Abstract
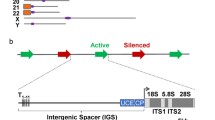
The genes coding for the rRNAs seem evolutionary conserved on the first glance, but astonish one with their variability in the structure and a variety of functions on closer examination. The non-coding parts of rDNA contain regulatory elements, protein binding sites, pseudogenes, repetitive sequences, and microRNA genes. Ribosomal intergenic spacers are not only in charge with the nucleolus morphology and functioning, namely, the rRNA expression and ribosome biogenesis, but also control nuclear chromatin formation thus mediating cell differentiation. The alterations in the expression of these non-coding regions of rDNA in response to environmental stimuli underlie the keen sense of a cell to various types of stressors. Malfunctioning of this process may result in a wide range of pathologies from oncology to neurodegenerative disease and mental illness. Here, we observe to-date materials on the structure and transcription of the ribosomal intergenic spacer in humans and its role in rRNA expression, in-born disease development, and cancer.




Similar content being viewed by others
REFERENCES
McStay B. 2016. Nucleolar organizer regions: genomic ‘dark matter’ requiring illumination. Genes Dev. 30 (14), 1598–1610.
Paredes S., Maggert K. 2009. Ribosomal DNA contributes to global chromatin regulation. Proc. Natl. Acad. Sci. U. S. A. 106 (42), 17829–17834.
Stępiński D. 2018. The nucleolus, an ally, and an enemy of cancer cells. Histochem. Cell Biol. 150 (6), 607–629.
Rajesh Y., Pal I., Banik P., Chakraborty S., Borkar S., Dey G., Mukherjee A., Mandal M. 2017. Insights into molecular therapy of glioma: current challenges and next generation blueprint. Acta Pharmacol. Sinica. 38 (5), 591–613.
Moore L., Kivinen V., Liu Y., Annala M., Cogdell D., Liu X., Liu C., Sawaya R., Yli-Harja O., Shmulevich I., Fuller G.N., Zhang W., Nykter M. 2013. Transcriptome and small RNA deep sequencing reveals deregulation of miRNA biogenesis in human glioma. J. Pathol. 229 (3), 449–459.
Brower J., Clark P., Lyon W., Kuo J. 2014. MicroRNAs in cancer: glioblastoma and glioblastoma cancer stem cells. Neurochem. Int. 77, 68–77.
Hosgood H., Hu W., Rothman N., Klugman M., Weinstein S., Virtamo J., Albanes D., Cawthon R., Lan Q. 2019. Variation in ribosomal DNA copy number is associated with lung cancer risk in a prospective cohort study. Carcinogenesis. 40 (8), 975–978.
Santoro R., Grummt I. 2005. Epigenetic mechanism of rRNA gene silencing: temporal order of NoRC-mediated histone modification, chromatin remodeling, and DNA methylation. Mol. Cell. Biol. 25 (7), 2539–2546.
Savić N., Bär D., Leone S., Frommel S., Weber F., Vollenweider E., Ferrari E., Ziegler U., Kaech A., Shakhova O., Cinelli P., Santoro R. 2014. LncRNA maturation to initiate heterochromatin formation in the nucleolus is required for exit from pluripotency in eSCS. Cell Stem Cell. 15 (6), 720–734.
McConkey E., Hopkins J. 1964. The relationship of the nucleolus to the synthesis of ribosomal RNA in HeLa cells. Proc. Natl. Acad. Sci. U. S. A. 51 (6), 1197–204.
Schwarzacher H., Wachtler F. 1993. The nucleolus. Anat. Embryol. 188 (6), 515–536.
Gonzalez I., Sylvester J. 1995. Complete sequence of the 43-kb human ribosomal DNA repeat: analysis of the intergenic spacer. Genomics. 27 (2), 320–328.
Gibbons J., Branco A., Godinho S., Yu S., Lemos B. 2015. Concerted copy number variation balances ribosomal DNA dosage in human and mouse genomes. Proc. Natl. Acad. Sci. U. S. A. 112 (8), 2485–2490.
Hall A., Turner T., Queitsch C. 2021. Thousands of high-quality sequencing samples fail to show meaningful correlation between 5S and 45S ribosomal DNA arrays in humans. Sci. Rep. 11 (1), 449.
