Abstract
Genetically engineered chymosin from the tree shrew (Tupaia belangeri chinensis) has been obtained and partially characterized for the first time. The target enzyme was produced in Escherichia coli, strain BL21(DE3). It was shown that tree shrew recombinant chymosin coagulates cow milk (Bos taurus). The total and specific milk-clotting activity of the obtained enzyme was 0.7–5.3 IMCU/mL and 8.8–16.6 IMCU/mg. The nonspecific proteolytic activity of tree shrew recombinant chymosin in relation to total bovine casein was 30 and 117% higher than that of recombinant chymosin of cow and of single-humped camel respectively. It was found that in comparison with most of the known genetically engineered chymosins, the tree shrew enzyme showed exceptionally low thermal stability. After heating at 45°C, the coagulation ability of tree shrew recombinant chymosin decreased by more than 40%, and at 50°C the enzyme lost more than 90% of the initial milk-clotting activity. The Michaelis constant (Km), enzyme turnover number (kcat), and catalytic efficiency (kcat/Km) for genetically engineered chymosin from the tree shrew were 6.3 ± 0.1 µM, 11 927 ± 3169 s–1 and 1968 ± 620 µM–1 s–1, respectively. Comparative analysis showed that the primary structure of the chymosin-sensitive site of cow kappa-casein and the supposed similar sequence of tree shrew kappa-casein differed by 75%. The ability of tree shrew recombinant chymosin to coagulate cow’s milk, along with a low thermal stability and high catalytic efficiency with respect to the substrate, imitating the chymosin-sensitive site of cow kappa-casein, suggests that this enzyme is of potential interest for cheese making.
Similar content being viewed by others
INTRODUCTION
Representatives of the class Mammalia possess a common adaptive mechanism, the ability to synthesize and secrete milk for feeding offspring. Neonatal milk-clotting proteinases, chymosins under the influence of which milk in the stomach of a newborn turns into a clot, which ensures its effective assimilation, probably arose in parallel with lactation [1]. Chymosins (EC 3.4.23.4) have exceptionally high specificity for the F105-M106 bond (or its analog) in the k-casein molecule (к-CN) and, at the same time, extremely low nonspecific proteolytic activity (PA). As a result, chymosin (Chn) effectively hydrolyzes the only peptide bond in the к-CN molecule, which causes the formation of a milk clot. At the same time, the enzyme does not attack immunoglobulins (Ig) and other protective proteins contained in colostrum and milk, which contributes to the effective transfer of passive immunity from mother to newborn [2].
Due to the ability to coagulate milk, milk-clotting enzymes (MCEs) and, in particular, Chn, play an important role in human practice, and any aspect of studying the biochemical properties of these enzymes is considered from the point of view of the possibility of their technological application [3].
Presumably, humanity learned to use MCE to make cheeses about 8000 years ago [4]. Since the appearance of industrial production, the best MCE for cheese making was considered to be bovine Chn— aspartic acid endopeptidase, synthesized by the cells of the gastric mucosa of a newborn calf [5]. Advances in genetic engineering have made it possible to introduce recombinant Chn (rChn) from cows (Bos taurus) into the practice of cheese making [6]. The complex of biochemical properties of bovine Chn was considered optimal for cheese making until 2006, when camel rChn was obtained and studied (Camelus dromedarius) [7]. Camel rChn is now widely used in cheese making along with cow rChn [3, 8–10].
Obtaining the rChn of the dromedary camel was the beginning of a new stage of research aimed at studying and understanding the mechanisms of the substrate specificity of neonatal gastric proteinases [9–14]. Moreover, the results of the study of C. dromedarius rChn continue to stimulate research aimed at the search and study of novel Chn species. Since B. taurus milk is the bulk of the raw material base for cheese making, the biochemical characterization of any new Chn begins with the determination of its milk-clotting activity (MCA) in relation to cow’s milk.
The answer to the question posed in the title of the article is not obvious due to the paradox “Bovine chymosin–camel milk”. Its essence is that cow Chn does not coagulate camel milk, which does not allow using cow enzyme for the production of cheese from camel milk [9, 15–20]. At the same time, camel rChn not only effectively coagulates cow milk, but also surpasses bovine rChn in specific MCA in relation to cow substrate [7]. It has been assumed that the reason for the paradox lies in the differences in the primary structure of bovine and camel к-CN in the 98–112 region [13, 14, 20]. The “bovine Chn–camel milk” paradox is the most famous, but not the only case of interspecies conflict between milk and Chn. Bovine chymosin is unable to coagulate horse (Equus caballus) milk, while bactrian camel (Camelus bactrianus) rChn successfully copes with this task [15, 21]. In addition to camel and horse milk, bovine Chn does not coagulate the milk of a representative of marsupial mammals, Fox Kuzu (Trichosurus vulpecula) [22]. Bovine Chn coagulates milk of rat (Rattus norvegicus), rabbit (Oryctolagus cuniculus), goat (Capra hircus), and reindeer (Rangifer tarandus) much slower than the homologous substrate [23]. The coagulation activity of rabbit rChn measured with the milk of donkey (Equus asinus), cow, goat, sheep (Ovis aries) and camel was 40.0, 10.0, 5.7, 3.1, and 2.7 IMCU/mL, respectively, i.e., the minimum and maximum values differ by more than an order of magnitude [24]. It can be expected that as novel species of Chn are obtained, the number of such conflicts will increase.
The “bovine Chn–camel milk” paradox allows conditionally dividing chymosins into two types: “universal”, capable of coagulating both homologous and heterologous milk equally effectively [7, 21, 25–28] and “specialized”, which showing the greatest activity in relation to homologous milk or milk of the closest phylogenetic relatives [7, 15, 23].
