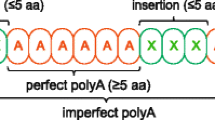
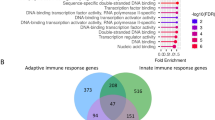
Abstract
Protein repeats are a source of rapid evolutionary and functional novelty. Repeats are crucial in development, neurogenesis, immunity, and disease. Repeat length variability and purity can alter the outcome of a pathway by altering the protein structure and affecting the protein−protein interaction affinity. Such rampant alterations can facilitate species to rapidly adapt to new environments or acquire various morphological/physiological features. With more than 11000 species, the avian clade is one of the most speciose vertebrate clades, with near-ubiquitous distribution globally. Explosive adaptive radiation and functional diversification facilitated the birds to occupy various habitats. High diversity in morphology, physiology, flight pattern, behavior, coloration, and life histories make birds ideal for studying protein repeats’ role in evolutionary novelty. Our results demonstrate a similar repeat diversity and proportion of repeats across all the avian orders considered, implying an essential role of repeats in necessary pathways. We detected positively selected sites (PSS) in the polyQ repeat of RUNX2 in the avian clade; and considerable repeat length contraction in the Psittacopasserae. The repeats show a species-wide bias towards a contraction in Galloanseriformes. Interestingly, we detected the length contrast of polyS repeat in PCDH20 between Galliformes and Anseriformes. We speculate the length variability of serine repeat and its interaction with β-catenin in the Wnt/β-catenin signaling pathway could have facilitated fowls to adapt to their respective environmental conditions. We believe our study emphasizes the role of protein repeats in functional/morphological diversification in birds. We also provide an extensive list of genes with considerable repeat length contrast to further explore the role of length volatility in evolutionary novelty and rapid functional diversification.




Similar content being viewed by others
REFERENCES
Marcotte E.M., Pellegrini M., Yeates T.O., Eisenberg D. 1999. A census of protein repeats. J. Mol. Biol. 293, 151–160.
Andrade M.A., Perez-Iratxeta C., Ponting C.P. 2001. Protein repeats: Structures, functions, and evolution. J. Struct. Biol. 134, 117–131.
Albà M.M., Tompa P., Veitia R.A. 2007. Amino acid repeats and the structure and evolution of proteins. In Gene and Protein Evolution. vol. 3. Basel: Karger, pp. 119–130.
Persi E., Wolf Y.I., Koonin E.V. 2016. Positive and strongly relaxed purifying selection drive the evolution of repeats in proteins. Nat. Commun. 7, 13570.
Karlin S., Brocchieri L., Bergman A., Mrázek J., Gentles A.J. 2002. Amino acid runs in eukaryotic proteomes and disease associations. Proc. Natl. Acad. Sci. U. S. A. 99, 333–338.
Toll-Riera M., Radó-Trilla N., Martys F., Albá M.M. 2012. Role of low-complexity sequences in the formation of novel protein coding sequences. Mol. Biol. Evol. 29, 883–886.
Schmitz-Linneweber C., Small I. 2008. Pentatricopeptide repeat proteins: A socket set for organelle gene expression. Trends Plant Sci. 13, 663–670.
Renault L., Nassar N., Vetter I., Becker J., Klebe C., Roth M., Wittinghofer A. 1998. The 1.7 Å crystal structure of the regulator of chromosome condensation (RCC1) reveals a seven-bladed propeller. Nature. 392 (6671), 97–101.
Jacobsen S.E., Binkowski K.A., Olszewski N.E. 1996. SPINDLY, a tetratricopeptide repeat protein involved in gibberellin signal transduction in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 93, 9292–9296.
Varela M., Diaz-Rosales P., Pereiro P., Forn-Cuní G., Costa M.M., Dios S., Romero A., Figueras A., Novoa B. 2014. Interferon-induced genes of the expanded IFIT family show conserved antiviral activities in non-mammalian species. PLoS One. 9, e100015.
Cerveny L., Straskova A., Dankova V., Hartlova A., Ceckova M., Staud F., Stulik J. 2013. Tetratricopeptide repeat motifs in the world of bacterial pathogens: Role in virulence mechanisms. Infect. Immun. 81, 629–635.
