kremlin, Kratk. Soobshch. Inst. Arkheol., 2021, vol. 265, pp. 209—308.
Sawyer, S., Renaud, G., Viola, B., et al., Nuclear and mitochondrial DNA sequences from two Denisovan individuals, Proc. Natl. Acad. Sci. U.S.A., 2015, vol. 112, no. 51, pp. 15696—15700. https://doi.org/10.1073/pnas.1519905112
Adler, C.J., Haak, W., Donlon, D., and Cooper, A., Survival and recovery of DNA from ancient teeth and bones, J. Archaeol. Sci., 2011, vol. 38, no. 5, pp. 956—964. https://doi.org/10.1016/j.jas.2010.11.010
Damgaard, P.B., Margaryan, A., Schroeder, H., et al., Improving access to endogenous DNA in ancient bones and teeth, Sci. Rep., 2015, vol. 5, no. 1, p. 11184. https://doi.org/10.1038/srep11184
Freeman, E., Periodontium, in Oral Histology: Development, Structure, and Function, Ten Cate, A.R., Ed., St. Louis: Mosby, 1994, pp. 276—312.
Lam, Y.M., Chen, X., and Pearson, O.M., Intertaxonomic variability in patterns of bone density and the differential representation of bovid, cervid, and equid elements in the archaeological record, Am. Antiq., 1999, vol. 64, no. 2, pp. 343—362. https://doi.org/10.2307/2694283
Hansen, H.B., Damgaard, P.B., Margaryan, A., et al., Comparing ancient DNA preservation in petrous bone and tooth cementum, PLoS One, 2017, vol. 12, no. 1. https://doi.org/10.1371/journal.pone.0170940
Pinhasi, R., Fernandes, D., Sirak, K., et al., Optimal ancient DNA yields from the inner ear part of the human petrous bone, PLoS One, 2015, vol. 10, no. 6. https://doi.org/10.1371/journal.pone.0129102
Pinhasi, R., Fernandes, D.M., Sirak, K., and Cheronet, O., Isolating the human cochlea to generate bone powder for ancient DNA analysis, Nat. Protoc., 2019, vol. 14, no. 4, pp. 1194—1205. https://doi.org/10.1038/s41596-019-0137-7
Sirak, K., Fernandes, D., Cheronet, O., et al., Human auditory ossicles as an alternative optimal source of ancient DNA, Genome Res., 2020, vol. 30, no. 3, pp. 427—436. https://doi.org/10.1101/gr.260141.119
Ponce de León, M.S., Koesbardiati, T., Weissmann, J.D., et al., Human bony labyrinth is an indicator of population history and dispersal from Africa, Proc. Natl. Acad. Sci. U.S.A., 2018, vol. 115, no. 16, pp. 4128—4133. https://doi.org/10.1073/pnas.1717873115
Parker, C., Rohrlach, A.B., Friederich, S., et al., A systematic investigation of human DNA preservation in medieval skeletons, Sci. Rep., 2020, vol. 10, no. 1. https://doi.org/10.1038/s41598-020-75163-w
Krings, M., Stone, A., Schmitz, R.W., et al., Neandertal DNA sequences and the origin of modern humans, Cell, 1997, vol. 90, no. 1, pp. 19—30. https://doi.org/10.1016/S0092-8674(00)80310-4
Lalueza, C., Pérez-Pérez, A., Prats, E., et al., Lack of founding Amerindian mitochondrial DNA lineages in extinct aborigines from Tierra del Fuego-Patagonia, Hum. Mol. Genet., 1997, vol. 6, no. 1, pp. 41—46. https://doi.org/10.1093/hmg/6.1.41
Prestuplenie veka: materialy sledstviya. Dokumental’no-arkhivnaya khronologiya sobytii, svyazannykh s gibel’yu Rossiiskogo imperatora Nikolaya II, ego sem’i i ikh priblizhennykh v 3 tomahk (Crime of the Century: Materials of the Investigation. Documentary and Archival Chronology of Events Related to the Perishing of the Russian Emperor Nicholas II, His Family and Their Entourage: in 3 Volumes), Moscow: Sledstvennyi Komitet RF, 2021, vol. 2.