It should be noted that apart from the rabbit (order Lagomorpha) [24] none of the mammalian species outside the order Cetartiodactyla has become a source of genes for obtaining and evaluating the biochemical properties of novel rChns. Partially characterized Chn of representatives of the order Primates (marmosets Callithrix jacchus) [29] and some other New World monkeys [30] stand apart because they have lost the important chymosin function of participation in the transfer of passive immunity factors from mother to newborn and are expressed in nature as pepsin-like enzymes with broad proteolytic specificity and high total PA.
Data on MCA and specificity of Chn with respect to heterologous substrates are necessary to understand the mechanism of action of these enzymes and to search for highly effective milk coagulants. In this regard, it is relevant to study the rChn of animals belonging to different orders of the class Mammalia.
The tree shrews (Scandentia) are the order of placental mammals that live in the tropical forests of Southeast Asia. The genome of the tupaia (Tupaia belangeri chinensis) or the Chinese tree shrew was sequenced in 2013. Phylogenetic analysis has shown that Tupaia belangeri chinensis (hereinafter referred to as T. belangeri) is a very distant relative of the cow and clusters within the Euarchonta clade, which also includes Dermoptera and Primates. At the same time, the structure of the tupaia genes has a high similarity with Primates and, in particular, with humans [31]. According to the NCBI, the amino-acid (a.a.) sequences of tupaia and bovine Chn have 81.6% identity. Tupaia rChn has not been studied so far.
The goal of the present work was to obtain the rChn of T. belangeri, to determine its coagulation activity on cow’s milk, and to study some biochemical properties important for cheese making.
MATERIALS AND METHODS
Construction of an expression vector. We used the nucleotide sequence of tupaia prochymosin (ProChn) found in the genome of T. chinensis (NW_006159751.1) by comparison with the reference sequence of the bovine Chn gene. The codon composition of the ProChn nucleotide sequence was optimized for expression in the Escherichia coli system using the Codon Optimization Tool service (Integrated DNA Technologies, United States). The synthesized nucleotide sequence was inserted into the pET21a expression vector (Novagen, Germany) at the unique BamHI and HindIII restriction sites. As a result, a plasmid vector pET21-CYM-Tup was obtained.
Obtaining tupaia rChn. Plasmid pET21-CYM-Tup was used for chemical transformation of E. coli BL21(DE3) cells. Colonies containing recombinant plasmids were cultured for a night using an orbital shaker (Biosan, Latvia) at 180 rpm in LB medium (AppliChem, United States) at 37°C. The inoculum at a ratio of 1 : 100 was transferred into Erlenmeyer flasks containing fresh LB medium and grown to an optical density of 0.8 (at λ = 600 nm). An inductor, isopropyl β-D-1-thiogalactopyranoside (IPTG) (Anatrace Products, United States), was added to the culture liquid to a final concentration of 1 mM and then the producer strain was additionally cultivated using a shaker (180 rpm) for 12 h at 25°C. A culture of the strain E. coli BL21 not transformed with the pET21-CYM-Tup plasmid was used as a control.
After completion of cultivation, the biomass was precipitated by centrifugation for 20 min at 5000 g and 4°C. The pellet was resuspended in STET buffer (8% sucrose, 50 mM Tris, 20 mM EDTA, 5% Triton X-100, pH 8.0) (20 mL of the buffer per 1 g of biomass) and incubated for a night at 4°C. At the end of the incubation, the cell suspension was sonicated (2000 W/L and 283 W/cm2) for 1 min, after which the mixture was cooled to 4°C. The procedure was repeated three times. The inclusion bodies were precipitated by centrifugation at 20 000 g for 20 min at 4°C.
Solubilization of inclusion bodies, refolding of recombinant ProChn, and subsequent activation of the zymogen were performed according to the procedures described in the work [26]. As a result of activation, a preparation of tupaia rChn (rChn-Tup) was obtained.
Determination of total and specific milk-clotting activity. Combined unpasteurized cow’s milk was used as a substrate, into which NaN3 was added to the concentration of 0.02% and the pH was adjusted to 6.5. The substrate (2.5 mL), heated in a water bath at 35°C for at least 10 min, was quickly mixed with 0.2 mL of the studied rChn, and the time of formation of the first coagulate flakes was recorded. A 0.5% aqueous solution of a dry commercial preparation of bovine rChn (Chr. Hansen, Denmark) with a declared MCA of 2201 IMCU/g (IMCU, International Milk Clotting Units) was used as a standard. All measurements were repeated at least three times (n ≥ 3). The total MA was expressed in IMCU/mL, which was calculated using the formula (1):
where MASt is the declared MCA of the standard (IMCU/g); 200 is the dilution factor (mL/g); T1 is the time (s) for substrate coagulation by the standard; and T2 is the time (s) of substrate coagulation by the solution of the studied enzyme.
A 0.5% aqueous solution of a dry commercial bovine rChn preparation (rChn-Bos) and 0.4% aqueous solution of dry commercial preparation of dromedary camel rChn (rChn-Cam) (Chr. Hansen, Denmark) were used as reference preparations.
The specific MCA of rChns was calculated after determining total MCA and protein concentration according to Bradford [32] and expressed in IMCU/mg.
Determination of nonspecific proteolytic activity. A 0.5% solution of bovine sodium caseinate (Sigma, United States) in 20 mM Na acetate buffer (pH 5.65) was used as a substrate. The substrate (2.0 mL) was incubated for 15 min in a water bath at 35°C, a solution of the studied rChn (0.5 mL) was added and quickly mixed, and the start time of incubation was noted. After 30, 90 and 180 min, 2.5 mL of 6% trichloroacetic acid (TCA) was added to 2.5 mL of the enzyme-substrate mixture, stirred, incubated for 30 min at room temperature, and filtered through a paper filter. The absorbance was determined in the filtrate at a wavelength of 280 nm (A280). To prepare the spectrophotometric control, the components of the enzyme-substrate mixture were added directly to 6% TCA. Nonspecific PA was expressed in A280 units. The curve of the dependence of A280 on the duration of heating was built.