Lynch V.J., Wagner G.P. 2008. Resurrecting the role of transcription factor change in developmental evolution. Evolution. 62, 2131–2154.
Emili A., Greenblatt J., Ingles C.J. 1994. Species-specific interaction of the glutamine-rich activation domains of Sp1 with the TATA box-binding protein. Mol. Cell. Biol. 14, 1582–1593.
Pelassa I., Corà D., Cesano F., Monje F.J., Montarolo P.G., Fiumara F. 2014. Association of polyalanine and polyglutamine coiled coils mediates expansion disease-related protein aggregation and dysfunction. Hum. Mol. Genet. 23, 3402–3420.
Gemayel R., Chavali S., Pougach K., Legendre M., Zhu B., Boeynaems S., van der Zande E., Gevaert K., Rousseau F., Schymkowitz J., Babu M.M., Verstrepen K.J. 2015. Variable glutamine-rich repeats modulate transcription factor activity. Mol. Cell. 59, 615–627.
Fiumara F., Fioriti L., Kandel E.R., Hendrickson W.A. 2010. Essential role of coiled coils for aggregation and activity of Q/N-rich prions and PolyQ proteins. Cell. 143, 1121–1135.
Thirunavukkarasu K., Mahajan M., McLarren K.W., Stifani S., Karsenty G. 1998. Two domains unique to osteoblast-specific transcription factor Osf2/Cbfa1 contribute to its transactivation function and its inability to heterodimerize with Cbfβ. Mol. Cell. Biol. 18, 4197–4208.
Brown L.Y., Brown S.A. 2004. Alanine tracts: The expanding story of human illness and trinucleotide repeats. Trends Genet. 20, 51–58.
Malik I., Kelley C.P., Wang E.T., Todd P.K. 2021. Molecular mechanisms underlying nucleotide repeat expansion disorders. Nat. Rev. Mol. Cell Biol. 22, 589–607.
Mularoni L., Guigó R., Albà M.M. 2006. Mutation patterns of amino acid tandem repeats in the human proteome. Genome Biol. 7 (4), R33.
Wren J.D., Forgacs E., Fondon J.W., 3rd, Pertsemlidis A., Cheng S.Y., Gallardo T., Williams R.S., Shohet R.V., Minna J.D., Garner H.R. 2000. Repeat polymorphisms within gene regions: Phenotypic and evolutionary implications. Am. J. Hum. Genet. 67, 345–356.
Newton A.H., Pask A.J. 2020. Evolution and expansion of the RUNX2 QA repeat corresponds with the emergence of vertebrate complexity. Commun. Biol. 3, 771.
King D.G., Soller M., Kashi Y. 1997. Evolutionary tuning knobs. Endeavour. 21, 36–40.
Hancock J.M., Simon M. 2005. Simple sequence repeats in proteins and their significance for network evolution. Gene. 345, 113–118.
Fondon J.W., Garner H.R. 2004. Molecular origins of rapid and continuous morphological evolution. Proc. Natl. Acad. Sci. U. S. A. 101, 18058–18063.
Mularoni L., Ledda A., Toll-Riera M., Albà M.M. 2010. Natural selection drives the accumulation of amino acid tandem repeats in human proteins. Genome Res. 20, 745–754.
Kashi Y., King D.G. 2006. Simple sequence repeats as advantageous mutators in evolution. Trends Genet. 22, 253–259.
Schüler A., Bornberg-Bauer E. 2016. Evolution of protein domain repeats in metazoa. Mol. Biol. Evol. 33, 3170–3182.
Teekas L., Sharma S., Vijay N. 2022. Lineage-specific protein repeat expansions and contractions reveal malleable regions of immune genes. Genes Immun. 23 (7), 218‒234.
Mier P., Alanis-Lobato G., Andrade-Navarro M.A. 2017. Context characterization of amino acid homorepeats using evolution, position, and order. Proteins Struct. Funct. Bioinform. 85, 709–719.