Margaryan, A., Hansen, H.B., Rasmussen, S., et al., Ancient pathogen DNA in human teeth and petrous bones, Ecol. Evol., 2018, vol. 8, no. 6, pp. 3534—3542. https://doi.org/10.1002/ece3.3924
Hajdinjak, M., Fu, Q., Hübner, A., et al., Reconstructing the genetic history of late Neanderthals, Nature, 2018, vol. 555, no. 7698, pp. 652—656. https://doi.org/10.1038/nature26151
Grigorenko, A.P., Borinskaya, S.A., Yankovsky, N.K., and Rogaev, E.I., Achievements and peculiarities in studies of ancient DNA and DNA from complicated forensic specimens, Acta Nat., 2009, vol. 1, no. 3, pp. 58—69.
Pálsdóttir, A.H., Bläuer, A., Rannamäe, E., et al., Not a limitless resource: ethics and guidelines for destructive sampling of archaeofaunal remains, R. Soc. Open Sci., 2019, vol. 6, no. 10. https://doi.org/10.1098/rsos.191059
Kemp, B.M., Winters, M., Monroe, C., and Barta, J.L., How much DNA is lost? Measuring DNA loss of short-tandem-repeat length fragments targeted by the PowerPlex 16® system using the Qiagen MinElute Purification Kit, Hum. Biol., 2014, vol. 86, no. 4, pp. 313—329. https://doi.org/10.13110/humanbiology.86.4.0313
Montiel, R., Malgosa, A., and Francalacci, P., Authenticating ancient human mitochondrial DNA, Hum. Biol., 2001, vol. 73, no. 5, pp. 689—713. https://doi.org/10.1353/hub.2001.0069
Ginther, C., Issel-Tarver, L., and King, M.C., Identifying individuals by sequencing mitochondrial DNA from teeth, Nat. Genet., 1992, vol. 2, no. 2, pp. 135—138. https://doi.org/10.1038/ng1092-135
Champlot, S., Berthelot, C., Pruvost, M., et al., An efficient multistrategy DNA decontamination procedure of PCR reagents for hypersensitive PCR applications, PLoS One, 2010, vol. 5, no. 9. https://doi.org/10.1371/journal.pone.0013042
Morales Colón, E., Hernández, M., Candelario, M., et al., Evaluation of a freezer mill for bone pulverization prior to DNA extraction: an improved workflow for STR analysis, J. Forensic Sci., 2018, vol. 63, no. 2, pp. 530—535. https://doi.org/10.1111/1556-4029.13551
Stone, A.C., Milner, G.R., Paäbo, S., and Stoneking, M., Sex determination of ancient human skeletons using DNA, Am. J. Phys. Anthropol., 1996, vol. 99, no. 2, pp. 231—238. https://doi.org/10.1002/(SICI)1096-8644(199602)9-9:2<231::AID-AJPA1>3.0.CO;2-1
Rohland, N., Siedel, H., and Hofreiter, M., Nondestructive DNA extraction method for mitochondrial DNA analyses of museum specimens, Biotechniques, 2004, vol. 36, no. 5, pp. 814—821. https://doi.org/10.2144/04365st05
Bolnick, D.A., Bonine, H.M., Mata-Míguez, J., et al., Nondestructive sampling of human skeletal remains yields ancient nuclear and mitochondrial DNA, Am. J. Phys. Anthropol., 2012, vol. 147, no. 2, pp. 293—300. https://doi.org/10.1002/ajpa.21647
Harney, É., Cheronet, O., Fernandes, D.M., et al., A minimally destructive protocol for DNA extraction from ancient teeth, Genome Res., 2021, vol. 31, no. 3, pp. 472—483. https://doi.org/10.1101/GR.267534.120
Loreille, O.M., Diegoli, T.M., Irwin, J.A., et al., High efficiency DNA extraction from bone by total demineralization, Forensic Sci. Int. Genet., 2007, vol. 1, no. 2, pp. 191—195. https://doi.org/10.1016/j.fsigen.2007.02.006