The coagulation specificity (proteolytic activity aimed only at hydrolysis of the Chn-sensitive bond in the к-CN molecule) was determined as the ratio of specific MCA and nonspecific PA (MCA/PA). To assess the coagulation specificity of rChn preparations, PA was taken as the A280 values of enzyme-substrate mixtures incubated for 180 min.
Determination of proteolytic activity in relation to isolated bovine α-, β- and κ-caseins. Determination of PA of rChn in relation to isolated caseins (CN) of cow milk was carried out according to the method described in the work [33], which was modified. To determine PA, tupaia rChn was concentrated by ultrafiltration to a total MCA of ≈ 8 IMCU/mL. The activity of the reference preparations (commercial rChn) was normalized according to the MCA of the tupaia enzyme.
Next, 0.1% solutions of α-, β-, and κ-CN substrates (Sigma, C6780, C6905, C0406, United States) in 20 mM Na-acetate buffer (pH 5.65) were prepared; 250 μL of the substrate solution was mixed with 5 μL of the studied rChn with an activity of 8.0 IMCU/mL. Control samples were supplemented with 5 μL of 20 mM Na-acetate buffer (pH 5.65) instead of rChn. Enzyme-substrate mixtures and control preparations were incubated at 35°C for 1 h. After the incubation, the polypeptide composition of the mixtures was studied by electrophoresis in the presence of sodium dodecyl sulfate (SDS) according to Laemmli [34]. The LMW-SDS Marker Kit (GE Healthcare, United States) was used as molecular-weight (MW) markers.
Thermal stability (TS). Solutions of the investigated MCE were heated at temperatures of 30–65°C for 30 min, quickly cooled to room temperature, and their residual MCA was determined. The MCA values obtained in samples heated at 30°C were taken as 100%. We plotted the dependence of the residual MCA on the heating temperature. The thermal inactivation threshold was considered the heating temperature at which rChn lost ≥20% of the initial MCA.
Determination of the parameters of the Michaelis–Menten kinetics. The Michaelis constant (Km), the rate constant of the hydrolysis reaction or the turnover number of the enzyme (kcat), and the catalytic efficiency (kcat/Km) were determined according to the procedure described in the work [35].
Statistical processing of the obtained data was carried out using Excel 2007 software (Microsoft Corporation, United States). The results of the determination of quantitative variables are presented as the arithmetic mean (M) with an indication of the standard deviation (±SD).
RESULTS AND DISCUSSION
Obtaining the tupaia rChn preparation. To obtain a recombinant analog of tupaia Chn, a system based on the strain E. coli BL21(DE3) and a plasmid expression vector of the pET21 series was used. The tupaia ProChn nucleotide sequence, with a codon composition optimized for expression in the E. coli system, was cloned into the pET21a expression vector in a such way that the N-terminal portion contained T7 expression tag sequence. The correctness of the inserted sequence was verified by sequencing the resulting recombinant plasmid pET21-CYM-Tup in the insertion region. After transformation of the competent E. coli BL21(DE3) cells with the constructed plasmid, a recombinant producer strain was obtained. To produce the target protein, a standard protocol was used, including the addition of an IPTG inducer. To determine the efficiency of the synthesis of recombinant tupaia ProChn (rProChn-Tup), as well as to determine its localization, electrophoretic analysis of various protein preparations obtained from cells of the recombinant producer strain was carried out (Fig. 1).
The results of SDS-electrophoresis of protein preparations obtained from the cells of the original strain and the producer strain: 1, biomass of the strain E. coli BL21 biomass after addition of the inducer (control); 2, biomass of producer E. coli BL21_rProChn-Tup cells after addition of the inductor (experiment); 3, soluble fraction of the biomass of the producer BL21_rProChn-Tup after processing with STET buffer; 4, insoluble fraction (inclusion bodies) after treatment with lysis buffer; 5, rChn-Tup after activation; 6, molecular weight markers.
Analysis of the protein composition of E. coli cells containing the pET21-CYM-Tup plasmid after the addition of the inductor showed a high protein content, the electrophoretic mobility of which coinciding with that calculated for rProChn-Tup (41 kDa). Its content was 50% or higher (Fig. 1, lane 2) of the total amount of cell proteins. It was shown that the soluble fraction of E. coli biomass after treatment with STET buffer and centrifugation (Fig. 1, lane 3) almost did not contain the target protein, while the fraction of inclusion bodies was almost completely represented by tupaia rProChn (Fig. 1, lane 4). Thus, the conditions for the expression of the tupaia Chn gene in the E. coli system led to overproduction of the target protein, the vast majority of which accumulated in an insoluble form in the inclusion bodies; therefore, the next step was the renaturation of rProChn by stepwise dialysis. Attempts to activate the zymogen before dialysis were unsuccessful: the preparations did not demonstrate coagulation activity. After dialysis, activated rProChn-Tup preparations showed enzymatic activity, which indicated that the zymogen restores the correct tertiary structure. After the activation, three preparations of rChn-Tup (series 1–3) were obtained, which differed in total and specific MCA (Table 1).
Milk-clotting activity. The assessment of the biochemical properties of any new MCE begins with the determination of its MCA in relation to cow milk, the main raw material for the production of rennet cheeses.