Wang M., Lemos B. 2017. Ribosomal DNA copy number amplification and loss in human cancers is linked to tumor genetic context, nucleolus activity, and proliferation. PLoS Genet. 13 (9), e1006994.
Smirnov E., Chmúrčiaková N., Liška F., Bažantová P., Cmarko D. 2021. Variability of human rDNA. Cells. 10 (2), 196.
Erickson J., Schmickel R. 1985. A molecular basis for discrete size variation in human ribosomal DNA. Am. J. Hum. Genet. 37 (2), 311–325.
Akamatsu Y., Kobayashi T. 2015. The human RNA polymerase I transcription terminator complex acts as a replication fork barrier that coordinates the progress of replication with rRNA transcription activity. Mol. Cell. Biol. 35 (10), 1871–1881.
Jacob M., Audas T., Mullineux S., Lee S. 2012. Where no RNA polymerase has gone before: novel functional transcripts derived from the ribosomal intergenic spacer. Nucleus. 3 (4), 315‒319.
Guetg C., Santoro R. 2012. Formation of nuclear heterochromatin: the nucleolar point of view. Epigenetics. 7 (8), 811–814.
van Koningsbruggen S., Gierlinski M., Schofield P., Martin D., Barton G., Ariyurek Y., den Dunnen J., Lamond A. 2010. High-resolution whole-genome sequencing reveals that specific chromatin domains from most human chromosomes associate with nucleoli. Mol. Biol. Cell. 21 (21), 3735–3748.
Németh A., Conesa A., Santoyo-Lopez J., Medina I., Montaner D., Péterfia B., Solovei I., Cremer T., Dopazo J., Längst G. 2010. Initial genomics of the human nucleolus. PLoS Genet. 6 (3), e1000889.
Yu S., Lemos B. 2018. The long-range interaction map of ribosomal DNA arrays. PLoS Genet. 14 (3), e1007258.
Strohner R., Nemeth A., Jansa P., Hofmann-Rohrer U., Santoro R., Längst G., Grummt I. 2001. NoRC—a novel member of mammalian ISWI-containing chromatin remodeling machines. EMBO J. 20 (17), 4892–4900.
Guetg C., Lienemann P., Sirri V., Grummt I., Hernandez-Verdun D., Hottiger M., Fussenegger M., Santoro R. 2010. The NoRC complex mediates the heterochromatin formation and stability of silent rRNA genes and centromeric repeats. EMBO J. 29 (13), 2135–2146.
Postepska-Igielska A., Krunic D., Schmitt N., Greulich-Bode K., Boukamp P., Grummt I. 2013. The chromatin remodelling complex NoRC safeguards genome stability by heterochromatin formation at telomeres and centromeres. EMBO Rep. 14 (8), 704–710.
Guetg C., Scheifele F., Rosenthal F., Hottiger M., Santoro R. 2012. Inheritance of silent rDNA chromatin is mediated by PARP1 via noncoding RNA. Mol. Cell. 45 (6), 790–800.
Krishnakumar R., Kraus W. 2010. PARP-1 regulates chromatin structure and transcription through a KDM5B-dependent pathway. Mol. Cell. 39 (5), 736–749.
Roper S., Chrysanthou, S., Senner C., Sienerth A., Gnan S., Murray A., Masutani M., Latos P., Hemberger M. 2014. ADP-ribosyltransferases Parp1 and Parp7 safeguard pluripotency of ES cells. Nucleic Acids Res. 42 (14), 8914–8927.
Schreiber V., Dantzer F., Amé J.C., de Murcia G. 2006. Poly(ADP-ribose): novel functions for an old molecule. Nat. Rev. 7 (7), 517–528.
Nalabothula N., Al-jumaily T., Eteleeb A., Flight R., Xiaorong S., Moseley H., Rouchka E., Fondufe-Mittendorf Y. 2015. Genome-wide profiling of PARP1 reveals an interplay with gene regulatory regions and DNA methylation. PLoS One. 10 (8), e0135410.
Mansuroglu Z., Benhelli-Mokrani H., Marcato V., Sultan A., Violet M., Chauderlier A., Delattre L., Loyens A., Talahari S., Bégard S., Nesslany F., Colin M., Souès S., Lefebvre B., Buée L., Galas M.C., Bonnefoy E. 2016. Loss of Tau protein affects the structure, transcription and repair of neuronal pericentromeric heterochromatin. Sci. Rep. 6, 33047.
