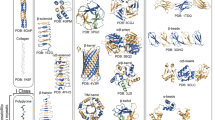
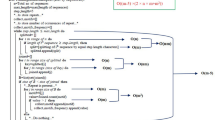
Abstract
The review summarizes and systematizes the data on the classification, taxonomic distribution, structural features, and functions of proteins with structural repeats. Modern approaches to the identification of structural repeats in proteins are considered. Features of specialized databases of protein domains are described. The review discusses the prospects of using repeat-containing proteins as scaffolds for drug design. The role of proteins with structural repeats in the pathogenesis of various diseases is considered. Various modern approaches to understanding the mechanisms of the evolutionary development of proteins with repeats are described and analyzed.



Similar content being viewed by others
REFERENCES
Marcotte E.M., Pellegrini M., Yeates T.O., Eisenberg D. 1999. A census of protein repeats. J. Mol. Biol. 293, 151–160. https://doi.org/10.1006/jmbi.1999.3136
Pellegrini M., Renda M.E., Vecchio A. 2012. Ab initio detection of fuzzy amino acid tandem repeats in protein sequences. BMC Bioinformatics. 13, S8. https://doi.org/10.1186/1471-2105-13-S3-S8
Pellegrini M., Marcotte E.M., Yeates T.O. 1999. A fast algorithm for genome-wide analysis of proteins with repeated sequences. Proteins. 35, 440–446. PMID:10382671
Jorda J., Kajava A.V. 2010. Protein homorepeats sequences, structures, evolution, and functions. Adv. Protein Chem. Struct. Biol. 79, 59–88. https://doi.org/10.1016/S1876-1623(10)79002-7
Schmitz-Linneweber C., Small I. 2008. Pentatricopeptide repeat proteins: A socket set for organelle gene expression. Trends Plant Sci. 13, 663–670. https://doi.org/10.1016/j.tplants.2008.10.001
Renault L., Nassar N., Vetter I., Becker J., Klebe C., Roth M., Wittinghofer A. 1998. The 1.7 A crystal structure of the regulator of chromosome condensation (RCC1) reveals a seven-bladed propeller. Nature. 392, 97–101. https://doi.org/10.1038/32204
Varela M., Diaz-Rosales P., Pereiro P., Forn-Cuní G., Costa M.M., Dios S., Romero A., Figueras A., Novoa B. 2014. Interferon-induced genes of the expanded IFIT family show conserved antiviral activities in non-mammalian species. PLoS One. 9, e100015. https://doi.org/10.1371/journal.pone.0100015
Cerveny L., Straskova A., Dankova V., Hartlova A., Ceckova M., Staud F., Stulik J. 2013. Tetratricopeptide repeat motifs in the world of bacterial pathogens: Role in virulence mechanisms. Infect. Immun. 81, 629–635. https://doi.org/10.1128/IAI.01035-12
Jacobsen S.E., Binkowski K.A., Olszewski N.E. 1996. SPINDLY, a tetratricopeptide repeat protein involved in gibberellin signal transduction in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 93, 9292–9296. https://doi.org/10.1073/pnas.93.17.9292
Baxa U., Cassese T., Kajava A.V., Steven A.C. 2006. Structure, function, and amyloidogenesis of fungal prions: Filament polymorphism and prion variants. Adv. Protein Chem. 73, 125–180. https://doi.org/10.1016/S0065-3233(06)73005-4
Kajava A.V, Squire J.M., Parry D.A.D. 2006. Beta-structures in fibrous proteins. Adv. Protein Chem. 73, 1–15. https://doi.org/10.1016/S0065-3233(06)73001-7
Darling A.L., Uversky V.N. 2017. Intrinsic disorder in proteins with pathogenic repeat expansions. Molecules. 22, 2027. https://doi.org/10.3390/molecules22122027
Sikorski P., Atkins E. 2005. New model for crystalline polyglutamine assemblies and their connection with amyloid fibrils. Biomacromolecules. 6, 425–432. https://doi.org/10.1021/bm0494388
Den Dunnen W.F.A. 2017. Trinucleotide repeat disorders. Handb. Clin. Neurol. 145, 383–391. https://doi.org/10.1016/B978-0-12-802395-2.00027-4
Shilova O.N., Deev S.M. 2019. Darpins: Promising targeted proteins for theranostics. Acta Naturae. 11, 42–53.
Mittl P.R., Ernst P., Plückthun A. 2020. Chaperone-assisted structure elucidation with DARPins. Curr. Opin. Struct. Biol. 60, 93–100. https://doi.org/10.1016/j.sbi.2019.12.009
Andrade M.A., Perez-Iratxeta C., Ponting C.P. 2001. Protein repeats: structures, functions, and evolution. J. Struct. Biol. 134, 117–131. https://doi.org/10.1006/jsbi.2001.4392
Ponting C.P., Russell R.B. 2000. Identification of distant homologues of fibroblast growth factors suggests a common ancestor for all beta-trefoil proteins. J. Mol. Biol. 302, 1041–1047. https://doi.org/10.1006/jmbi.2000.4087
Apic G., Huber W., Teichmann S.A. 2003. Multi-domain protein families and domain pairs: Comparison with known structures and a random model of domain recombination. J. Struct. Funct. Genomics. 4, 67–78. https://doi.org/10.1023/a:1026113408773
Ye Y., Godzik A. 2004. Comparative analysis of protein domain organization. Genome Res. 14, 343–353. https://doi.org/10.1101/gr.1610504
Moore A.D., Bornberg-Bauer E. 2012. The dynamics and evolutionary potential of domain loss and emergence. Mol. Biol. Evol. 29, 787–796. https://doi.org/10.1093/molbev/msr250
Kersting A.R., Bornberg-Bauer E., Moore A.D., Grath S. 2012. Dynamics and adaptive benefits of protein domain emergence and arrangements during plant genome evolution. Genome Biol. Evol. 4, 316–329. https://doi.org/10.1093/gbe/evs004
Kummerfeld S.K., Teichmann S.A. 2005. Relative rates of gene fusion and fission in multi-domain proteins. Trends Genet. 21, 25–30. https://doi.org/10.1016/j.tig.2004.11.007
Weiner J., Bornberg-Bauer E. 2006. Evolution of circular permutations in multidomain proteins. Mol. Biol. Evol. 23, 734–743. https://doi.org/10.1093/molbev/msj091