The total MCA of the resulting rChn-Tup preparations varied within 0.7–5.3 IMCU/mL. The protein concentration in the rChn-Tup preparations was, on average, approximately an order of magnitude higher than in the reference preparations. As a result, in terms of specific MCA, rChn-Tup was 50–60 times inferior to the commercial rChn of cows and dromedaries (Table 1).
The low specific MA may be explained by inefficient refolding of rChn-Tup. It is known that the restoration of the correct three-dimensional structure is a problematic step in obtaining rChn in E. coli expression systems [36–39]. Some rChn-Tup molecules probably remained enzymatically inactive after renaturation, which led to a decrease in the specific MCA of the enzyme.
Thus, it was found that rChn-Tup obtained in the E. coli expression system coagulated a heterologous biological substrate, cow’s milk, but in terms of specific MCA it was inferior to B. taurus and C. dromedarius rChn synthesized in the production system of the higher mold fungus Aspergillus niger var. Awamori.
The trigger initiating the enzymatic coagulation of cow milk was the hydrolysis of the Chn-sensitive bond F105-M106 in the κ-CN molecule and the removal of the glycomacropeptide from the surface of casein micelles. The ability of rChn-Tup to coagulate B. taurus milk indicates that the enzyme is specific to at least one of the peptide bonds (presumably F105-M106) in the к-CN molecule.
Nonspecific proteolytic activity and the MCA/PA ratio. The proteolytic activity of Chn is the basis of its ability to coagulate milk. Conventionally, the PA of enzymes that coagulate milk can be divided into specific (milk-clotting) and nonspecific one. The specific PA (i.e., MCA) provides hydrolysis of the Chn-sensitive bond F105-M106 in the κ-CN molecule, which leads to destabilization of casein micelles and formation of a milk clot. The nonspecific PA characterizes the ability of MCE to hydrolyze any peptide bonds for which it is specific, except for the Chn-sensitive bond of κ-CN.
In scientific literature devoted to the problems of studying and practical application of MCE, the ratio of its MCA and nonspecific PA (MCA/PA) is used to assess the technological efficiency of a milk coagulant [7, 28]. The ratio characterizes the activity aimed only at the hydrolysis of the Chn-sensitive bond in the κ-CN molecule and leading to milk coagulation. To avoid confusion with enzymatic specificity in the present work, we have designated the MCA/PA ratio by the term coagulation specificity. An ideal milk coagulant for cheese making should exhibit maximum MCA with minimum total PA [5]. The higher the coagulation specificity, the more versatile the MCE and the wider the range of cheeses for which it can be used.
Since one of the objectives of this study was to assess the industrial prospects of rChn-Tup, a substrate of bovine origin was used to determine its nonspecific PA (as well as to determine MCA). The nonspecific PA was assessed by A280 of supernatants obtained after incubation and sedimentation of the components of enzyme-substrate mixtures with 3% TCA. The pool of TCA-soluble components also contains the product of hydrolysis of the Chn-sensitive bond - glycomacropeptide k-CN (f.106-169), which does not affect the result of the determination of nonspecific PA, since it does not contain a.a. residues absorbing at 280 nm [7].
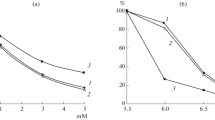
At all stages of incubation of enzyme–substrate mixtures, rChn-Tup showed a higher PA in relation to bovine CN than camel and bovine rChn (Fig. 2).
After 180 min of incubation, the nonspecific PA rChn of tupaia, cow, and camel was 0.544, 0.418, and 0.056 A280 units, respectively. According to the dynamics of the increase in the concentration of products of nonspecific proteolysis, bovine and tupaia rChn were similar and differed markedly from the rChn of the camel. The observed low nonspecific PA rChn of the dromedary camel is its characteristic (marker) biochemical property and has been confirmed by other studies [7, 27].
To compare the coagulation specificity of rChn-Tup and commercial rChn, data on their specific MCA (%) and nonspecific PA (%) were used, and bovine rChn values were taken as 100% (Table 2). It was shown that, in terms of the MCA/PA ratio, the rChn-Tup preparation with the highest specific MCA (series 1) was catastrophically inferior to the reference enzymes.
The low coagulation specificity of the tupaia enzyme was the result of a low specific MCA, which may be due to incomplete refolding of its zymogen isolated from inclusion bodies. It is possible that the use of a eukaryotic expression system instead of a prokaryotic one may increase the yield of active rChn-Tup, its specific MCA, and, as a consequence, the MCA/PA ratio.
Thus, the nonspecific PA of rChn-Tup obtained in the prokaryotic production system was 30 and 117% higher than that of rChn-Bos and rChn-Cam, respectively. The high nonspecific PA limits the use of rChn-Tup to the production of fast-ripening cheeses with short (no more than 14 days) shelf lives.
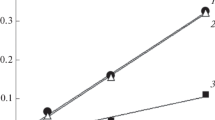
Proteolytic activity against bovine α-, β-, and κ-caseins. As follows from the data presented in Fig. 3, rChn- was able to hydrolyze isolated bovine α-, β-, and κ-CN. Under the experimental conditions, the genetically engineered tupaia Chn effectively attacked not only bovine κ-CN, but also its hydrolysis product, para-κ-CN (band with MM ≈ 16 kDa). Therefore, components with MM < 14 kDa accumulated in the enzyme-substrate mixture (Fig. 3, lane 6). Recombinant Chn of B. taurus completely degraded κ-CN and partially degraded para-κ-CN. In this case, a product with MM ≈ 14 kDa appeared in the enzyme-substrate mixture (Fig. 3, lane 7). In contrast to the tupaia and cow, PA of C. dromedarius rChn was directed mainly to κ-CN and almost did not affect para-κ-CN (Figs. 3, lane 8).