Schweizer M., Liu Y. 2018. Avian Diversity and Distributions and Their Evolution Through Space and Time. Cham: Springer, pp. 129–145.
de Oliveira T.D., Kretschmer R., Bertocchi N.A., Degrandi T.M., de Oliveira E.H.C., Cioffi M. de B., Garnero A.D.V., Gunski R.J. 2017. Genomic organization of repetitive DNA in woodpeckers (Aves, Piciformes): Implications for karyotype and ZW sex chromosome differentiation. Lustig A.J., Ed. PLoS One. 12, e0169987.
Zhang G., Li C., Li Q., Li B., Larkin D.M., Lee C., Storz J.F., Antunes A., Greenwold M.J., Meredith R.W., Ödeen A., Cui J., Zhou Q., Xu L., Pan H., Wang Z., Jin L., Zhang P., Hu H., Yang W., Hu J., Xiao J., Yang Z., Liu Y., Xie Q., Yu H., Lian J., Wen P., Zhang F., Li H., Zeng Y., Xiong Z., Liu S., Zhou L., Huang Z., An N., Wang J., Zheng Q., Xiong Y., Wang G., Wang B., Wang J., Fan Y., da Fonseca R.R., Alfaro-Núñez A., Schubert M., Orlando L., Mourier T., Howard J.T., Ganapathy G., Pfenning A., Whitney O., Rivas M.V., Hara E., Smith J., Farré M., Narayan J., Slavov G., Romanov M.N., Borges R., Machado J.P., Khan I., Springer M.S., Gatesy J., Hoffmann F.G., Opazo J.C., Håstad O., Sawyer R.H., Kim H., Kim K.W., Kim H.J., Cho S., Li N., Huang Y., Bruford M.W., Zhan X., Dixon A., Bertelsen M.F., Derryberry E., Warren W., Wilson R.K., Li S., Ray D.A., Green R.E., O’Brien S.J., Griffin D., Johnson W.E., Haussler D., Ryder O.A., Willerslev E., Graves G.R., Alström P., Fjeldså J., Mindell D.P., Edwards S.V., Braun E.L., Rahbek C., Burt D.W., Houde P., Zhang Y., Yang H., Wang J.; Avian Genome Consortium; Jarvis E.D., Gilbert M.T., Wang J. 2014. Comparative genomics reveals insights into avian genome evolution and adaptation. Science. 346 (6215), 1311‒1320.
Andrews C.B., Mackenzie S.A., Gregory T.R. 2009. Genome size and wing parameters in passerine birds. Proc. R. Soc. B. 276, 55–61.
Schmid M., Nanda I., Guttenbach M., Steinlein C., Hoehn M., Schartl M., Haaf T., Weigend S., Fries R., Buerstedde J.M., Wimmers K., Burt D.W., Smith J., A’Hara S., Law A., Griffin D.K., Bumstead N., Kaufman J., Thomson P.A., Burke T., Groenen M.A., Crooijmans R.P., Vignal A., Fillon V., Morisson M., Pitel F., Tixier-Boichard M., Ladjali-Mohammedi K., Hillel J., Mäki-Tanila A., Cheng H.H., Delany M.E., Burnside J., Mizuno S. 2000. First report on chicken genes and chromosomes 2000. Cytogenet. Cell Genet. 90, 169–218.
Brusatte S.L., O’Connor J.K., Jarvis E.D. 2015. The origin and diversification of birds. Curr. Biol. 25, R888–R898.
Moen D., Morlon H. 2014. From dinosaurs to modern bird diversity: Extending the time scale of adaptive radiation. PLoS Biol. 12, e1001854.