Weiner J., Beaussart F., Bornberg-Bauer E. 2006. Domain deletions and substitutions in the modular protein evolution. FEBS J. 273, 2037–2047. https://doi.org/10.1111/j.1742-4658.2006.05220.x
Wang M., Caetano-Anollés G. 2009. The evolutionary mechanics of domain organization in proteomes and the rise of modularity in the protein world. Structure. 17, 66–78. https://doi.org/10.1016/j.str.2008.11.008
Zmasek C.M., Godzik A. 2011. Strong functional patterns in the evolution of eukaryotic genomes revealed by the reconstruction of ancestral protein domain repertoires. Genome Biol. 12, R4. https://doi.org/10.1186/gb-2011-12-1-r4
Zmasek C.M., Godzik A. 2012. This Déjà vu feeling: Analysis of multidomain protein evolution in eukaryotic genomes. PLoS Comput. Biol. 8, e1002701. https://doi.org/10.1371/journal.pcbi.1002701
Forslund S.K., Kaduk M., Sonnhammer E.L.L. 2019. Evolution of protein domain architectures. Methods Mol. Biol. 1910, 469–504. https://doi.org/10.1007/978-1-4939-9074-0_15
Moore A.D., Grath S., Schüler A., Huylmans A.K., Bornberg-Bauer E. 2013. Quantification and functional analysis of modular protein evolution in a dense phylogenetic tree. Biochim. Biophys. Acta. 1834, 898–907. https://doi.org/10.1016/j.bbapap.2013.01.007
Garrido-Ramos M.A. 2017. Satellite DNA: An evolving topic. Genes (Basel). 8, 230. https://doi.org/10.3390/genes8090230
Björklund A.K., Ekman D., Elofsson A. 2006. Expansion of protein domain repeats. PLoS Comput. Biol. 2, e114. https://doi.org/10.1371/journal.pcbi.0020114
Buard J., Vergnaud G. 1994. Complex recombination events at the hypermutable minisatellite CEB1 (D2S90). EMBO J. 13, 3203–3210. https://doi.org/10.1002/j.1460-2075.1994.tb06619.x
Djian P. 1998. Evolution of simple repeats in DNA and their relation to human disease. Cell. 94, 155–160. https://doi.org/10.1016/s0092-8674(00)81415-4
Ellegren H. 2000. Microsatellite mutations in the germline: Implications for evolutionary inference. Trends Genet. 16, 551–558. https://doi.org/10.1016/s0168-9525(00)02139-9
Kruglyak S., Durrett R.T., Schug M.D., Aquadro C.F. 1998. Equilibrium distributions of microsatellite repeat length resulting from a balance between slippage events and point mutations. Proc. Natl. Acad. Sci. U. S. A. 95, 10774–10778. https://doi.org/10.1073/pnas.95.18.10774
Björklund A.K., Light S., Sagit R., Elofsson A. 2010. Nebulin: A study of protein repeat evolution. J. Mol. Biol. 402, 38–51. https://doi.org/10.1016/j.jmb.2010.07.011
Deryusheva E.I., Machulin A. V., Selivanova O.M., Galzitskaya O.V. 2017. Taxonomic distribution, repeats, and functions of the S1 domain-containing proteins as members of the OB-fold family. Proteins. 85, 602–613. https://doi.org/10.1002/prot.25237
Machulin A. V, Deryusheva E.I., Selivanova O.M., Galzitskaya O.V. 2019. The number of domains in the ribosomal protein S1 as a hallmark of the phylogenetic grouping of bacteria. PLoS One. 14, e0221370. https://doi.org/10.1371/journal.pone.0221370
Sokol D., Benson G., Tojeira J. 2007. Tandem repeats over the edit distance. Bioinformatics. 23, e30-5. https://doi.org/10.1093/bioinformatics/btl309
Kajava A.V. 2012. Tandem repeats in proteins: From sequence to structure. J. Struct. Biol. 179, 279–88. https://doi.org/10.1016/j.jsb.2011.08.009
Perutz M.F. 1999. Glutamine repeats and neurodegenerative diseases: Molecular aspects. Trends Biochem. Sci. 24, 58–63. https://doi.org/10.1016/s0968-0004(98)01350-4
Fan X. 2001. Oligomerization of polyalanine expanded PABPN1 facilitates nuclear protein aggregation that is associated with cell death. Hum. Mol. Genet. 10, 2341–2351. https://doi.org/10.1093/hmg/10.21.2341
Strømme P., Mangelsdorf M.E., Shaw M.A., Lower K.M., Lewis S.M.E., Bruyere H., Lütcherath V., Gedeon A.K., Wallace R.H., Scheffer I.E., Turner G., Partington M., Frints S.G.M., Fryns J.-P., Sutherland G.R., et al. 2002. Mutations in the human ortholog of Aristaless cause X-linked mental retardation and epilepsy. Nat. Genet. 30, 441–445. https://doi.org/10.1038/ng862
Orr H.T., Zoghbi H.Y. 2007. Trinucleotide repeat disorders. Annu. Rev. Neurosci. 30, 575–621. https://doi.org/10.1146/annurev.neuro.29.051605.113042
Mosbach V., Poggi L., Richard G.-F. 2019. Trinucleotide repeat instability during double-strand break repair: From mechanisms to gene therapy. Curr. Genet. 65, 17–28. https://doi.org/10.1007/s00294-018-0865-1
Mosbach V., Poggi L., Viterbo D., Charpentier M., Richard G.-F. 2018. TALEN-induced double-strand break repair of CTG trinucleotide repeats. Cell Rep. 22, 2146–2159. https://doi.org/10.1016/j.celrep.2018.01.083
Faux N.G., Bottomley S.P., Lesk A.M., Irving J.A., Morrison J.R., de la Banda M.G., Whisstock J.C. 2005. Functional insights from the distribution and role of homopeptide repeat-containing proteins. Genome Res. 15, 537–551. https://doi.org/10.1101/gr.3096505
Jorda J., Xue B., Uversky V.N., Kajava A.V. 2010. Protein tandem repeats – the more perfect, the less structured. FEBS J. 277, 2673–2682. https://doi.org/10.1111/j.1742-464X.2010.07684.x
Healy E.F., Little C., King P.J. 2014. A model for small heat shock protein inhibition of polyglutamine aggregation. Cell Biochem. Biophys. 69, 275–281. https://doi.org/10.1007/s12013-013-9795-1
Gruber A., Hornburg D., Antonin M., Krahmer N., Collado J., Schaffer M., Zubaite G., Lüchtenborg C., Sachsenheimer T., Brügger B., Mann M., Baumeister W., Hartl F.U., Hipp M.S., Fernández-Busnadiego R. 2018. Molecular and structural architecture of polyQ aggregates in yeast. Proc. Natl. Acad. Sci. U. S. A. 115, E3446–E3453. https://doi.org/10.1073/pnas.1717978115