Yang, D.Y., Eng, B., Waye, J.S., et al., Technical note: improved DNA extraction from ancient bones using silica-based spin columns, Am. J. Phys. Anthropol., 1998, vol. 105, no. 4, pp. 539—543. https://doi.org/10.1002/(SICI)1096-8644(199804)1-05:4<539::AID-AJPA10>3.0.CO;2-1
Höss, M. and Pääbo, S., DNA extraction from pleistocene bones by a silica-based purification method, Nucleic Acids Res., 1993, vol. 21, no. 16, pp. 3913—3914. https://doi.org/10.1093/nar/21.16.3913
Voong, C.P., Spencer, P.S., Navarrete, C.V., et al., HLA-DR genotyping and mitochondrial DNA analysis reveal the presence of family burials in a fourth century Romano-British Christian cemetery, Front. Genet., 2017, vol. 8, p. 182. https://doi.org/10.3389/fgene.2017.00182
Kalmár, T., Bachrati, C.Z., Marcsik, A., and Raskó, I., A simple and efficient method for PCR amplifiable DNA extraction from ancient bones, Nucleic Acids Res., 2000, vol. 28, no. 12, p. 67. https://doi.org/10.1093/nar/28.12.e67
Hofreiter, M., Rabeder, G., Jaenicke-Després, V., et al., Evidence for reproductive isolation between Cave Bear populations, Curr. Biol., 2004, vol. 14, no. 1, pp. 40—43. https://doi.org/10.1016/j.cub.2003.12.035
Leonard, J.A., Wayne, R.K., and Cooper, A., Population genetics of Ice Age brown bears, Proc. Natl. Acad. Sci. U.S.A., 2000, vol. 97, no. 4, pp. 1651—1654. https://doi.org/10.1073/pnas.040453097
Scheible, M., Loreille, O., Just, R., and Irwin, J., Short tandem repeat typing on the 454 platform: strategies and considerations for targeted sequencing of common forensic markers, Forensic Sci. Int. Genet., 2014, vol. 12, pp. 107—119. https://doi.org/10.1016/j.fsigen.2014.04.010
Boom, R., Sol, C.J.A., Salimans, M.M.M., et al., Rapid and simple method for purification of nucleic acids, J. Clin. Microbiol., 1990, vol. 28, no. 3, pp. 495–503. https://doi.org/10.1128/jcm.28.3.495-503.1990
Gamba, C., Hanghøj, K., Gaunitz, C., et al., Comparing the performance of three ancient DNA extraction methods for high-throughput sequencing, Mol. Ecol. Resour., 2016, vol. 16, no. 2, pp. 459—469. https://doi.org/10.1111/1755-0998.12470
Vigilant, L., Hofreiter, M., Siedel, H., and Boesch, C., Paternity and relatedness in wild chimpanzee communities, Proc. Natl. Acad. Sci. U.S.A., 2001, vol. 98, no. 23, pp. 12890—12895. https://doi.org/10.1073/pnas.231320498
Hänni, C., Brousseau, T., Laudet, V., and Stehelin, D., Isopropanol precipitation removes PCR inhibitors from ancient bone extracts, Nucleic Acids Res., 1995, vol. 23, no. 5, pp. 881—882. https://doi.org/10.1093/nar/23.5.881
Richards, M.B., Sykes, B.C., and Hedges, R.E.M., Authenticating DNA extracted from ancient skeletal remains, J. Archaeol. Sci., 1995, vol. 22, no. 2, pp. 291—299. https://doi.org/10.1006/jasc.1995.0031
Keyser-Tracqui, C., Crubézy, E., and Ludes, B., Nuclear and mitochondrial DNA analysis of a 2000-year-old necropolis in the Egyin Gol valley of Mongolia, Am. J. Hum. Genet., 2003, vol. 73, no. 2, pp. 247—260. https://doi.org/10.1086/377005