Proteolytic activity of tupaia, bovine, and camel rChn in relation to bovine α-, β-, and κ-CN: 1, α-CN; 2, β-CN; 3, κ-CN; 4, MW markers; 5, κ-CN (control); 6, κ-CN + rChn-Tup; 7, κ-CN + rChn-Bos; 8, κ-CN + rChn-Cam; 9, α-CN (control); 10, α-CN + rChn-Tup; 11, α-CN + rChn-Bos; 12, α-CN + rChn-Cam; 13, β-CN (control); 14, β-CN + rChn-Tup; 15, β-CN + rChn-Bos; 16, β-CN + rChn-Cam. The arrows indicate the position of para-κ-CN (MW ≈ 16 kDa) on the lanes 6–8.
All studied enzymes showed PA in relation to isolated bovine α-CN. The main α-CN band was retained after 60 min of incubation with all enzymes, but in the case of rChn-Tup, its intensity was the lowest (Fig. 3, lanes 10–12). This indicated a higher PA of the tupaia enzyme in relation to isolated α-CN, as compared to commercial rChn.
All studied rChn differed in the intensity of proteolysis of free bovine β-CN. Recombinant tupaia and bovine chymosins completely hydrolyzed β-CN, with the formation of polypeptides with MM in the range of 20–26 kDa, while camel rChn had the least proteolytic activity in relation of this substrate (Fig. 3, lanes 14–16).
The electrophoresis method has already been used to study the PA of bovine and camel rChn in relation to individual fractions of bovine CN [24, 33, 40]. According to [33, 40], bovine and camel rChn obtained in the P. pastoris expression system had PA only with respect to κ-CN, but did not hydrolyze bovine α- and β-CN. Such a contradiction with our data can be explained by differences in the design of experiments to assess the specificity of rChn (purity and MCA of rChn, ratio of enzyme and substrate, composition of buffers, pH, temperature and duration of incubation of enzyme-substrate mixtures). In general, our results are in agreement with the data of [24], whose authors studied the substrate specificity of commercial bovine and camel rChn (Chr. Hansen, Denmark) obtained in the A. niger expression system.
Thus, compared to commercial genetically engineered chymosins, rChn-Tup causes a deeper proteolytic degradation of isolated bovine CN fractions. According to the intensity of the proteolytic action on bovine α-, β-, and κ-CN, the enzymes can be arranged as follows: rChn-Tup > rChn-Bos > rChn-Cam.
It can be assumed that when using total (micellar) CN or cow milk preparations rather than isolated CN fractions as substrates, the pattern of proteolysis and assessment of rChn specificity may differ. If the substrate is in the form of micelles, outwardly exposed C-terminal regions of κ-CN forming a hairy layer are easily accessible for proteolytic attack by Chn (or other MCEs). On the contrary, it is much more difficult for MCE to attack the N-terminal regions of κ-CN, as well as α- and β-CN molecules located under the hairy layer and in the bulk of the micelle. In this regard, informativeness of the results of determining the PA of chymosins using isolated α-, β-, and κ-CN is questionable. Perhaps, for a more adequate assessment of the new rChn, it is advisable to study its specificity using a milk substrate, in which CN are in the micellar form.
Thermal stability. Thermal stability is the most important characteristic of any industrial enzyme. At the same time, enzyme preparations with different TC ranges are required for different technological processes. For example, for the production of cheeses with a high temperature of the second heating and long periods of maturation and storage, milk coagulants that are stable at temperatures of 50°C or less are in demand. A milk-clotting enzyme with high TC, which remains active after cheese grain is heated to 52–58°C, can exhibit undesirable PA in cheese and worsen its physicochemical and organoleptic parameters [41, 42].
The scope of application of milk coagulants that remain PA at temperatures higher than 60°C is limited and applies mainly to the production of cheeses with short maturation and storage periods. For the production of semi-hard and hard cheeses that require long periods of ripening and storage, thermolabile MCEs are preferred. Reducing the TC of a promising milk coagulant may require the development of special technological methods. For example, in order to maximally adapt natural milk-clotting endopeptidases (mucorpepsins) of mold fungi of the genus Rhizomucor for the industrial cheese making, methods for their chemical modification were specially developed to reduce its initially high TC [43, 44].
Modern technological rChns differ in TS. According to [7, 45], the rChn of the dromedary camel is more thermally stable than the bovine rChn and retains a higher residual MCA in the temperature range of 50–60°C. The melting points of cow and camel rChn are 57.7 and 60.7°C, respectively [9]. A clear demonstration of the importance of differences of 3°C between the thermal denaturation thresholds of industrial MCE was presented in [41]. It was shown that an increase in the curd heating temperature from 50 to 56°C led to a significant decrease in the concentration of αs1-CN proteolysis products in Reggianito-type cheeses produced using bovine and camel rChn, due to a deeper thermal inactivation of MCE. At the same time, in cheeses produced with a curd heating temperature of 56°C and using more thermostable camel rChn, the intensity of proteolysis was higher than when using bovine rChn [41]. Thus, camel rChn, which has a four times lower level of total PA than bovine rChn but exceeds it in TC, showed a higher nonspecific PA in ripening and stored cheeses.
Compared to commercial and experimental genetically engineered chymosins [25], rChn-Tup has an unusually low TC (Fig. 4). The threshold of thermal inactivation of rChn-Tup was 45°C. At this temperature, its coagulation ability decreased by more than 40%, and at 50°C, the enzyme lost more than 90% of the initial MCA. For commercial coagulants, the thermal inactivation threshold was 10°C higher and amounted to 55°C. At the same time, rChn-Bos was completely inactivated at 60°C, while rChn-Cam was completely inactivated at 65°C.