Lyubchenko Y.L., Krasnoslobodtsev A. V, Luca S. 2012. Fibrillogenesis of huntingtin and other glutamine containing proteins. Subcell. Biochem. 65, 225–251. https://doi.org/10.1007/978-94-007-5416-4_10
Christie N.T.M., Lee A.L., Fay H.G., Gray A.A., Kikis E.A. 2014. Novel polyglutamine model uncouples proteotoxicity from aging. PLoS One. 9, e96835. https://doi.org/10.1371/journal.pone.0096835
Sorushanova A., Delgado L.M., Wu Z., Shologu N., Kshirsagar A., Raghunath R., Mullen A.M., Bayon Y., Pandit A., Raghunath M., Zeugolis D.I. 2019. The collagen suprafamily: From biosynthesis to advanced biomaterial development. Adv. Mater. 31, e1801651. https://doi.org/10.1002/adma.201801651
Lupas A.N., Bassler J., Dunin-Horkawicz S. 2017. The structure and topology of α-helical coiled coils. Subcell. Biochem. 82, 95–129. https://doi.org/10.1007/978-3-319-49674-0_4
Hennet T. 2019. Collagen glycosylation. Curr. Opin. Struct. Biol. 56, 131–138. https://doi.org/10.1016/j.sbi.2019.01.015
Berisio R., Vitagliano L., Mazzarella L., Zagari A. 2009. Crystal structure of the collagen triple helix model [(Pro-Pro-Gly)10]3. Protein Sci. 11, 262–270. https://doi.org/10.1110/ps.32602
Gordon M.K., Hahn R.A. 2010. Collagens. Cell Tissue Res. 339, 247–257. https://doi.org/10.1007/s00441-009-0844-4
Lupas A.N., Gruber M. 2005. The structure of alpha-helical coiled coils. Adv. Protein Chem. 70, 37–78. https://doi.org/10.1016/S0065-3233(05)70003-6
Gromiha M.M., Parry D.A. 2004. Characteristic features of amino acid residues in coiled-coil protein structures. Biophys. Chem. 111, 95–103. https://doi.org/10.1016/j.bpc.2004.05.001
Kobe B., Kajava A.V. 2000. When protein folding is simplified to protein coiling: The continuum of solenoid protein structures. Trends Biochem. Sci. 25, 509–515. https://doi.org/10.1016/s0968-0004(00)01667-4
Groves M.R., Barford D. 1999. Topological characteristics of helical repeat proteins. Curr. Opin. Struct. Biol. 9, 383–389. https://doi.org/10.1016/s0959-440x(99)80052-9
Kajava A.V., Steven A.C. 2006. Beta-rolls, beta-helices, and other beta-solenoid proteins. Adv. Protein Chem. 73, 55–96. https://doi.org/10.1016/S0065-3233(06)73003-0
Hennetin J., Jullian B., Steven A.C., Kajava A.V. 2006. Standard conformations of beta-arches in beta-solenoid proteins. J. Mol. Biol. 358, 1094–1105. https://doi.org/10.1016/j.jmb.2006.02.039
Kobe B., Deisenhofer J. 1996. Mechanism of ribonuclease inhibition by ribonuclease inhibitor protein based on the crystal structure of its complex with ribonuclease A. J. Mol. Biol. 264, 1028–1043. https://doi.org/10.1006/jmbi.1996.0694
Peters J.W., Stowell M.H., Rees D.C. 1996. A leucine-rich repeat variant with a novel repetitive protein structural motif. Nat. Struct. Biol. 3, 991–994. https://doi.org/10.1038/nsb1296-991
Huizinga E.G., Tsuji S., Romijn R.A.P., Schiphorst M.E., de Groot P.G., Sixma J.J., Gros P. 2002. Structures of glycoprotein Ibalpha and its complex with von Willebrand factor A1 domain. Science. 297, 1176–1179. https://doi.org/10.1126/science.107355
Liou Y.C., Tocilj A., Davies P.L., Jia Z. 2000. Mimicry of ice structure by surface hydroxyls and water of a beta-helix antifreeze protein. Nature. 406, 322–324. https://doi.org/10.1038/35018604
Fournier D., Palidwor G.A., Shcherbinin S., Szengel A., Schaefer M.H., Perez-Iratxeta C., Andrade-Navarro M.A. 2013. Functional and genomic analyses of alpha-solenoid proteins. PLoS One. 8, e79894. https://doi.org/10.1371/journal.pone.0079894
Cho U.S., Xu W. 2007. Crystal structure of a protein phosphatase 2A heterotrimeric holoenzyme. Nature. 445, 53–57. https://doi.org/10.1038/nature05351
Xing Y., Takemaru K.-I., Liu J., Berndt J.D., Zheng J.J., Moon R.T., Xu W. 2008. Crystal structure of a full-length beta-catenin. Structure. 16, 478–487. https://doi.org/10.1016/j.str.2007.12.021
Hast M.A., Beese L.S. 2008. Structure of protein geranylgeranyltransferase-I from the human pathogen Candida albicans complexed with a lipid substrate. J. Biol. Chem. 283, 31933–31940. https://doi.org/10.1074/jbc.M805330200
Mitraki A., Papanikolopoulou K., Van Raaij M.J. 2006. Natural triple beta-stranded fibrous folds. Adv. Protein Chem. 73, 97–124. https://doi.org/10.1016/S0065-3233(06)73004-2
Schrag J.D., Bergeron J.J.M., Li Y., Borisova S., Hahn M., Thomas D.Y., Cygler M. 2001. The structure of calnexin, an ER chaperone involved in quality control of protein folding. Mol. Cell. 8, 633–644. https://doi.org/10.1016/s1097-2765(01)00318-5
Ellgaard L., Riek R., Herrmann T., Güntert P., Braun D., Helenius A., Wüthrich K. 2001. NMR structure of the calreticulin P-domain. Proc. Natl. Acad. Sci. U. S. A. 98, 3133–3138. https://doi.org/10.1073/pnas.051630098
Makabe K., Biancalana M., Yan S., Tereshko V., Gawlak G., Miller-Auer H., Meredith S.C., Koide S. 2008. High-resolution structure of a self-assembly-competent form of a hydrophobic peptide captured in a soluble beta-sheet scaffold. J. Mol. Biol. 378, 459–467. https://doi.org/10.1016/j.jmb.2008.02.051
Alvarez M., Zeelen J.P., Mainfroid V., Rentier-Delrue F., Martial J.A., Wyns L., Wierenga R.K., Maes D. 1998. Triose-phosphate isomerase (TIM) of the psychrophilic bacterium Vibrio marinus. Kinetic and structural properties. J. Biol. Chem. 273, 2199–2206. https://doi.org/10.1074/jbc.273.4.2199