Lalueza-Fox, C., Calderón, F.L., Calafell, F., et al., MtDNA from extinct Tainos and the peopling of the Caribbean, Ann. Hum. Genet., 2001, vol. 65, no. 2, pp. 137—151. https://doi.org/10.1046/j.1469-1809.2001.6520137.x
Palmirotta, R., Verginelli, F., Di Tota, G., et al., Use of a multiplex polymerase chain reaction assay in the sex typing of DNA extracted from archaeological bone, Int. J. Osteoarchaeol., 1997, vol. 7, no. 6, pp. 605—609. https://doi.org/10.1002/(sici)1099-1212(199711/12)-7:6<605::aid-oa365>3.0.co;2-r
Korlević, P. and Meyer, M., Pretreatment: removing DNA contamination from ancient bones and teeth using sodium hypochlorite and phosphate, Methods Mol. Biol., 2019, vol. 1963, pp. 15—19. https://doi.org/10.1007/978-1-4939-9176-1_2
Korlević, P., Gerber, T., and Gansauge, M.-T. et al. Reducing microbial and human contamination in DNA extractions from ancient bones and teeth, Biotechniques, 2015, vol. 59, no. 2, pp. 87—93. https://doi.org/10.2144/000114320
Weiner, S. and Price, P.A., Disaggregation of bone into crystals, Calcif. Tissue Int., 1986, vol. 39, no. 6, pp. 365—375. https://doi.org/10.1007/BF02555173
Salamon, M., Tuross, N., Arensburg, B., and Weiner, S., Relatively well preserved DNA is present in the crystal aggregates of fossil bones, Proc. Natl. Acad. Sci. U.S.A., 2005, vol. 102, no. 39, pp. 13783—13788. https://doi.org/10.1073/pnas.0503718102
Hayatsu, H., Pan, S., and Ukita, T., Reaction of sodium hypochlorite with nucleic acids and their constituents, Chem. Pharm. Bull., 1971, vol. 19, no. 10, pp. 2189—2192. https://doi.org/10.1248/cpb.19.2189
Prince, A.M. and Andrus, L., PCR: how to kill unwanted DNA, Biotechniques, 1992, vol. 12, no. 3.
Rohland, N., Glocke, I., Aximu-Petri, A., and Meyer, M., Extraction of highly degraded DNA from ancient bones, teeth and sediments for high-throughput sequencing, Nat. Protoc., 2018, vol. 13, no. 11, pp. 2447—2461. https://doi.org/10.1038/s41596-018-0050-5
Ginolhac, A., Vilstrup, J., Stenderup, J., et al., Improving the performance of true single molecule sequencing for ancient DNA, BMC Genomics, 2012, vol. 13, no. 1. https://doi.org/10.1186/1471-2164-13-177
Orlando, L., Ginolhac, A., Raghavan, M., et al., True single-molecule DNA sequencing of a Pleistocene horse bone, Genome Res., 2011, vol. 21, no. 10, pp. 1705—1719. https://doi.org/10.1101/gr.122747.111
Meyer, M., Fu, Q., Aximu-Petri, A., et al., A mitochondrial genome sequence of a hominin from Sima de los Huesos, Nature, 2014, vol. 505, no. 7483, pp. 403—406. https://doi.org/10.1038/nature12788
de Sousa, S.M.G. and Silva, T.L., Efeito do EDTA, EGTA, CDTA e ácido cítrico na desmineralização da dentina radicular: estudo comparativo, Braz. Oral Res., 2005, vol. 19, no. 3, pp. 188—192. https://doi.org/10.1590/S1806-83242005000300006
Medina Cardenas, M.E., Calvo Pérez, V., and Sánchez Planells, U., Comparison of the chelating capacity of EDTA and EGTA, a new demineralized agent, on molars in vitro, Rev. Dent. Chile, 1989, vol. 80, no. 1, pp. 4—10.