Maina M., Bailey L., Wagih S., Biasetti L., Pollack S., Quinn J., Thorpe J., Doherty A., Serpell L. 2018. The involvement of Tau in nucleolar transcription and the stress response. Acta Neuropathol. Commun. 6 (1), 70.
Kuhn A., Grummt I. 1992. Dual role of the nucleolar transcription factor UBF: trans-activator and antirepressor. Proc. Natl. Acad. Sci. U. S. A. 89 (16), 7340–7344.
Sanij E., Poortinga G., Sharkey K., Hung S., Holloway T., Quin J., Robb E., Wong L., Thomas W., Stefanovsky V., Moss T., Rothblum L., Hannan K.M., McArthur G.A., Pearson R.B., Hannan R.D. 2008. UBF levels determine the number of active ribosomal RNA genes in mammals. J. Cell. Biol. 183 (7), 1259–1274.
Hannan K., Hannan R., Rothblum L. 1998. Transcription by RNA polymerase I. Front. Biosci. 3, d376–d398.
Grummt I. 1999. Regulation of mammalian ribosomal gene transcription by RNA polymerase I. Prog. Nucleic Acids Res. Mol. Biol. 62, 109–154.
Grummt I. 2010. Wisely chosen paths—regulation of rRNA synthesis. FEBS J. 277 (22), 4626–4639.
Tanaka Y., Tsuneoka M. 2018. Control of ribosomal RNA transcription by nutrients. In: Gene Expression and Regulation in Mammalian Cells—Transcription toward the Establishment of Novel Therapeutics. Uchiumi F., Ed. London: IntechOpen. Ch. 2.
Matthews D., Olson M. 2006. What is new in the nucleolus?: Workshop on the nucleolus: new perspectives. EMBO Rep. 7 (9), 870–873.
Zhao Z., Senturk N., Song C., Grummt I. 2018. l-ncRNA PAPAS tethered to the rDNA enhancer recruits hypophosphorylated CHD4/NuRD to repress rRNA synthesis at elevated temperatures. Genes Dev. 32 (11–12), 836–848.
Grummt I., Ladurner A. 2008. A metabolic throttle regulates the epigenetic state of rDNA. Cell. 133 (4), 577–580.
Zentner G., Saiakhova A., Manaenkov P., Adams M., Scacheri P. 2011. Integrative genomic analysis of human ribosomal DNA. Nucleic Acids Res. 39 (12), 4949–4960.
Srivastava R., Srivastava R., Ahn S. 2016. The epigenetic pathways to ribosomal DNA silencing. Microbiol. Mol. Biol. Rev. 80 (3), 545–563.
Li J., Längst G., Grummt I. 2006. NoRC-dependent nucleosome positioning silences rRNA genes. EMBO J. 25 (24), 5735–5741.
Li J., Santoro R., Koberna K., Grummt I. 2005. The chromatin remodeling complex NoRC controls replication timing of rRNA genes. EMBO J. 24 (1), 120–127.
Wang M., Lemos B. 2019. Ribosomal DNA harbors an evolutionarily conserved clock of biological aging. Genome Res. 29 (3), 325–333.
Németh A., Guibert S., Tiwari V., Ohlsson R., Längst G. 2008. Epigenetic regulation of TTF-I-mediated promoter–terminator interactions of rRNA genes. EMBO J. 27 (8), 1255–1265.
Xie W., Ling T., Zhou Y., Feng W., Zhu Q., Stunnenberg H., Grummt I., Tao W. 2012. The chromatin remodeling complex NuRD establishes the poised state of rRNA genes characterized by bivalent histone modifications and altered nucleosome positions. Proc. Natl. Acad. Sci. U. S. A. 109 (21), 8161–8166.
Yuan X., Feng W., Imhof A., Grummt I., Zhou Y. 2007. Activation of RNA polymerase I transcription by cockayne syndrome group B protein and histone methyltransferase G9a. Mol. Cell. 27 (4), 585–595.
Salifou K., Ray S., Verrier L., Aguirrebengoa M., Trouche D., Panov K., Vandromme M. 2016. The histone demethylase JMJD2A/KDM4A links ribosomal RNA transcription to nutrients and growth factors availability. Nat. Commun. 7, 10174.