It is possible that the extraordinary thermolability of rCh-Tup is associated with the peculiarities of thermoregulation in animals of this group of mammals. According to [46], the average body temperature of T. belangeri is 39.6°C. It is also known that tupaia is characterized by diurnal fluctuations in body temperature, synchronized with the rhythms of activity. The amplitude of oscillations is unusually high and is about 5°C, which is much greater than in most other mammals studied. According to telemetry data, the minimum body temperature of the tupaia is 35–36°C, while the maximum is 40–41°C [47]. In this regard, the extremely low TS of tupaia Chn may be an element of protection of passive immunity factors from excessive PA of this gastric proteinase. It can be assumed that after the activation of the zymogen in the stomach lumen of the newborn, the thermolabile tupaia Chn has time to coagulate the milk contained in it, but is further thermally inactivated and does not have a deep proteolytic effect on Ig and other protective proteins. As a result, passive immunity factors contained in milk enter the bloodstream and provide immunological protection for the newborn due to the mechanisms of direct transcellular transfer in the intestinal epithelium.
Thus, compared with bovine and camel rChn, genetically engineered tupaia Chn is an exclusively thermolabile MCE. As for the cheese-making prospects of rChn-Tup, it is possible that its low TS can, at least partially, compensate for the negative effect of high PA and allow more widespread use of this enzyme. For example, in the production of cheeses with maturation and storage periods of 30–45 days.
Michaelis–Menten kinetics parameters. To determine the main parameters of enzymatic kinetics, the most active among 3 rChn-Tup preparations was used (series 1).
The main parameters of the Michaelis–Menten kinetics of Chn-Tup differ markedly from the values of the kinetic constants obtained for commercial rChn (Table 3). Compared to rChn-Bos and rChn-Cam, the enzyme of T. belangeri demonstrated the lowest Km values, indicating a higher affinity for the chromogenic substrate used. In contrast to the reference enzymes, reaction products dissociated from rChn-Tup 2.3–2.5 times faster and the enzyme bound the substrate again, as indicated by higher values of the enzyme turnover number (kcat) and its catalytic efficiency (kcat/Km). Thus, the kinetic constants of rChn-Tup indicated that its enzymatic efficiency with respect to a synthetic fluorogenic substrate that mimics the Chn-sensitive site of bovine κ-CN is higher than that of modern commercial rChn.
In order to understand the reasons for the high catalytic efficiency of the genetically engineered tupaia Chn, primary structures of B. taurus, C. dromedarius, and T. belangeri κ-CN were compared. A mature bovine κ-CN consists of 169 a.a. (Chn-sensitive bond is located in the position F105-M106), camel κ-CN consists of 162 a.a. (Chn-sensitive bond is in the position F97-I98), while tupaia κ-CN contains 164 a.a. (suggested Chn-sensitive bond is in the position L98-K99). Surprisingly, despite the 81.6% identity of tupaia and bovine Chn, the similarity of the primary structure of κ-CN of these species was one of the lowest among those already determined, and amounted to only 41.9%. In turn, the a.a. sequences of bovine and camel κ-CN were 58.4% identical. Thus, the κ-CN of cow, tupaia, and camel demonstrated significant interspecies differences.
The amino-acid sequences of the Chn-sensitive regions of tupaia and bovine α-CN coincided by 25%. They differed in 18 out of 24 amino-acid residues, including two substitutions at the key peptide bond site: F105 (B. taurus) → L98 (T. belangeri), where the nonpolar aromatic radical was replaced by a nonpolar aliphatic one, and M106 (B. taurus) → Lys99 (T. belangeri), where hydrophobic sulfur-containing radical was replaced by hydrophilic positively charged a.a. residue (Fig. 5).
Amino acid sequences of Chn-sensitive regions of bovine, camel, and tupaia κ-CN. The parentheses after the species name indicate the degree of homology of the primary structure of the κ-CN of the given species in relation to the bovine κ-CN. Sequence 105–106 in the molecule of bovine κ-CN is marked in green, amino acid substitutions in tupaia and camel κ-CN compared to bovine κ-CN are marked in yellow, a.a. residues that coincide in all three sequences are marked in blue. Arrows indicate peptide bonds hydrolyzed by homologous and heterologous chymosins. Sign ? indicate the putative Chn-sensitive peptide bond of the tupaia κ-CN. The primary structures of k-caseins are from the GenBank database (T. belangeri – XM_006142970; B. taurus – AY380228.1; C. dromedarius – NP_001290489.1).
By comparison, the identity of 96-119 regions of bovine and cow κ-CN was 62.5%. It could be expected that the significant differences in the primary structure of similar regions of bovine and tupaia κ-CN would lead to a decrease in the affinity of T. belangeri rChn with the peptide that mimics the Chn-sensitive region of the heterologous substrate. However, in practice, we observed the opposite situation: in terms of the rate of saturation with the substrate, tupaia rChn exceeded bovine and camel enzymes by 1.9 and 3.2 times, respectively. It can be assumed that other subsites of specificity were involved in the binding of rChn-Tup and bovine κ-CN than in the case of bovine and camel rChn.
Since the structures of the Chn-sensitive regions of the κ-CN of the cow and the tupaia were significantly different, the question of the site of hydrolysis of the bovine substrate remains open. It is known that MCE endothiapepsin (EC 3.4.23.22), which is used to a limited extent in cheese making, does not hydrolyze the F105-M106 key bond in bovine κ-CN, but the preceding S104-F105 sequence, where the uncharged polar R-group is in position P1. This was despite the fact that the specificity of endothiapepsin was close to the specificity of porcine pepsin, i.e., hydrophobic R-groups of amino acids in position P1 and P1' were preferable for hydrolysis [48, 49]. Therefore, based on the analysis of the kinetics of the hydrolysis of the synthetic substrate, it can only be stated that rChn-Tup attacked bovine κ-CN in the region 96–119 so effectively that it caused milk coagulation. Additional studies are needed to accurately determine the site of hydrolysis of bovine κ-CN by rChn-Tup.