Koebnik R., Locher K.P., Van Gelder P. 2000. Structure and function of bacterial outer membrane proteins: Barrels in a nutshell. Mol. Microbiol. 37, 239–253. https://doi.org/10.1046/j.1365-2958.2000.01983.x
Wierenga R.K. 2001. The TIM-barrel fold: A versatile framework for efficient enzymes. FEBS Lett. 492, 193–198. https://doi.org/10.1016/s0014-5793(01)02236-0
Goldman A.D., Beatty J.T., Landweber L.F. 2016. The TIM barrel architecture facilitated the early evolution of protein-mediated metabolism. J. Mol. Evol. 82, 17–26. https://doi.org/10.1007/s00239-015-9722-8
Chen C.K.-M., Chan N.-L., Wang A.H.-J. 2011. The many blades of the β-propeller proteins: Conserved but versatile. Trends Biochem. Sci. 36, 553–561. https://doi.org/10.1016/j.tibs.2011.07.004
Pitt J.J., Da Silva E., Gorman J.J. 2000. Determination of the disulfide bond arrangement of Newcastle Disease virus hemagglutinin neuraminidase. J. Biol. Chem. 275, 6469–6478. https://doi.org/10.1074/jbc.275.9.6469
Schapira M., Tyers M., Torrent M., Arrowsmith C.H. 2017. WD40 repeat domain proteins: A novel target class? Nat. Rev. Drug Discov. 16, 773–786. https://doi.org/10.1038/nrd.2017.179
Jain B.P., Pandey S. 2018. WD40 repeat proteins: Signalling scaffold with diverse functions. Protein J. 37, 391–406. https://doi.org/10.1007/s10930-018-9785-7
Kumar V., Yadav A.N., Verma P., Sangwan P., Saxena A., Kumar K., Singh B. 2017. β-Propeller phytases: Diversity, catalytic attributes, current developments and potential biotechnological applications. Int. J. Biol. Macromol. 98, 595–609. https://doi.org/10.1016/j.ijbiomac.2017.01.134
Murzin A.G., Lesk A.M., Chothia C. 1992. beta-Trefoil fold. Patterns of structure and sequence in the Kunitz inhibitors interleukins-1 beta and 1 alpha and fibroblast growth factors. J. Mol. Biol. 223, 531–543. https://doi.org/10.1016/0022-2836(92)90668-a
Gosavi S., Whitford P.C., Jennings P.A., Onuchic J.N. 2008. Extracting function from a beta-trefoil folding motif. Proc. Natl. Acad. Sci. U. S. A. 105, 10384–19389. https://doi.org/10.1073/pnas.0801343105
Bendre A.D., Ramasamy S., Suresh C.G. 2018. Analysis of Kunitz inhibitors from plants for comprehensive structural and functional insights. Int. J. Biol. Macromol. 113, 933–943. https://doi.org/10.1016/j.ijbiomac.2018.02.148
Zhou J., Li C., Chen A., Zhu J., Zou M., Liao H., Yu Y. 2020. Structural and functional relationship of Cassia obtusifolia trypsin inhibitor to understand its digestive resistance against Pieris rapae. Int. J. Biol. Macromol. 148, 908–920. https://doi.org/10.1016/j.ijbiomac.2020.01.193
Giri Rao V.V.H., Gosavi S. 2015. Structural perturbations present in the folding cores of interleukin-33 and interleukin-1β correlate to differences in their function. J. Phys. Chem. B. 119, 11203–11214. https://doi.org/10.1021/acs.jpcb.5b03111
Hailey K.L., Capraro D.T., Barkho S., Jennings P.A. 2013. Allosteric switching of agonist/antagonist activity by a single point mutation in the interluekin-1 receptor antagonist, IL-1Ra. J. Mol. Biol. 425, 2382–2392. https://doi.org/10.1016/j.jmb.2013.03.016
Liao J.-H., Chien C.-T.H., Wu H.-Y., Huang K.-F., Wang I., Ho M.-R., Tu I.-F., Lee I.-M., Li W., Shih Y.-L., Wu C.-Y., Lukyanov P.A., Hsu S.-T.D., Wu S.-H. 2016. A multivalent marine lectin from Crenomytilus grayanus possesses anti-cancer activity through recognizing globotriose Gb3. J. Am. Chem. Soc. 138, 4787–4795. https://doi.org/10.1021/jacs.6b00111
Bensen D.C., Rodriguez S., Nix J., Cunningham M.L., Tari L.W. 2012. Structure of MurA (UDP-N-acetylglucosamine enolpyruvyl transferase. from Vibrio fischeri in complex with substrate UDP-N-acetylglucosamine and the drug fosfomycin. Acta Crystallogr. F: Struct. Biol. Cryst. Commun. 68, 382–385. https://doi.org/10.1107/S1744309112006720
Pautsch A., Schulz G.E. 1998. Structure of the outer membrane protein A transmembrane domain. Nat. Struct. Biol. 5, 1013–1017. https://doi.org/10.1038/2983
Kim K., Kim K.-P., Choi J., Lim J.-A., Lee J., Hwang S., Ryu S. 2010. Outer membrane proteins A (OmpA) and X (OmpX) are essential for basolateral invasion of Cronobacter sakazakii. Appl. Environ. Microbiol. 76, 5188–5198. https://doi.org/10.1128/AEM.02498-09
Balasubramaniam D., Arockiasamy A., Kumar P.D., Sharma A., Krishnaswamy S. 2012. Asymmetric pore occupancy in crystal structure of OmpF porin from Salmonella typhi. J. Struct. Biol. 178, 233–244. https://doi.org/10.1016/j.jsb.2012.04.005
Kim B.-H., Andersen C., Kreth J., Ulmke C., Schmid K., Benz R. 2002. Site-directed mutagenesis within the central constriction site of ScrY (sucroseporin): Effect on ion transport and comparison of maltooligosaccharide binding to LamB of Escherichia coli. J. Membr. Biol. 187, 239–253. https://doi.org/10.1007/s00232-001-0167-1
Ferguson A.D., Deisenhofer J. 2002. TonB-dependent receptors: Structural perspectives. Biochim. Biophys. Acta – Biomembr. 1565, 318–332. https://doi.org/10.1016/S0005-2736(02)00578-3
Oteiza P.I., Mackenzie G.G. 2005. Zinc, oxidant-triggered cell signaling, and human health. Mol. Aspects Med. 26, 245–255. https://doi.org/10.1016/j.mam.2005.07.012
García C.C., Damonte E.B. 2007. Zn finger containing proteins as targets for the control of viral infections. Infect. Disord. Drug Targets. 7, 204–212. https://doi.org/10.2174/187152607782110004
Kusunoki H., Minasov G., Macdonald R.I., Mondragón A. 2004. Independent movement, dimerization and stability of tandem repeats of chicken brain alpha-spectrin. J. Mol. Biol. 344, 495–511. https://doi.org/10.1016/j.jmb.2004.09.019