Murayama A., Ohmori K., Fujimura A., Minami H., Yasuzawa-Tanaka K., Kuroda T., Oie S., Daitoku H., Okuwaki M., Nagata K., Fukamizu A., Kimura K., Shimizu T., Yanagisawa J. 2008. Epigenetic control of rDNA loci in response to intracellular energy status. Cell. 133 (4), 627–639.
Kumazawa T., Nishimura K., Kuroda T., Ono W., Yamaguchi C., Katagiri N., Tsuchiya M., Masumoto H., Nakajima Y., Murayama A., Kimura K., Yanagisawa J. 2011. Novel nucleolar pathway connecting intracellular energy status with p53 activation. J. Biol. Chem. 286 (23), 20861–20869.
Yan Q., Zhu C., Guang S., Feng X. 2019. The functions of non-coding RNAs in rRNA regulation. Front. Genet. 10, 290.
Jacob M., Audas T., Uniacke J., Trinkle-Mulcahy L., Lee S. 2013. Environmental cues induce a long noncoding RNA-dependent remodeling of the nucleolus. Mol. Biol. Cell. 24 (18), 2943–2953.
Bierhoff H., Schmitz K., Maass F., Ye J., Grummt I. 2010. Noncoding transcripts in sense and antisense orientation regulate the epigenetic state of ribosomal RNA genes. Cold Spring Harb. Symp. Quant. Biol. 75, 357–364.
Schmitz K., Mayer C., Postepska A., Grummt I. 2010. Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev. 24 (20), 2264–2269.
Mayer C., Neubert M., Grummt I. 2008. The structure of NoRC-associated RNA is crucial for targeting the chromatin remodelling complex NoRC to the nucleolus. EMBO Rep. 9 (8), 774–780.
Bierhoff H., Dammert M., Brocks D., Dambacher S., Schotta G., Grummt I. 2014. Quiescence-induced LncRNAs trigger H4K20 trimethylation and transcriptional silencing. Mol. Cell. 54 (4), 675–682.
Li D., Zhang J., Wang M., Li X., Gong H., Tang H., Chen L., Wan L., Liu Q. 2018. Activity dependent LoNA regulates translation by coordinating rRNA transcription and methylation. Nat. Commun. 9 (1), 1726–1739.
Caudron-Herger M., Pankert T., Seiler J., Németh A., Voit R., Grummt I., Rippe K. 2015. Alu element-containing RNAs maintain nucleolar structure and function. EMBO J. 34 (22), 2758–2774.
Xing Y., Yao R., Zhang Y., Guo C., Jiang S., Xu G., Dong R., Yang L., Chen L. 2017. SLERT regulates DDX21 rings associated with Pol I transcription. Cell. 169 (4), 664–678.
Morgan G., Reeder R., Bakken A. 1983. Transcription in cloned spacers of Xenopus laevis ribosomal DNA. Proc. Natl. Acad. Sci. U. S. A. 80 (21), 6490–6494.
Kuhn A., Grummt I. 1987. A novel promoter in the mouse rDNA spacer is active in vivo and in vitro. EMBO J. 6 (11), 3487–3492.
Agrawa S., Ganley A. 2018. The conservation landscape of the human ribosomal RNA gene repeats. PLoS One. 13 (12), e0207531.
Li Y., Wang H., Wan F., Liu F., Liu J., Zhang N., Jin S., Li J. 2012. Deep sequencing analysis of small non-coding RNAs reveals the diversity of microRNAs and piRNAs in the human epididymis. Gene. 497 (2), 330–335.
Ma X., Liu H., Zheng Y., Dai Y., Lingling E., Zhang R., Zhang S. 2022. Genome-wide screening of different expressed genes and its potential associations with aging dental pulp stem cells. Comb. Chem. High Throughput Screen. https://doi.org/10.2174/1386207325666220705120904
Pirogov S., Gvozdev V., Klenov M. 2019. Long noncoding RNAs and stress response in the nucleolus. Cells. 8 (7), 668.
Mayer C., Schmitz K., Li J., Grummt I., Santoro R. 2006. Intergenic transcripts regulate the epigenetic state of rRNA genes. Mol. Cell. 22 (3), 351–361.