The features of the a.a. sequences of κ-CN of T. belangeri also raised a number of other questions related to the substrate specificity of Chn of different species, for example: what peptide bond is hydrolyzed by homologous and heterologous Chns in the tupaia κ-CN molecule and what is the specificity of rChn-Tup with respect to the Chn-sensitive κ-CN sites in animals belonging to different orders of mammals?
CONCLUSIONS
Thus, in the E. coli expression system a genetically engineered tupaia Chn was obtained and partially characterized for the first time. It was shown that despite significant differences in the structure of B. taurus and T. belangeri κ-CN, especially in the region of the Chn-sensitive sequence, rChn-Tup was able to coagulate cow milk. The nonspecific PA of the tupaia enzyme in relation to the total fraction of bovine κ-CN was 30 and 117% higher than that of bovine and camel rChn, respectively. Compared with commercial genetically engineered chymosins (C. dromedarius and B. taurus), tupaia rChn showed increased PA relative to isolated α-, β-, and κ-CN of cow milk. The thermal inactivation threshold of rChn-Tup was 45°C, which was significantly lower than that of the vast majority of commercial and experimental rChns. The reasons for the extraordinary thermal lability of rChn-Tup should be the subject of a separate study, since its results may be of importance for the engineering of industrial MCE, for directed changes in their thermal resistance. The ability of genetically engineered tupaia Chn to coagulate cow’s milk, along with high catalytic efficiency with respect to a peptide that mimics the Chn-sensitive site of bovine κ-CN, makes it possible to classify this enzyme as a universal milk coagulant that may be of potential interest for cheese making. For a complete biochemical characterization of rChn-Tup, an extended study of its specificity with respect to homologous and heterologous κ-CN is required.
The authors hope that the results of this work will intensify the search and study of novel universal chymosins among species that do not belong to the order Cetartiodactyla.
REFERENCES
Foltmann, B., Scand. J. Clin. Lab. Invest., 1992, vol. 52, pp. 65–79.
Lopes-Marques, M., Ruivo, R., Fonseca, E., Teixeira, A., and Castro, L.F.C., Mol. Phylogenet. Evol., 2017, vol. 116, pp. 78–86.
Bansal, N., Drake, M.A., Piraino, P., Broe, M.L., Harboe, M., Fox, P.F., et al., Int. Dairy J., 2009, vol. 19, no. 9, pp. 510–517.
Fox, P.F. and McSweeney, P.L.H., Cheese: Chem., Phys. Microbiol., 2004, vol. 1, pp. 1–18.
Harboe, M., Broe, M.L., and Qvist, K.B., Technol. Cheesemaking, 2010, pp. 98–129.
Flamm, E.L., Nat. Biotechnol., 1991, vol. 9, no. 4, p. 349.
Kappeler, S.R., Rahbek-Nielsen, H., Farah, Z., Puhan, Z., Hansen, E.B., and Johansen, E., Biochem. Biophys. Res. Commun., 2006, vol. 342, no. 2, pp. 647–654.
Borsting, M.W., Qvist, K.B., Rasmussen, M., Vindelov, J., Vogensen, F.K., and Ardo, Y., Dairy Sci. Technol., 2012, vol. 92, no. 5, pp. 593–612.
Jensen, J.L., Molgaard, A., Poulsen, J.-C.N., Harboe, M.K., Simonsen, J.B., Lorentzen, A.M., et al., Acta Crystallogr., Sect. D: Biol. Crystallogr., 2013, vol. 69, no. 5, pp. 901–913.
Jensen, J.L., Jacobsen, J., Moss, M.L., Rasmussen, F., Qvist, K.B., Larsen, S., et al., J. Dairy Sci., 2015, vol. 98, no. 5, pp. 2853–2860.
Ansari, S.M., Coletta, A., Skeby, K.K., Sorensen, J., Schiott, B., and Palmer, D.S., J. Phys. Chem. B, vol. 120, no. 40, pp. 10453–10462.
Ansari, S.M., Sorensen, J., Schiott, B., and Palmer, D.S., Proteins Struct. Funct. Bioinf., 2018, vol. 86, no. 1, pp. 75–87.
Sorensen, J., Palmer, D.S., Qvist, K.B., and Schiott, B., J. Agric. Food Chem., 2011, vol. 59, no. 10, pp. 5636–5647.
Sorensen, J., Palmer, D.S., and Schiott, B., J. Agric. Food Chem., 2013, vol. 61, no. 33, pp. 7949–7959.
Uniacke-Lowe, T. and Fox, P.F., Cheese, 2017, 4th ed., pp. 69–113.
Bayoumi, S., Kieler Milchwirtsch Forschungsberichte, 1990, vol. 42, no. 1, pp. 3–8.
Wangoh, J., Farah, Z., and Puhan, Z., Milchwissenschaft, 1993, vol. 48, no. 6, pp. 322–325.
Saliha, B.H., Louis, L.C., Farida, M.M., Saliha, S.A., Nasma, M., Elkhir, S.O., et al., Emirates J. Food Agric., 2011, vol. 23, no. 4, pp. 301–310.
Farah, Z. and Bachmann, M.R., Milchwissenschaft, 1987, vol. 42, no. 11, pp. 689–692.
Kappeler, S., Farah, Z., and Puhan, Z., J. Dairy Res., 1998, vol. 65, no. 2, pp. 209–222.