Tanaka Y., Sakamoto S., Kuroda M., Goda S., Gao Y.-G., Tsumoto K., Hiragi Y., Yao M., Watanabe N., Ohta T., Tanaka I. 2008. A helical string of alternately connected three-helix bundles for the cell wall-associated adhesion protein Ebh from Staphylococcus aureus. Structure. 16, 488–496. https://doi.org/10.1016/j.str.2007.12.018
Zheng N., Schulman B.A., Song L., Miller J.J., Jeffrey P.D., Wang P., Chu C., Koepp D.M., Elledge S.J., Pagano M., Conaway R.C., Conaway J.W., Harper J.W., Pavletich N.P. 2002. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature. 416, 703–709. https://doi.org/10.1038/416703a
Lukacik P., Roversi P., White J., Esser D., Smith G.P., Billington J., Williams P.A., Rudd P.M., Wormald M.R., Harvey D.J., Crispin M.D.M., Radcliffe C.M., Dwek R.A., Evans D.J., Morgan B.P., Smith R.A.G., Lea S.M. 2004. Complement regulation at the molecular level: The structure of decay-accelerating factor. Proc. Natl. Acad. Sci. U. S. A. 101, 1279–1284. https://doi.org/10.1073/pnas.0307200101
Harrison O.J., Jin X., Hong S., Bahna F., Ahlsen G., Brasch J., Wu Y., Vendome J., Felsovalyi K., Hampton C.M., Troyanovsky R.B., Ben-Shaul A., Frank J., Troyanovsky S.M., Shapiro L., Honig B. 2011. The extracellular architecture of adherens junctions revealed by crystal structures of type I cadherins. Structure. 19, 244–256. https://doi.org/10.1016/j.str.2010.11.016
Van Bibber N.W., Haerle C., Khalife R., Xue B., Uversky V.N. 2020. Intrinsic disorder in tetratricopeptide repeat proteins. Int. J. Mol. Sci. 21, 3709. https://doi.org/10.3390/ijms21103709
Machulin A., Deryusheva E., Lobanov M., Galzitskaya O. 2019. Repeats in S1 proteins: Flexibility and tendency for intrinsic disorder. Int. J. Mol. Sci. 20, 2377. https://doi.org/10.3390/ijms20102377
Aachmann F.L., Svanem B.I.G., Güntert P., Peter-sen S.B., Valla S., Wimmer R. 2006. NMR structure of the R-module: A parallel beta-roll subunit from an Azotobacter vinelandii mannuronan C-5 epimerase. J. Biol. Chem. 281, 7350–7356. https://doi.org/10.1074/jbc.M510069200
Apic G., Gough J., Teichmann S.A. 2001. Domain combinations in archaeal, eubacterial and eukaryotic proteomes. J. Mol. Biol. 310, 311–325. https://doi.org/10.1006/jmbi.2001.4776
Ekman D., Björklund A.K., Frey-Skött J., Elofsson A. 2005. Multi-domain proteins in the three kingdoms of life: Orphan domains and other unassigned regions. J. Mol. Biol. 348, 231–243. https://doi.org/10.1016/j.jmb.2005.02.007
Delucchi M., Schaper E., Sachenkova O., Elofsson A., Anisimova M. 2020. A new census of protein tandem repeats and their relationship with intrinsic disorder. Genes (Basel). 11, 407. https://doi.org/10.3390/genes11040407
Schaper E., Korsunsky A., Pečerska J., Messina A., Murri R., Stockinger H., Zoller S., Xenarios I., Anisimova M. 2015. TRAL: Tandem repeat annotation library. Bioinformatics. 31, 3051–3053. https://doi.org/10.1093/bioinformatics/btv306
Tørresen O.K., Star B., Mier P., Andrade-Navarro M.A., Bateman A., Jarnot P., Gruca A., Grynberg M., Kajava A.V., Promponas V.J., Anisimova M., Jakobsen K.S., Linke D. 2019. Tandem repeats lead to sequence assembly errors and impose multi-level challenges for genome and protein databases. Nucleic Acids Res. 47, 10994–11006. https://doi.org/10.1093/nar/gkz841
Bilgin Sonay T., Koletou M., Wagner A. 2015. A survey of tandem repeat instabilities and associated gene expression changes in 35 colorectal cancers. BMC Genomics. 16, 702. https://doi.org/10.1186/s12864-015-1902-9
Theriot J.A. 2013. Why are bacteria different from eukaryotes? BMC Biol. 11, 119. https://doi.org/10.1186/1741-7007-11-119
Schaper E., Gascuel O., Anisimova M. 2014. Deep conservation of human protein tandem repeats within the eukaryotes. Mol. Biol. Evol. 31, 1132–1148. https://doi.org/10.1093/molbev/msu062
Schaper E., Kajava A.V., Hauser A., Anisimova M. 2012. Repeat or not repeat?–Statistical validation of tandem repeat prediction in genomic sequences. Nucleic Acids Res. 40, 10005–10017. https://doi.org/10.1093/nar/gks726
Galzitskaya O.V., Lobanov M.Y. 2015. Phyloproteomic analysis of 11780 six-residue-long motifs occurrences. Biomed. Res. Int. 2015, 208346. https://doi.org/10.1155/2015/208346
Lobanov M.Y., Galzitskaya O.V. 2011. Disordered patterns in clustered Protein Data Bank and in eukaryotic and bacterial proteomes. PLoS One. 6, e27142. https://doi.org/10.1371/journal.pone.0027142
Lobanov M.Y., Galzitskaya O.V. 2012. Occurrence of disordered patterns and homorepeats in eukaryotic and bacterial proteomes. Mol. Biosyst. 8, 327–337. https://doi.org/10.1039/c1mb05318c
Kajava A.V. 2001. Review: Proteins with repeated sequence–structural prediction and modeling. J. Struct. Biol. 134, 132–144. https://doi.org/10.1006/jsbi.2000.4328
Jernigan K.K., Bordenstein S.R. 2015. Tandem-repeat protein domains across the tree of life. Peer. J. 3, e732. https://doi.org/10.7717/peerj.732
D’Andrea L.D., Regan L. 2003. TPR proteins: The versatile helix. Trends Biochem. Sci. 28, 655–662. https://doi.org/10.1016/j.tibs.2003.10.007
Gruber M., Söding J., Lupas A.N. 2005. REPPER-repeats and their periodicities in fibrous proteins. Nucleic Acids Res. 33, W239–W243. https://doi.org/10.1093/nar/gki405
Taylor W.R., Heringa J., Baud F., Flores T.P. 2002. A Fourier analysis of symmetry in protein structure. Protein Eng. 15, 79–89. https://doi.org/10.1093/protein/15.2.79