McStay B., Grummt I. 2008. The epigenetics of rRNA genes: from molecular to chromosome biology. Annu. Rev. Cell Dev. Biol. 24, 131–157.
Mars J.C., Sabourin-Felix M., Tremblay M., Moss T. 2017. A deconvolution protocol for ChIP-seq reveals analogous enhancer structures on the mouse and human ribosomal RNA genes. G3. 8 (1), 303–314.
Shiao Y., Lupascu S., Gu Y., Kasprzak W., Hwang C., Fields J., Leighty R., Quiñones O., Shapiro B., Alvord W., Anderson L. 2009. An intergenic non-coding RNA correlated with expression of the rRNA and frequency of an rRNA single nucleotide polymorphism in lung cancer cells. PLoS One. 4 (10), e7505.
Vacík T., Kereïche S., Raška I., Cmarko D., Smirnov E. 2019. Life time of some RNA products of rDNA intergenic spacer in HeLa cells. Histochem. Cell. Biol. 152 (4), 271–280.
Todd M., Huh M., Picketts D. 2016. The sub-nucleolar localization of PHF6 defines its role in rDNA transcription and early processing events. Eur. J. Hum. Genet. 24 (10), 1453–1459.
Sadova A., Kupriyanova N., Pavlova G. 2020. Mapping and quantification of non-coding RNA originating from the rDNA in human glioma cells. Cancers. 12 (8), 2090.
Sadova A., Panteleev D., Pavlova G. 2021. Zooming in: PAGE-northern blot helps to analyze anti-sense transcripts originating from human rIGS under transcriptional stress. Noncoding RNA. 7 (3), 50.
Abraham K., Khosraviani N., Chan J., Gorthi A., Samman A., Zhao D.Y., Wang M., Bokros M., Vidya E., Ostrowski L.A., Oshidari R., Pietrobon V., Patel P.S., Algouneh A., Singhania R., Liu Y., Yerlici V.T., De Carvalho D.D., Ohh M., Dickson B.C., Hakem R., Greenblatt J.F., Lee S., Bishop A.J.R., Mekhail K. 2020. Nucleolar RNA polymerase II drives ribosome biogenesis. Nature. 585 (7824), 298–302.
Warmerdam D., Wolthuis R. 2019. Keeping ribosomal DNA intact: a repeating challenge. Chromosome Res. 27 (1–2), 57–72.
Machwe A., Orren D.K., Bohr V.A. 2000. Accelerated methylation of ribosomal RNA genes during the cellular senescence of Werner syndrome fibroblasts. FASEB J. 14 (12), 1715–1724.
Zeng J., Libien J., Shaik F., Wolk J., Hernández A. 2016. Nucleolar PARP-1 expression is decreased in Alzheimer’s disease: consequences for epigenetic regulation of rDNA and cognition. Neural Plasticity. 2016, 8987928.
Pietrzak M., Rempala G., Nelson P., Zheng J., Hetman M. 2011. Epigenetic silencing of nucleolar rRNA genes in Alzheimer’s disease. PLoS One. 6 (7), e22585.
Teschler S., Gotthardt J., Dammann G., Dammann R. 2016. Aberrant DNA methylation of rDNA and PRIM-A1 in borderline personality disorder. Int. J. Mol. Sci. 17 (1), E67.
McGowan P., Sasaki A., Huang T., Unterberger A., Suderman M., Ernst C., Meaney M., Turecki G., Szyf M. 2008. Promoter-wide hypermethylation of the ribosomal RNA gene promoter in the suicide brain. PLoS One. 3 (5), e2085.
Hallgren J., Pietrzak M., Rempala G., Nelson P., Hetman M. 2014. Neurodegeneration-associated instability of ribosomal DNA. Biochim. Biophys. Acta. 1842 (6), 860–868.
Dastidar S., Nair D. 2022. A ribosomal perspective on neuronal local protein synthesis. Front. Mol. Neurosci. 15, 823135.
Allen K., Regier M., Hsieh C., Tsokas P., Barnard M., Phatarpekar S., Wolk J., Sacktor T., Fenton A., Hernández A. 2018. Learning-induced ribosomal RNA is required for memory consolidation in mice—evidence of differentially expressed rRNA variants in learning and memory. PLoS One. 13 (10), e020337.