Akishev, Z., Kiribayeva, A., Mussakhmetov, A., Baltin, K., Ramankulov, Y., and Khassenov, B., Heliyon, 2021, vol. 7, no. 5, p. e07137. https://doi.org/10.1016/j.heliyon.2021.e07137
Stasiuk, S.J., Summers, E.L., and Demmer, J., Reprod. Fertil. Dev., 2000, vol. 12, no. 4, pp. 215–222.
Kotts, C. and Jenness, R., J. Dairy Sci., 1976, vol. 59, no. 5, pp. 816–822.
Alihanoglu, S., Ektiren, D., and Karaaslan, M., Protein Expr. Purif., 2021, vol. 183, p. 105874. https://doi.org/10.1016/j.pep.2021.105874
Belenkaya, S.V., Balabova, D.V., Belov, A.N., Koval, A.D., Shcherbakov, D.N., and Elchaninov, V.V., Appl. Biochem. Microbiol., 2020, vol. 56, no. 4, pp. 363–372.
Belenkaya, S.V., Shcherbakov, D.N., Balabova, D.V., Belov, A.N., Koval, A.D., and Elchaninov, V.V., Appl. Biochem. Microbiol., 2020, vol. 56, no. 6, pp. 647–656.
Belenkaya, S.V., Rudometov, A.P., Shcherbakov, D.N., Balabova, D.V., Kriger, A.V., Belov, A.N., et al., Appl. Biochem. Microbiol., 2018, vol. 54, no. 6, pp. 569–576.
Filkin, S.Y., Chertova, N.V., Zatsepin, S.S., Sadykhov, E.G., Fedorov, A.N., and Lipkin, A.V., Appl. Biochem. Microbiol., 2021, vol. 57, no. 3, pp. 297–302.
Kageyama, T., Biochemistry, 2004, vol. 43, no. 48, pp. 15122–15130.
Kageyama, T., J. Biochem., 2000, vol. 127, no. 5, pp. 761–770.
Fan, Y., Huang, Z-Y., Cao, C-C., Chen, C-S., Chen, Y-X., Fan, D-D., et al., Nat. Commun., 2013, vol. 5, no. 1, p. 1426.
Bradford, M.M., Anal. Biochem., 1976, vol. 72, nos. 1–2, pp. 248–254.
Wang, N., Wang, K.Y., Li, G., Guo, W., and Liu, D., Protein Expr. Purif., 2015, no. 111, pp. 75–81.
Laemmli, U.K., Nature, 1970, vol. 227, no. 5259, pp. 680–685.
Belenkaya, S.V., Bondar, A.A., Kurgina, T.A., Elchaninov, V.V., Bakulina, A.Y., Rukhlova, et al., Biochemistry (Moscow), 2020, vol. 85, no. 7, pp. 781–791.
Wei, C., Tang, B., Zhang, Y., and Yang, K., Biochem. J., 1999, vol. 340, no. 1, pp. 345–351.
Chen, H., Zhang, G., Zhang, Y., Dong, Y., and Yang, K., Biochemistry, 2000, vol. 39, no. 40, pp. 12140–12148.
Wei, C., Zhang, Y., and Yang, K., J. Protein Chem., 2000, vol. 19, no. 6, pp. 449–456.
Eskandari, M.H., Hosseini, A., Alasvand Zarasvand, S., and Aminlari, M., Food Biotechnol., 2012, vol. 26, no. 2, pp. 143–153.
Jiang, X.P., Yin, M.L., Chen, P., and Yang, Q., J. Microbiol. Biotechnol., 2012, vol. 28, no. 5, pp. 2087–2093.
Costabel, L.M., Bergamini, C.V., Pozza, L., Cuffia, F., Candioti, M.C., and Hynes, E., J. Dairy Res., 2015, vol. 82, no. 3, pp. 375–384.
Upadhyay, V.K., McSweeney, P.L.H., Magboul, A.A.A., and Fox, P.F., Cheese: Chem., Phys. Microbiol., 2004, vol. 1, pp. 391–434.
Feijoo-Siota, L., Blasco, L., Rodriguez-Rama, J.L., Barros-Velazquez, J., de Miguel, T., Sanchez-Perez, A., et al., Recent Adv. DNA Gene Seq., 2014, vol. 8, no. 1, pp. 44–55.
UK Patent no. 4348482, 1982.
Vallejo, J.A., Ageitos, J.M., Poza, M., and Villa, T.G., J. Dairy Sci., 2012, vol. 95, no. 2, pp. 609–613.
Wang, J., Xu, X-L., Ding, Z-Y., Mao, R-R., Zhou, Q-X., Lv, L-B., et al., Zool. Res., vol. 34, no. E2, pp. E69–E74.
Refinetti, R. and Menaker, M., J. Exp. Zool., 1992, vol. 263, no. 4, pp. 453–457.
Bailey, D., Cooper, J.B., Veerapandian, B., Blundell, T.L., Atrash, B., Jones, D.M., et al., Biochem. J., 1993, vol. 289, no. 2, pp. 363–371.
Whitaker, J.R., Methods Enzymol., 1970, vol. 19, pp. 436–445.
Funding
The work was carried out within the framework of the state task of the Ministry of Science and Higher Education of the Russian Federation (FZMW-2020-0002, Development of producers of recombinant enzymes for cheese making).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The authors declare that they have no conflicts of interest. This article does not contain any studies involving animals or human participants performed by any of the authors.
Additional information
Translated by A. Bulaev
Rights and permissions
Open Access. This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Balabova, D.V., Belenkaya, S.V., Volosnikova, E.A. et al. Can Recombinant Tree Shrew (Tupaia belangeri chinensis) Chymosin Coagulate Cow (Bos taurus) Milk?. Appl Biochem Microbiol 58, 761–770 (2022). https://doi.org/10.1134/S0003683822060023
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0003683822060023