Newman A.M., Cooper J.B. 2007. XSTREAM: A practical algorithm for identification and architecture modeling of tandem repeats in protein sequences. BMC Bioinformatics. 8, 382. https://doi.org/10.1186/1471-2105-8-382
Jorda J., Kajava A.V. 2009. T-REKS: Identification of tandem REpeats in sequences with a K-meanS based algorithm. Bioinformatics. 25, 2632–2638. https://doi.org/10.1093/bioinformatics/btp482
Heger A., Holm L. 2000. Rapid automatic detection and alignment of repeats in protein sequences. Proteins. 41, 224–237. https://doi.org/10.1002/1097-0134(20001101)41:2<224::aid-prot70>3.0.co;2-z
Szklarczyk R., Heringa J. 2004. Tracking repeats using significance and transitivity. Bioinformatics. 20 (Suppl 1), i311–i317. https://doi.org/10.1093/bioinformatics/bth911
Bucher P., Karplus K., Moeri N., Hofmann K. 1996. A flexible motif search technique based on generalized profiles. Comput. Chem. 20, 3–23. https://doi.org/10.1016/s0097-8485(96)80003-9
Biegert A., Söding J. 2008. De novo identification of highly diverged protein repeats by probabilistic consistency. Bioinformatics. 24, 807–814. https://doi.org/10.1093/bioinformatics/btn039
Bliven S.E., Lafita A., Rose P.W., Capitani G., Prlić A., Bourne P.E. 2019. Analyzing the symmetrical arrangement of structural repeats in proteins with CE-Symm. PLoS Comput. Biol. 15, e1006842. https://doi.org/10.1371/journal.pcbi.1006842
Chakrabarty B., Parekh N. 2014. Identifying tandem ankyrin repeats in protein structures. BMC Bioinformatics. 15, 6599. https://doi.org/10.1186/s12859-014-0440-9
Sabarinathan R., Basu R., Sekar K. 2010. ProSTRIP: A method to find similar structural repeats in three-dimensional protein structures. Comput. Biol. Chem. 34, 126–130. https://doi.org/10.1016/j.compbiolchem.2010.03.006
Abraham A.-L., Rocha E.P.C., Pothier J. 2008. Swelfe: A detector of internal repeats in sequences and structures. Bioinformatics. 24, 1536–1537. https://doi.org/10.1093/bioinformatics/btn234
Do Viet P., Roche D.B., Kajava A.V. 2015. TAPO: A combined method for the identification of tandem repeats in protein structures. FEBS Lett. 589, 2611–2619. https://doi.org/10.1016/j.febslet.2015.08.025
Fankhauser N., Nguyen-Ha T.-M., Adler J., Mäser P. 2007. Surface antigens and potential virulence factors from parasites detected by comparative genomics of perfect amino acid repeats. Proteome Sci. 5, 20. https://doi.org/10.1186/1477-5956-5-20
Parra R.G., Espada R., Sánchez I.E., Sippl M.J., Ferreiro D.U. 2013. Detecting repetitions and periodicities in proteins by tiling the structural space. J. Phys. Chem. B. 117, 12887–12897. https://doi.org/10.1021/jp402105j
Mistry J., Chuguransky S., Williams L., Qureshi M., Salazar G.A., Sonnhammer E.L.L., Tosatto S.C.E., Paladin L., Raj S., Richardson L.J., Finn R.D., Bateman A. 2021. Pfam: The protein families database in 2021. Nucleic Acids Res. 49, D412–D419. https://doi.org/10.1093/nar/gkaa913
Letunic I., Khedkar S., Bork P. 2021. SMART: Recent updates, new developments and status in 2020. Nucleic Acids Res. 49, D458–D460. https://doi.org/10.1093/nar/gkaa937
Blum M., Chang H.-Y., Chuguransky S., Grego T., Kandasaamy S., Mitchell A., Nuka G., Paysan-Lafosse T., Qureshi M., Raj S., Richardson L., Salazar G.A., Williams L., Bork P. Bridge A., et al. 2021. The InterPro protein families and domains database: 20 years on. Nucleic Acids Res. 49, D344–D354. https://doi.org/10.1093/nar/gkaa977
Sigrist C.J.A., De Castro E., Cerutti L., Cuche B.A., Hulo N., Bridge A., Bougueleret L., Xenarios I. 2013. New and continuing developments at PROSITE. Nucleic Acids Res. 41, D344–D347. https://doi.org/10.1093/nar/gks1067
Pandurangan A.P., Stahlhacke J., Oates M.E., Smi-thers B., Gough J. 2019. The SUPERFAMILY 2.0 database: A significant proteome update and a new webserver. Nucleic Acids Res. 47, D490–D494. https://doi.org/10.1093/nar/gky1130
UniProt Consortium. 2021. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 49, D480–D489. https://doi.org/10.1093/nar/gkaa1100
Jorda J., Baudrand T., Kajava A.V. 2012. PRDB: Protein Repeat DataBase. Proteomics. 12, 1333–1336. https://doi.org/10.1002/pmic.201100534
Paladin L., Bevilacqua M., Errigo S., Piovesan D., Mičetić I., Necci M., Monzon A.M., Fabre M.L., Lopez J.L., Nilsson J.F., Rios J., Menna P.L., Cabrera M., Buitron M.G., Kulik M.G., et al. 2021. RepeatsDB in 2021: Improved data and extended classification for protein tandem repeat structures. Nucleic Acids Res. 49, D452–D457. https://doi.org/10.1093/nar/gkaa1097
Burley S.K., Bhikadiya C., Bi C., Bittrich S., Chen L., Crichlow G. V, Christie C.H., Dalenberg K., Di Costanzo L., Duarte J.M., Dutta S., Feng Z., Ganesan S., Goodsell D.S., Ghosh S., et al. 2021. RCSB Protein Data Bank: Powerful new tools for exploring 3D structures of biological macromolecules for basic and applied research and education in fundamental biology, biomedicine, biotechnology, bioengineering and energy sciences. Nucleic Acids Res. 49, D437–D451. https://doi.org/10.1093/nar/gkaa1038
Offord V., Werling D. 2013. LRRfinder2.0: A webserver for the prediction of leucine-rich repeats. Innate Immun. 19, 398–402. https://doi.org/10.1177/1753425912465661
Lobanov M.Y., Sokolovskiy I.V, Galzitskaya O.V. 2014. HRaP: Database of occurrence of HomoRepeats and patterns in proteomes. Nucleic Acids Res. 42, D273–D278. https://doi.org/10.1093/nar/gkt927
Deryusheva E. I., Machulin, A.V., Selivanova O.M., Serdyuk, I.N. 2010. The S1 ribosomal protein family contains a unique conservative domain. Mol. Biol. (Moscow). 44 (4), 642–647.