Allen K., Gourov A., Harte C., Gao P., Lee C., Sylvain D., Splett J., Oxberry W., van de Nes P., Troy-Regier M., Wolk J., Alarcon J.M., Hernández A.I. 2014. Nucleolar integrity is required for the maintenance of long-term synaptic plasticity. PLoS One. 9 (8), e104364.
Lyapunova N., Porokhovnik L., Kosyakova N., Mandron I., Tsvetkova T. 2017. Effects of the copy number of ribosomal genes (genes for rRNA) on viability of subjects with chromosomal abnormalities. Gene. 611, 47–53.
Ravaioli F., Zampieri M., Morandi L., Pirazzini C., Pellegrini C., De Fanti S., Gensous N., Pirazzoli G., Sambati L., Ghezzo A., Ciccarone F., Reale A., Monti D., Salvioli S., Caiafa P., Capri M., Bürkle A., Moreno-Villanueva M., Garagnani P., Franceschi C., Bacalini M.G. 2022. DNA methylation analysis of ribosomal DNA in adults with Down syndrome. Front. Genet. 13, 792165.
Chestkov I.V., Jestkova E.M., Ershova E.S., Golimbet V.E., Lezheiko T.V., Kolesina N.Y., Porokhovnik L.N., Lyapunova N.A., Izhevskaya V.L., Kutsev S.I., Veiko N.N., Kostyuk S.V. 2018. Abundance of ribosomal RNA gene copies in the genomes of schizophrenia patients. Schizophr. Res. 197, 305–314.
Ershova E.S., Malinovskaya E.M., Golimbet V.E., Lezheiko T.V., Zakharova N.V., Shmarina G.V., Veiko R.V., Umriukhin P.E., Kostyuk G.P., Kutsev S.I., Izhevskaya V.L., Veiko N.N., Kostyuk S.V. 2020. Copy number variations of satellite III (1q12) and ribosomal repeats in health and schizophrenia. Schizophr. Res. 223, 199–212.
Umriukhin P., Ershova E., Filev A., Agafonova O., Martynov A., Zakharova N., Veiko R., Porokhovnik L., Kostyuk G., Kutsev S., Veiko N., Kostyuk S. 2022. The psychoemotional stress-induced changes in the abundance of SatIII (1q12) and telomere repeats, but not ribosomal DNA, in human leukocytes. Genes (Basel). 13 (2), 343.
Kondratyeva E.I., Ershova E.S., Voronkova A.Yu., Shmarina G.V., Krasovsky S.A., Zhekaite E.K., Petrova N.V., Melyanovskaya Yu.L., Odinaeva N.D., Veiko N.N., Kostyuk S.V. 2021. Variation in the number of copies of ribosomal genes in the genomes of patients with cystic fibrosis. Med. Genet. 20 (2), 49–60.
Veiko N., Shubaeva N., Tsvetkova T., Mandron I., Malinovskaya T., Speransky A., Lyapunova N. 2005. Quantitative characteristics of ribosomal gene complex in patients with severe forms of rheumatoid arthritis. Med. Genetics. 4 (4), 74.
Zamanpoor M., Ghaedi H., Omrani M. 2020. The genetic basis for the inverse relationship between rheumatoid arthritis and schizophrenia. Mol. Genet. Genom. Med. 8 (11), e1483.
Porokhovnik L., Lyapunova N. 2019. Dosage effects of human ribosomal genes (rDNA) in health and disease. Chromosome Res.: Int. J. Mol., Supramol. Evol. Aspects Chromosome Biol. 27 (1–2), 5–17.
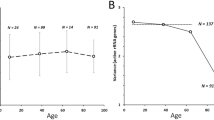
Malinovskaya E., Ershova E., Golimbet V., Porokhovnik L., Lyapunova N., Kutsev S., Veiko N., Kostyuk S. 2018. Copy number of human ribosomal genes with aging: unchanged. Front. Genet. 9, 306.
Veiko N., Ershova E., Veiko R., Umriukhin P., Kurmyshev M., Kostyuk G., Kutsev S., Kostyuk S. 2022. Mild cognitive impairment is associated with low copy number of ribosomal genes in the genomes of elderly people. Front. Genet. 13, 967448.