Howe K.L., Achuthan P., Allen J., Allen J., Alvarez-Jarreta J., Amode M.R., Armean I.M., Azov A.G., Bennett R., Bhai J., Billis K., Boddu S., Charkhchi M., Cummins C., Da Rin Fioretto L., Davidson C., Dodiya K., El Houdaigui B., Fatima R., Gall A., Garcia Giron C., Grego T., Guijarro-Clarke C., Haggerty L., Hemrom A., Hourlier T., Izuogu O.G., Juettemann T., Kaikala V., Kay M., Lavidas I., Le T., Lemos D., Gonzalez Martinez J., Marugán J.C., Maurel T., Mc-Mahon A.C., Mohanan S., Moore B., Muffato M., Oheh D.N., Paraschas D., Parker A., Parton A., Prosovetskaia I., Sakthivel M.P., Salam A.I.A., Schmitt B.M., Schuilenburg H., Sheppard D., Steed E., Szpak M., Szuba M., Taylor K., Thormann A., Threadgold G., Walts B., Winterbottom A., Chakiachvili M., Chaubal A., De Silva N., Flint B., Frankish A., Hunt S.E., IIsley G.R., Langridge N., Loveland J.E., Martin F.J., Mudge J.M., Morales J., Perry E., Ruffier M., Tate J., Thybert D., Trevanion S.J., Cunningham F., Yates A.D., Zerbino D.R., Flicek P. 2021. Ensembl 2021. Nucleic Acids Res. 49, D884–D891.
Sayers E.W., Beck J., Brister J.R., Bolton E.E., Canese K., Comeau D.C., Funk K., Ketter A., Kim S., Kimchi A., Kitts P.A., Kuznetsov A., Lathrop S., Lu Z., McGarvey K., Madden T.L., Murphy T.D., O’Leary N., Phan L., Schneider V.A., Thibaud-Nissen F., Trawick B.W., Pruitt K.D., Ostell J. 2020. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 48 (D1), D9‒D16.
Jetz W., Thomas G.H., Joy J.B., Hartmann K., Mooers A.O. 2012. The global diversity of birds in space and time. Nature. 491 (7424), 444−448.
Harrison P.M. 2021. fLPS 2.0: Rapid annotation of compositionally-biased regions in biological sequences. PeerJ. 9, e12363.
Penn O., Privman E., Ashkenazy H., Landan G., Graur D., Pupko T. 2010. GUIDANCE: A web server for assessing alignment confidence scores. Nucleic Acids Res. 38, 23–28.
Edgar R.C. 2004. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinf. 5, 113.
Ge S.X., Jung D., Jung D., Yao R. 2020. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics. 36, 2628–2629.
Conway J.R., Lex A., Gehlenborg N. 2017. UpSetR: An R package for the visualization of intersecting sets and their properties. Hancock J., Ed. Bioinformatics. 33, 2938–2940.
R Core Team. 2021. R: A Language and Environment for Statistical Computing. https://search.gesis.org/publication/zis-RCoreTeam2021R.
Felsenstein J. 1985. Phylogenies and the comparative method. Am. Soc. Nat. 125, 1–15.
Revell L.J. 2012. Phytools: An R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 3, 217–223.
Yang Z. 2007. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591.
Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Žídek A., Potapenko A., Bridgland A., Meyer C., Kohl S.A.A., Ballard A.J., Cowie A., Romera-Pa-redes B., Nikolov S., Jain R., Adler J., Back T., Petersen S., Reiman D., Clancy E., Zielinski M., Steinegger M., Pacholska M., Berghammer T., Bodenstein S., Silver D., Vinyals O., Senior A.W., Kavukcuoglu K., Kohli P., Hassabis D. 2021. Highly accurate protein structure prediction with AlphaFold. Nature. 596, 583–589.
Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F.T., de Beer T.A.P., Rempfer C., Bordoli L., Lepore R., Schwede T. 2018. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303.
Goddard T.D., Huang C.C., Meng E.C., Pettersen E.F., Couch G.S., Morris J.H., Ferrin T.E. 2018. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25.
Selvatti A.P., Gonzaga L.P., Russo C.A.M. 2015. A Paleogene origin for crown passerines and the diversification of the Oscines in the New World. Mol. Phylogenet. Evol. 88, 1–15.
Ericson P.G.P., Irestedt M., Johansson U.S. 2003. Evolution, biogeography, and patterns of diversification in passerine birds. J. Avian Biol. 34, 3–15.