Tripodi, D., D’Ercole, S., De Fazio, P., and Spoto, G., Demineralizing action of EGTA in endodontics, Int. J. Immunopathol. Pharmacol., 2007, vol. 20, no. 1, suppl. 1, pp. 93—96. https://doi.org/10.1177/039463200702001s18
Simpson, T.A. and Smith, R.J.H., Amplification of mitochondrial DNA from archival temporal bone specimens, Laryngoscope, 1995, vol. 105, no. 1, pp. 28—34. https://doi.org/10.1288/00005537-199501000-00009
Hagelberg, E. and Clegg, J.B., Isolation and characterization of DNA from archaeological bone, Proc. R. Soc. London, Ser. B, 1991, vol. 244, no. 1309, pp. 45—50. https://doi.org/10.1098/rspb.1991.0049
Higgins, D., Kaidonis, J., Townsend, G., and Austin, J.J., Evaluation of carrier RNA and low volume demineralization for recovery of nuclear DNA from human teeth, Forensic Sci. Med. Pathol., 2014, vol. 10, no. 1, pp. 56—61. https://doi.org/10.1007/s12024-013-9519-2
Aksyuchits, V., Pokayanie: materialy Pravitel’stvennoi komissii po izucheniyu voprosov, svyazannykh s issledovaniem i perezakhoroneniem ostankov Rossiiskogo Imperatora Nikolaya II i chlenov ego sem’i (Repentance: Materials of the Russian Governmental Commission Responsible for the Study and Reburial of the Remains of the Russian Emperor Nicholas II and of Members of His Family), Moscow: Vybor, 1998.
Katevatis, C., Fan, A., and Klapperich, C.M., Low concentration DNA extraction and recovery using a silica solid phase, PLoS One, 2017, vol. 12, no. 5. https://doi.org/10.1371/journal.pone.0176848
Bouwman, A.S. and Brown, T.A., Comparison between silica-based methods for the extraction of DNA from human bones from 18th to mid-19th century London, Anc. Biomol., 2002, vol. 4, no. 4, pp. 173—178. https://doi.org/10.1080/1358612021000028470
Rohland, N., Siedel, H., and Hofreiter, M., A rapid column-based ancient DNA extraction method for increased sample throughput, Mol. Ecol. Resour., 2010, vol. 10, no. 4, pp. 677–683. https://doi.org/10.1111/j.1755-0998.2009.02824.x
Dabney, J., Knapp, M., Glocke, I., et al., Complete mitochondrial genome sequence of a Middle Pleistocene cave bear reconstructed from ultrashort DNA fragments, Proc. Natl. Acad. Sci. U.S.A., 2013, vol. 110, no. 39, pp. 15758—15763. https://doi.org/10.1073/pnas.1314445110
Amory, S., Huel, R., Bilić, A., et al., Automatable full demineralization DNA extraction procedure from degraded skeletal remains, Forensic Sci. Int. Genet., 2012, vol. 6, no. 3, pp. 398—406. https://doi.org/10.1016/j.fsigen.2011.08.004
Marshall, P.L., Stoljarova, M., Schmedes, S.E., et al., A high volume extraction and purification method for recovering DNA from human bone, Forensic Sci. Int. Genet., 2014, vol. 12, pp. 155—160. https://doi.org/10.1016/j.fsigen.2014.06.011
Jakubowska, J., Maciejewska, A., and Pawłowski, R, Comparison of three methods of DNA extraction from human bones with different degrees of degradation, Int. J. Legal Med., 2012, vol. 126, no. 1, pp. 173—178. https://doi.org/10.1007/s00414-011-0590-5
Hasap, L., Chotigeat, W., Pradutkanchana, J., et al., A novel, 4-h DNA extraction method for STR typing of casework bone samples, Int. J. Legal Med., 2020. https://doi.org/10.1007/s00414-019-02232-9
Duijs, F.E. and Sijen, T., A rapid and efficient method for DNA extraction from bone powder, Forensic Sci. Int. Genet. Rep., 2020, vol. 2, p. 100099. https://doi.org/10.1016/j.fsir.2020.100099
Gansauge, M.T. and Meyer, M., Single-stranded DNA library preparation for the sequencing of ancient or damaged DNA, Nat. Protoc., 2013, vol. 8, no. 4, pp. 737—748. https://doi.org/10.1038/nprot.2013.038
Jónsson, H., Ginolhac, A., Schubert, M., et al., mapDamage2.0: fast approximate Bayesian estimates of ancient DNA damage parameters, Bioinformatics, 2013, vol. 29, no. 13, pp. 1682—1684. https://doi.org/10.1093/bioinformatics/btt193