Holdt L., Stahringer A., Sass K., Pichler G., Kulak N., Wilfert W., Kohlmaier A., Herbst A., Northoff B., Nicolaou A., Gäbel G., Beutner F., Scholz M., Thiery J., Musunuru K., Krohn K., Mann M., Teupser D. 2016. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat. Commun. 7, 12429.
Narla A., Ebert B. 2010. Ribosomopathies: human disorders of ribosome dysfunction. Blood. 115 (16), 3196–3205.
Mills E., Green R. 2017. Ribosomopathies: there’s strength in numbers. Science. 358 (6363), eaan2755.
Calo E., Gu B., Bowen M., Aryan F., Zalc A., Liang J., Flynn R., Swigut T., Chang H., Attardi L., Wysocka J. 2018. Tissue-selective effects of nucleolar stress and rDNA damage in developmental disorders. Nature. 554 (7690), 112–117.
Von Walden F., Gantelius S., Liu C., Borgström H., Björk L., Gremark O., Stål P., Nader G., Ponté N. 2018. Muscle contractures in patients with cerebral palsy and acquired brain injury are associated with extracellular matrix expansion, pro-inflammatory gene expression, and reduced rRNA synthesis. Muscle Nerve. 58 (2), 277–285.
Hwang Y., Han D., Kim K., Min S., Kowall N., Yang L., Lee J., Kim Y., Ryu H. 2014. ESET methylates UBF at K232/254 and regulates nucleolar heterochromatin plasticity and rDNA transcription. Nucleic Acids Res. 42 (3), 1628–1643.
Xie Q., Li C., Song X., Wu L., Jiang Q., Qiu Z., Cao H., Yu K., Wan C., Li J., Yang F., Huang Z., Niu B., Jiang Z., Zhang T. 2017. Folate deficiency facilitates recruitment of upstream binding factor to hot spots of DNA double-strand breaks of rRNA genes and promotes its transcription. Nucleic Acids Res. 45 (5), 2472–2489.
Hetman M., Slomnicki L. 2019. Ribosomal biogenesis as an emerging target of neurodevelopmental pathologies. J. Neurochem. 148 (3), 325–347.
Smirnov E., Chmúrčiaková N., Cmarko D. 2021. Human rDNA and cancer. Cells. 10 (12), 3452.
Valori V., Tus K., Laukaitis C., Harris D., LeBeau L., Maggert K. 2019. Human rDNA copy number is unstable in metastatic breast cancers. Epigenetics. 15 (1–2), 85–106.
Xu B., Li H., Perry J., Singh V., Unruh J., Yu Z., Zakari M., McDowell W., Li L., Gerton J. 2017. Ribosomal DNA copy number loss and sequence variation in cancer. PLoS Genet. 13 (6), e1006771.
Baskaran S., Mayrhofer M., Kultima H., Bergström T., Elfineh L., Cavelier L., Isaksson A., Nelander S. 2018. Primary glioblastoma cells for precision medicine: a quantitative portrait of genomic (in)stability during the first 30 passages. Neuro-Oncol. 20 (8), 1080–1091.
Belin S., Beghin A., Solano-Gonzàlez E., Bezin L., Brunet-Manquat S., Textoris J., Prats A., Mertani H., Dumontet C., Diaz J. 2009. Dysregulation of ribosome biogenesis and translational capacity is associated with tumor progression of human breast cancer cells. PLoS One. 4 (9), e7147.
Rajput P., Shukla S., Kumar V. 2015. The HBx oncoprotein of hepatitis B virus potentiates cell transformation by inducing c-Myc-dependent expression of the RNA polymerase I transcription factor UBF. Virol. J. 12, 62.
Grandori C., Gomez-Roman N., Felton-Edkins Z., Ngouenet C., Galloway D., Eisenman R., White R. 2005. c-Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nat. Cell. Biol. 7 (3), 311–318.
Hannan K., Hannan R., Smith S., Jefferson L., Lun M., Rothblum L. 2000. Rb and p130 regulate RNA polymerase I transcription: Rb disrupts the interaction between UBF and SL-1. Oncogene. 19 (43), 4988–4999.
Frescas D., Guardavaccaro D., Bassermann F., Koyama-Nasu R., Pagano M. 2007. JHDM1B/FBXL10 is a nucleolar protein that represses transcription of ribosomal RNA genes. Nature. 450 (7167), 309–313.