Vuckovic D., Dawson S., Scheffer D.I., Rantanen T., Morgan A., Di Stazio M., Vozzi D., Nutile T., Concas M.P., Biino G., Nolan L., Bahl A., Loukola A., Viljanen A., Davis A., Ciullo M., Corey D.P., Pirastu M., Gasparini P., Girotto G. 2015. Genome-wide association analysis on normal hearing function identifies PCDH20 and SLC28A3 as candidates for hearing function and loss. Hum. Mol. Genet. 24, 5655–5664.
Lees A.C., Haskell L., Allinson T., Bezeng S.B., Burfield I.J., Renjifo L.M., Rosenberg K. V, Viswanathan A., Butchart S.H.M. 2022. State of the World’s Birds. Ann-u. Rev. Environ. Resour. 47, 231–260.
Dyke G.J. 2001. The evolutionary radiation of modern birds: Systematics and patterns of diversification. Geol. J. 36, 305–315.
Tobias J.A., Ottenburghs J., Pigot A.L. 2020. Avian diversity: Speciation, macroevolution, and ecological function. Annu. Rev. Ecol. Evol. Syst. 51, 533–560.
Albà M.M., Guigó R. 2004. Comparative analysis of amino acid repeats in rodents and humans. Genome Res. 14, 549–554.
Coletta A., Pinney J.W., Solís D.Y.W., Marsh J., Pettifer S.R., Attwood T.K. 2010. Low-complexity regions within protein sequences have position-dependent roles. BMC Syst. Biol. 4, 43.
Inohara N., Chamaillard M., McDonald C., Nuñez G. 2005. NOD-LRR proteins: Role in host-microbial interactions and inflammatory disease. Annu. Rev. Biochem. 74, 355–383.
Meylan E., Tschopp J., Karin M. 2006. Intracellular pattern recognition receptors in the host response. Nature. 442, 39–44.
Cohn M. 2002. The immune system: A weapon of mass destruction invented by evolution to even the odds during the war of the DNAs. Immunol. Rev. 185, 24–38.
Kȩdzierski Ł., Montgomery J., Curtis J., Handman E. 2004. Leucine-rich repeats in host-pathogen interactions. Arch. Immunol. Ther. Exp. (Warsz.). 52, 104–112.
Lavoie H., Debeane F., Trinh Q.D., Turcotte J.F., Corbeil-Girard L.P., Dicaire M.J., Saint-Denis A., Pagé M., Rouleau G.A., Brais B. 2003. Polymorphism, shared functions and convergent evolution of genes with sequences coding for polyalanine domains. Hum. Mol. Genet. 12, 2967–2979.
Cocquet J., De Baere E., Caburet S., Veitia R.A. 2003. Compositional biases and polyalanine runs in humans. Genetics. 165, 1613–1617.
Caburet S., Demarez A., Moumné L., Fellous M., De Baere E., Veitia R.A. 2004. A recurrent polyalanine expansion in the transcription factor FOXL2 induces extensive nuclear and cytoplasmic protein aggregation. J. Med. Genet. 41, 932–936.
Lynch V.J., Wagner G.P. 2021. Cooption of polyalanine tract into a repressor domain in the mammalian transcription factor HoxA11. J. Exp. Zool., Part B. https://doi.org/10.1002/jez.b.23063
Eldon E., Kooyer S., D’Evelyn D., Duman M., Lawinger P., Botas J., Bellen H. 1994. The Drosophila 18 wheeler is required for morphogenesis and has striking similarities to Toll. Development. 120, 885–899.
Kobe B., Deisenhofer J. 1995. Proteins with leucine-rich repeats. Curr. Opin. Struct. Biol. 5, 409–416.
Staahl B.T., Tang J., Wu W., Sun A., Gitler A.D., Yoo A.S., Crabtree G.R. 2013. Kinetic analysis of npBA-F to nBAF switching reveals exchange of SS18 with CREST and integration with neural developmental pathways. J. Neurosci. 33, 10348–10361.
Hargreaves D.C., Crabtree G.R. 2011. ATP-dependent chromatin remodeling: Genetics, genomics and mechanisms. Cell Res. 21, 396–420.