Zavala, E.I., Jacobs, Z., Vernot, B., et al., Pleistocene sediment DNA reveals hominin and faunal turnovers at Denisova Cave, Nature, 2021. https://doi.org/10.1038/s41586-021-03675-0
Vernot, B., Zavala, E.I., Gómez-Olivencia, A., et al., Unearthing Neanderthal population history using nuclear and mitochondrial DNA from cave sediments, Science, 2021, vol. 372, no. 6542. https://doi.org/10.1126/science.abf1667
Bokelmann, L., Hajdinjak, M., Peyrégne, S., et al., A genetic analysis of the Gibraltar Neanderthals, Proc. Natl. Acad. Sci. U.S.A., 2019, vol. 116, no. 31, pp. 15610—15615. https://doi.org/10.1073/pnas.1903984116
Axelsson, E., Willerslev, E., Gilbert, M.T.P., and Nielsen, R., The effect of ancient DNA damage on inferences of demographic histories, Mol. Biol. Evol., 2008, vol. 25, no. 10, pp. 2181—2187. https://doi.org/10.1093/molbev/msn163
Briggs, A.W., Stenzel, U., Meyer, M., et al., Removal of deaminated cytosines and detection of in vivo methylation in ancient DNA, Nucleic Acids Res., 2009, vol. 38, no. 6. https://doi.org/10.1093/nar/gkp1163
Rohland, N., Harney, E., Mallick, S., et al., Partial uracil-DNA-glycosylase treatment for screening of ancient DNA, Philos. Trans. R. Soc., B, 2015, vol. 370, no. 1660. https://doi.org/10.1098/rstb.2013.0624
Gorden, E.M., Sturk-Andreaggi, K., and Marshall, C., Repair of DNA damage caused by cytosine deamination in mitochondrial DNA of forensic case samples, Forensic Sci. Int. Genet., 2018, vol. 34, pp. 257—264. https://doi.org/10.1016/j.fsigen.2018.02.015
Segawa, T., Yonezawa, T., Mori, H., et al., Ancient DNA reveals multiple origins and migration waves of extinct Japanese brown bear lineages, R. Soc. Open Sci., 2021, vol. 8, no. 8. https://doi.org/10.1098/rsos.210518
Mouttham, N., Klunk, J., Kuch, M., et al., Surveying the repair of ancient DNA from bones via high-throughput sequencing, Biotechniques, 2015, vol. 59, no. 1, pp. 19—25. https://doi.org/10.2144/000114307
Andreeva, T., Manakhov, A., Kunizheva, S., and Rogaev, E.I., Genetic evidence of authenticity of a hair shaft relic from the portrait of Tsesarevich Alexei, the son of the last Russian Emperor, Biochemistry (Moscow), 2021, vol. 86, no. 12, pp. 1572—1578.
Orlando, L., Gilbert, M.T.P., and Willerslev, E., Reconstructing ancient genomes and epigenomes, Nat. Rev. Genet., 2015, vol. 16, no. 7, pp. 395—408. https://doi.org/10.1038/nrg3935
Gansauge, M.-T., Gerber, T., Glocke, I., et al., Single-stranded DNA library preparation from highly degraded DNA using T4 DNA ligase, Nucleic Acids Res., 2017, vol. 45, no. 10. e79. https://doi.org/10.1093/nar/gkx033
Gansauge, M.-T. and Meyer, M., Selective enrichment of damaged DNA molecules for ancient genome sequencing, Genome Res., 2014, vol. 24, no. 9, pp. 1543—1549. https://doi.org/10.1101/gr.174201.114
ACKNOWLEDGMENTS
We thank M.V. Dobrovolskaya, N.A. Makarov (Institute of Archaeology, Russian Academy of Sciences) and A.P. Buzhilova (Research Institute and Museum of Anthropology, Moscow State University) for providing bone specimens.
Funding
This study was supported by the project of the Russian Ministry of Education and Science, system no. 075-10-2020-116 (grant no. 13.1902.21.0023).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
The authors declare that they have no conflicts of interest. This article does not contain any studies involving animals or human participants performed by any of the authors.
Additional information
Translated by N. Maleeva
Rights and permissions
About this article
Cite this article
Andreeva, T.V., Malyarchuk, A.B., Soshkina, A.D. et al. Methodologies for Ancient DNA Extraction from Bones for Genomic Analysis: Approaches and Guidelines. Russ J Genet 58, 1017–1035 (2022). https://doi.org/10.1134/S1022795422090034
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1022795422090034