Lozzio C., Lozzio B. 1975. Human chronic myelogenous leukemia cell-line with positive Philadelphia chromosome. Blood. 45 (3), 321–334.
Foltankova V., Legartova S., Kozubek S., Bartova E. 2012. Tumor-specific histone signature and DNA methylation in multiple myeloma and leukemia cells. Neoplasma. 59 (4), 450–462.
Giard D., Aaronson S., Todaro G., Arnstein P., Kersey J., Dosik H., Parks W. 1973. In vitro cultivation of human tumors: establishment of cell lines derived from a series of solid tumors. J. Natl. Cancer Inst. 51 (5), 1417–1423.
Soule H., Vazguez J., Long A., Albert S., Brennan M. 1973. A human cell line from a pleural effusion derived from a breast carcinoma. J. Natl. Cancer Inst. 51 (5), 1409–1416.
Lee A., Oesterreich S., Davidson N. 2015. MCF-7 cells—changing the course of breast cancer research and care for 45 years. J. Natl. Cancer Inst. 107 (7), djv073.
Johnston R., D’Costa Z., Ray S., Gorski J., Harkin D., Mullan P., Panov K. 2016. The identification of a novel role for BRCA1 in regulating RNA polymerase I transcription. Oncotarget. 7 (42), 68097–68110.
Kaighn M., Narayan K., Ohnuki Y., Lechner J., Jones L. 1979. Establishment and characterization of a human prostatic carcinoma cell line (PC-3). Invest. Urol. 17 (1), 16–23.
Zhang D., Park D., Zhong Y., Lu Y., Rycaj K., Gong S., Chen X., Liu X., Chao H., Whitney P., Calhoun-Davis T., Takata Y., Shen J., Iyer V.R., Tang, D.G. 2016. Stem cell and neurogenic gene-expression profiles link prostate basal cells to aggressive prostate cancer. Nat. Commun. 7, 10798.
Yan Y., Chen Z., Xiao Y., Wang X., Qian K. 2019. Long non-coding RNA SNHG6 is upregulated in prostate cancer and predicts poor prognosis. Mol. Biol. Rep. 46 (3), 2771–2778.
Holmberg Olausson K., Nister M., Lindstrom M. 2014. Loss of nucleolar histone chaperone NPM1 triggers rearrangement of heterochromatin and synergizes with a deficiency in DNA methyltransferase DNMT3a to drive ribosomal DNA transcription. J. Biol. Chem. 289 (50), 34601–34619.
Kobayashi T. 2008. A new role of the rDNA and nucleolus in the nucleus—rDNA instability maintains genome integrity. Bioessays. 30 (3), 267–272.
O'Sullivan J., Pai D., Cridge A., Engelke D., Ganley A. 2013. The nucleolus: a raft adrift in the nuclear sea or the keystone in nuclear structure? Biomol. Concepts. 4 (3), 277–286.
Audas T., Jacob M., Lee S. 2012. Immobilization of proteins in the nucleolus by ribosomal intergenic spacer noncoding RNA. Mol. Cell. 45 (2), 147–157.
Funding
The work was funded by the Ministry of Science and Higher Education of the Russian Federation, grant no. 075-15-2020-809 (13.1902.21.0030).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
COMPLIANCE WITH ETHICAL STANDARDS
Authors declare no conflicts of interests. The article does not contain the investigation with human participants or animals as objects.
ADDITIONAL INFORMATION
The article was translated by the authors.
Additional information
Translated by A. A. Sadova
Abbreviations: ETS, external transcribed spacer; IGS-RNA, RNAs, transcribed from rIGS; ITS, internal transcribed spacer; lncRNA, long non-coding RNA; ncRNA, non-coding RNA; NOR, nucleolus organizer region; NoRC, nucleolar remodeling complex; NuRD, nucleosome remodeling and deacetylation; PAPAS, promoter and pre-rRNA antisense RNA; rIGS, ribosomal intergenic spacer.
Rights and permissions
About this article
Cite this article
Sadova, A.A., Panteleev, D.Y. & Pavlova, G.V. The Structure, Expression, and Non-Canonical Functions of Human rDNA: The Role of Non-Coding Regions. Mol Biol 57, 398–411 (2023). https://doi.org/10.1134/S002689332303007X
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S002689332303007X