Chi T. 2004. A BAF-centred view of the immune system. Nat. Rev. Immunol. 4, 965–977.
Chi T.H., Wan M., Zhao K., Taniuchi I., Chen L., Uttman D.R., Crabtree G.R. 2002. Reciprocal regulation of CD4/CD8 expression by SWI/SNF-like BAF complexes. Nature. 418, 195–199.
Pattenden S.G., Klose R., Karaskov E., Bremner R. 2002. Interferon-γ-induced chromatin remodeling at the CIITA locus is BRG1 dependent. EMBO J. 21, 1978–1986.
Eo S.H., Bininda-Emonds O.R.P., Carroll J.P. 2009. A phylogenetic supertree of the fowls (Galloanserae, Aves). Zool. Scr. 38, 465–481.
Chen C.K., Chuang H.F., Wu S.M., Li W.H. 2019. Feather evolution from precocial to altricial birds. Zool. Stud. 58, e24.
Meiri S., Murali G., Zimin A., Shak L., Itescu Y., Caetano G., Roll U. 2021. Different solutions lead to similar life history traits across the great divides of the amniote tree of life. J. Biol. Res. 28, 3.
Rennert J., Coffman J.A., Mushegian A.R., Robertson A.J. 2003. The evolution of Runx genes I. A comparative study of sequences from phylogenetically diverse model organisms. BMC Evol. Biol. 3, 1–11.
Mevel R., Draper J.E., Lie-a-Ling M., Kouskoff V., Lacaud G. 2019. RUNX transcription factors: Orchestrators of development. Development. 146 (17), dev148296.
Galant R., Carroll S.B. 2002. Evolution of a transcriptional repression domain in an insect Hox protein. Nature. 415, 910–913.
Gerber H., Seipel K., Georgiev O., Höfferer M., Hug M., Rusconi S., Schaffner W. 1994. Transcriptional activation modulated by homopolymeric glutamine and proline stretches. Science. 263, 808–811.
Redies C., Vanhalst K., van Roy F. 2005. δ-Protocadherins: Unique structures and functions. Cell. Mol. Life Sci. 62, 2840–2852.
Ren D., Zhu X., Kong R., Zhao Z., Sheng J., Wang J., Xu X., Liu J., Cui K., Zhang X.H., Zhao H., Wong S.T.C. 2018. Targeting brain-adaptive cancer stem cells prohibits brain metastatic colonization of triple-negative breast cancer. Cancer Res. 78, 2052–2064.
Yang X., Chen M.W., Terry S., Vacherot F., Chopin D.K., Bemis D.L., Kitajewski J., Benson M.C., Guo Y., Buttyan R. 2005. A human- and male-specific protocadherin that acts through the Wnt signaling pathway to induce neuroendocrine transdifferentiation of prostate cancer cells. Cancer Res. 65, 5263–5271.
Pancho A., Aerts T., Mitsogiannis M.D., Seuntjens E. 2020. Protocadherins at the crossroad of signaling pathways. Front. Mol. Neurosci. 13, 117.
Rose A., Meier I. 2004. Scaffolds, levers, rods and springs: Diverse cellular functions of long coiled-coil proteins. Cell. Mol. Life Sci. 61, 1996–2009.
Wu Y., Zheng S., Yao J., Li M., Yang G., Zhang N., Zhang S., Zhong B. 2017. Decreased expression of protocadherin 20 is associated with poor prognosis in hepatocellular carcinoma. Oncotarget. 8, 3018–3028.
Gong Z., Hu G. 2019. PCDH20 acts as a tumour-suppressor gene through the Wnt/β-catenin signalling pathway in hypopharyngeal squamous cell carcinoma. Cancer Biomarkers. 26, 209–217.
Lv J., Zhu P., Yang Z., Li M., Zhang X., Cheng J., Chen X., Lu F. 2015. PCDH20 functions as a tumour-suppressor gene through antagonizing the Wnt/β-catenin signalling pathway in hepatocellular carcinoma. J. Viral Hepatitis. 22, 201–211.