Orafidiya F.A., McEwan I.J. 2015. Trinucleotide repeats and protein folding and disease: The perspective from studies with the androgen receptor. Futur. Sci. OA. 1, FSO47. https://doi.org/10.4155/fso.15.47
Walcott J.L., Merry D.E. 2002. Trinucleotide repeat disease. The androgen receptor in spinal and bulbar muscular atrophy. Vitam. Horm. 65, 127–147. https://doi.org/10.1016/s0083-6729(02)65062-9
McEwan I.J. 2001. Structural and functional alterations in the androgen receptor in spinal bulbar muscular atrophy. Biochem. Soc. Trans. 29, 222–227. https://doi.org/10.1042/0300-5127:0290222
Hor C.H.H., Tang B.L. 2019. Beta-propeller protein-associated neurodegeneration (BPAN) as a genetically simple model of multifaceted neuropathology resulting from defects in autophagy. Rev. Neurosci. 30, 261–277. https://doi.org/10.1515/revneuro-2018-0045
Mollereau B., Walter L. 2019. Is WDR45 the missing link for ER stress-induced autophagy in beta-propeller associated neurodegeneration?. Autophagy. 15, 2163–2164. https://doi.org/10.1080/15548627.2019.1668229
Pons T., Gómez R., Chinea G., Valencia A. 2003. Beta-propellers: Associated functions and their role in human diseases. Curr. Med. Chem. 10, 505–524. https://doi.org/10.2174/0929867033368204
Matsushima N., Takatsuka S., Miyashita H., Kretsinger R.H. 2019. Leucine rich repeat proteins: Sequences, mutations, structures and diseases. Protein Pept. Lett. 26, 108–131. https://doi.org/10.2174/0929866526666181208170027
Matsushima N., Tachi N., Kuroki Y., Enkhbayar P., Osaki M., Kamiya M., Kretsinger R.H. 2005. Structural analysis of leucine-rich-repeat variants in proteins associated with human diseases. Cell. Mol. Life Sci. 62, 2771–2791. https://doi.org/10.1007/s00018-005-5187-z
Hugot J.P., Chamaillard M., Zouali H., Lesage S., Cézard J.P., Belaiche J., Almer S., Tysk C., O’Morain C.A., Gassull M., Binder V., Finkel Y., Cortot A., Modigliani R., Laurent-Puig P., et al. 2001. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature. 411, 599–603. https://doi.org/10.1038/35079107
Shimizu T. 2013. Structural basis for β-galactosidase associated with lysosomal disease. Yakugaku Zasshi. 133, 509–517. https://doi.org/10.1248/yakushi.13-00001-1
Ohto U., Usui K., Ochi T., Yuki K., Satow Y., Shimizu T. 2012. Crystal structure of human β-galactosidase: Structural basis of Gm1 gangliosidosis and morquio B diseases. J. Biol. Chem. 287, 1801–1812. https://doi.org/10.1074/jbc.M111.293795
Ishiguro N., Motoi T., Osaki M., Araki N., Minamizaki T., Moriyama M., Ito H., Yoshida H. 2005. Immunohistochemical analysis of a muscle ankyrin-repeat protein, Arpp, in paraffin-embedded tumors: Evaluation of Arpp as a tumor marker for rhabdomyosarcoma. Hum. Pathol. 36, 620–625. https://doi.org/10.1016/j.humpath.2005.04.014
Ishiguro N., Baba T., Ishida T., Takeuchi K., Osaki M., Araki N., Okada E., Takahashi S., Saito M., Watanabe M., Nakada C., Tsukamoto Y., Sato K., Ito K., Fukayama M., et al. 2002. Carp, a cardiac ankyrin-repeated protein, and its new homologue, Arpp, are differentially expressed in heart, skeletal muscle, and rhabdomyosarcomas. Am. J. Pathol. 160, 1767–1778. https://doi.org/10.1016/S0002-9440(10)61123-6
Tee J.-M., Peppelenbosch M.P. 2010. Anchoring skeletal muscle development and disease: The role of ankyrin repeat domain containing proteins in muscle physiology. Crit. Rev. Biochem. Mol. Biol. 45, 318–330. https://doi.org/10.3109/10409238.2010.488217
Crist R.C., Roth J.J., Baran A.A., McEntee B.J., Siracusa L.D., Buchberg A.M. 2010. The armadillo repeat domain of Apc suppresses intestinal tumorigenesis. Mamm. Genome. 21, 450–457. https://doi.org/10.1007/s00335-010-9288-0
Li D., Song H., Mei H., Fang E., Wang X., Yang F., Li H., Chen Y., Huang K., Zheng L., Tong Q. 2018. Armadillo repeat containing 12 promotes neuroblastoma progression through interaction with retinoblastoma binding protein 4. Nat. Commun. 9, 2829. https://doi.org/10.1038/s41467-018-05286-2
Topaz O., Shurman D.L., Bergman R., Indelman M., Ratajczak P., Mizrachi M., Khamaysi Z., Behar D., Petronius D., Friedman V., Zelikovic I., Raimer S., Metzker A., Richard G., Sprecher E. 2004. Mutations in GALNT3, encoding a protein involved in O-linked glycosylation, cause familial tumoral calcinosis. Nat. Genet. 36, 579–581. https://doi.org/10.1038/ng1358
Duncan E.L., Danoy P., Kemp J.P., Leo P.J., McCloskey E., Nicholson G.C., Eastell R., Prince R.L., Eisman J.A., Jones G., Sambrook P.N., Reid I.R., Dennison E.M., Wark J., Richards J.B., et al. 2011. Genome-wide association study using extreme truncate selection identifies novel genes affecting bone mineral density and fracture risk. PLoS Genet. 7, e1001372. https://doi.org/10.1371/journal.pgen.1001372
Esapa C.T., Head R.A., Jeyabalan J., Evans H., Hough T.A., Cheeseman M.T., McNally E.G., Carr A.J., Thomas G.P., Brown M.A., Croucher P.I., Brown S.D.M., Cox R.D., Thakker R.V. 2012. A mouse with an N-Ethyl-N-nitrosourea (ENU. Induced Trp589Arg Galnt3 mutation represents a model for hyperphosphataemic familial tumoural calcinosis. PLoS One. 7, e43205. https://doi.org/10.1371/journal.pone.0043205
Lorenz V., Cejas R.B., Bennett E.P., Nores G.A., Irazoqui F.J. 2017. Functional control of polypeptide GalNAc-transferase 3 through an acetylation site in the C-terminal lectin domain. Biol. Chem. 398, 1237–1246. https://doi.org/10.1515/hsz-2017-0130
Percival J.M. 2018. Perspective: Spectrin-like repeats in dystrophin have unique binding preferences for syntrophin adaptors that explain the mystery of how nNOSμ localizes to the sarcolemma. Front. Physiol. 9, 1369. https://doi.org/10.3389/fphys.2018.01369