Carraro R.S., Nogueira G.A., Sidarta-Oliveira D., Gaspar R.S., Dragano N.R., Morari J., Bobbo V.C.D., Araujo E.P., Mendes N.F., Zanesco A.M., Tobar N., Ramos C.D., Toscaro J.M., Bajgelman M.C., Velloso L.A. 2021. Arcuate nucleus overexpression of NHLH2 reduces body mass and attenuates obesity-associated anxiety/depression-like behavior. J. Neurosci. 41, 10004–10022.
Vella K.R., Good D.J. 2010. Nhlh2 is a cold-responsive gene. Open Neuroendocrinol. J. 3, 38–44.
Good D.J., Porter F.D., Mahon K.A., Parlow A.F., Westphal H., Kirsch I.R. 1997. Hypogonadism and obesity in mice with a targeted deletion of the Nhlh2 gene. Nat. Genet. 15, 397–401.
Johnson S.A., Marín-Bivens C.L., Miele M., Coyle C.A., Fissore R., Good D.J. 2004. The Nhlh2 transcription factor is required for female sexual behavior and reproductive longevity. Horm. Behav. 46, 420–427.
Tremblay L.O., Herscovics A. 2000. Characterization of a cDNA encoding a novel human Golgi α1,2-mannosidase (IC) involved in N-glycan biosynthesis. J. Biol. Chem. 275, 31655–31660.
Gonzalez-Andrades M., Jalimarada S.S., Rodriguez-Benavente M., Feeley M.N., Woodward A.M., AbuSamra D.B., Argüeso P. 2020. Golgi α1,2-mannosidase I induces clustering and compartmentalization of CD147 during epithelial cell migration. Cell Adhes. Migr. 14, 96–105.
Li H., Wang G., Yu Y., Jian W., Zhang D., Wang Y., Wang T., Meng Y., Yuan C., Zhang C. 2018. α-1,2-mannosidase MAN1C1 inhibits proliferation and invasion of clear cell renal cell carcinoma. J. Cancer. 9, 4618–4626.
Tu H.C., Hsiao Y.C., Yang W.Y., Tsai S.L., Lin H.K., Liao C.Y., Lu J.W., Chou Y.T., Wang H.D., Yuh C.H. 2017. Up-regulation of golgi α-mannosidase IA and down-regulation of golgi α-mannosidase IC activates unfolded protein response during hepatocarcinogenesis. Hepatol. Commun. 1, 230–247.
Taniguchi N., Honke K., Fukuda M., Narimatsu H., Yamaguchi Y., Angata T. 2014. Handbook of Glycosyltransferases and Related Genes, vols. 1–2. Taniguchi N., Honke K., Fukuda M., Narimatsu H., Yamaguchi Y., Angata T., Eds. Tokyo: Springer.
Mast S.W., Diekman K., Karaveg K., Davis A., Sifers R.N., Moremen K.W. 2005. Human EDEM2, a novel homolog of family 47 glycosidases, is involved in ER-associated degradation of glycoproteins. Glycobiology. 15, 421–436.
Funding
We thank the Ministry of Human Resource Development for fellowship to LT and SS. Computational analyses were done on the Har Gobind Khorana Computational Biology cluster established and maintained by combining funds from the Indian Institute os Science Education and Research Bhopal under Grant # INST/BIO/2017/019, IYBA 2018 from the Department of Biotechnology (Grant no. BT/11/IYBA/2018/03), and ECRA from Science and Engineering Research Board (Grant no. ECR/2017/001430).
Author information
Authors and Affiliations
Contributions
SS and LT wrote the manuscript with NV. SS and LT analyzed the data and generated the results. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
COMPLIANCE WITH ETHICAL STANDARDS
The article does not contain any studies involving humans or animals in experiments performed by any of the authors.
CONFLICT OF INTEREST
The authors declare that they have no conflicts of interest.
Additional information
Publisher’s Note.
Pleiades Publishing remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
About this article
Cite this article
Sharma, S., Teekas, L. & Vijay, N. Protein Repeats Show Clade-Specific Volatility in Aves. Mol Biol 57, 1199–1211 (2023). https://doi.org/10.1134/S0026893324010163
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0026893324010163