Thomas G.D. 2013. Functional muscle ischemia in Duchenne and Becker muscular dystrophy. Front. Physiol. 4, 381. https://doi.org/10.3389/fphys.2013.00381
Dušková L., Nohelová L., Loja T., Fialová J., Zapletalová P., Réblová K., Tichý L., Freiberger T., Fajkusová L. 2020. Low density lipoprotein receptor variants in the beta-propeller subdomain and their functional impact. Front. Genet. 11, 691. https://doi.org/10.3389/fgene.2020.00691
Cogo S., Manzoni C., Lewis P.A., Greggio E. 2020. Leucine-rich repeat kinase 2 and lysosomal dyshomeostasis in Parkinson disease. J. Neurochem. 152, 273–283. https://doi.org/10.1111/jnc.14908
Lee J.-M., Correia K., Loupe J., Kim K.-H., Barker D., Hong E.P., Chao M.J., Long J.D., Lucente D., Vonsattel J.P.G., Pinto R.M., Abu Elneel K., Ramos E.M., Mysore J.S., Gillis T., et al. 2019. CAG repeat not polyglutamine length determines timing of Huntington’s disease onset. Cell. 178, 887–900, e14. https://doi.org/10.1016/j.cell.2019.06.036
Bates G.P., Dorsey R., Gusella J.F., Hayden M.R., Kay C., Leavitt B.R., Nance M., Ross C.A., Scahill R.I., Wetzel R., Wild E.J., Tabrizi S.J. 2015. Huntington disease. Nat. Rev. Dis. Prim. 1, 15005. https://doi.org/10.1038/nrdp.2015.5
Prat C., Lemaire O., Bret J., Zabraniecki L., Fournié B. 2008. Morquio syndrome: Diagnosis in an adult. Joint. Bone Spine. 75, 495–498. https://doi.org/10.1016/j.jbspin.2007.07.021
Bley A.E., Giannikopoulos O.A., Hayden D., Kubilus K., Tifft C.J., Eichler F.S. 2011. Natural history of infantile G(M2. gangliosidosis. Pediatrics. 128, e1233–1241. https://doi.org/10.1542/peds.2011-0078
Saravanan K.M., Ponnuraj K. 2019. Sequence and structural analysis of fibronectin-binding protein reveals importance of multiple intrinsic disordered tandem repeats. J. Mol. Recognit. 32, e2768. https://doi.org/10.1002/jmr.2768
Li X., Tao Y., Murphy J.W., Scherer A.N., Lam T.T., Marshall A.G., Koleske A.J., Boggon T.J. 2017. The repeat region of cortactin is intrinsically disordered in solution. Sci. Rep. 7, 16696. https://doi.org/10.1038/s41598-017-16959-1
Roberts S., Dzuricky M., Chilkoti A. 2015. Elastin-like polypeptides as models of intrinsically disordered proteins. FEBS Lett. 589, 2477–2486. https://doi.org/10.1016/j.febslet.2015.08.029
Lobanov M.Y., Galzitskaya O.V. 2015. How common is disorder? Occurrence of disordered residues in four domains of life. Int. J. Mol. Sci. 16, 19490–19507. https://doi.org/10.3390/ijms160819490
Lobanov M.Y., Klus P., Sokolovsky I.V., Tartaglia G.G., Galzitskaya O.V. 2016. Non-random distribution of homo-repeats: Links with biological functions and human diseases. Sci. Rep. 6, 26941. https://doi.org/10.1038/srep26941
Lobanov M.Y., Furletova E.I., Bogatyreva N.S., Roytberg M.A., Galzitskaya O.V (2010. Library of disordered patterns in 3D protein structures. PLoS Comput. Biol. 6, e1000958. https://doi.org/10.1371/journal.pcbi.1000958
Forrer P., Binz H.K., Stumpp M.T., Plückthun A. 2004. Consensus design of repeat proteins. ChemBioChem. 5, 183–189. https://doi.org/10.1002/cbic.200300762
Forrer P., Stumpp M.T., Binz H.K., Plückthun A. 2003. A novel strategy to design binding molecules harnessing the modular nature of repeat proteins. FEBS Lett. 539, 2–6. https://doi.org/10.1016/s0014-5793(03)00177-7
Main E.R.G., Jackson S.E., Regan L. 2003. The folding and design of repeat proteins: Reaching a consensus. Curr. Opin. Struct. Biol. 13, 482–489. https://doi.org/10.1016/s0959-440x(03)00105-2
Main E.R.G., Lowe A.R., Mochrie S.G.J., Jackson S.E., Regan L. 2005. A recurring theme in protein engineering: The design, stability and folding of repeat proteins. Curr. Opin. Struct. Biol. 15, 464–471. https://doi.org/10.1016/j.sbi.2005.07.003
Javadi Y., Itzhaki L.S. 2013. Tandem-repeat proteins: Regularity plus modularity equals design-ability. Curr. Opin. Struct. Biol. 23, 622–631. https://doi.org/10.1016/j.sbi.2013.06.011
Stumpp M.T., Forrer P., Binz H.K., Pluckthun A. 2015. Repeat protein from collection of repeat proteins comprising repeat modules. US Patent No. 9,006,389. https://patents.google.com/patent/US9006389B2/en
Glasgow A.A., Huang Y.-M., Mandell D.J., Thompson M., Ritterson R., Loshbaugh A.L., Pellegrino J., Krivacic C., Pache R.A., Barlow K.A., Ollikainen N., Jeon D., Kelly M.J.S., Fraser J.S., Kortemme T. 2019. Computational design of a modular protein sense-response system. Science. 366, 1024–1028. https://doi.org/10.1126/science.aax8780
Sawyer N., Chen J., Regan L. 2013. All repeats are not equal: A module-based approach to guide repeat protein design. J. Mol. Biol. 425, 1826–1838. https://doi.org/10.1016/j.jmb.2013.02.013
Parmeggiani F., Huang P.-S., Vorobiev S., Xiao R., Park K., Caprari S., Su M., Seetharaman J., Mao L., Janjua H., Montelione G.T., Hunt J., Baker D. 2015. A general computational approach for repeat protein design. J. Mol. Biol. 427, 563–575. https://doi.org/10.1016/j.jmb.2014.11.005
Leaver-Fay A., Tyka M., Lewis S.M., Lange O.F., Thompson J., Jacak R., Kaufman K., Renfrew P.D., Smith C.A., Sheffler W., Davis I.W., Cooper S., Treuille A., Mandell D.J., Richter F., et al. 2011. ROSETTA3: An object-oriented software suite for the simulation and design of macromolecules. Methods Enzymol. 487, 545–574. https://doi.org/10.1016/B978-0-12-381270-4.00019-6
Kajander T., Cortajarena A.L., Mochrie S., Regan L. 2007. Structure and stability of designed TPR protein superhelices: Unusual crystal packing and implications for natural TPR proteins. Acta Crystallogr. D: Biol. Crystallogr. 63, 800–811. https://doi.org/10.1107/S0907444907024353
Mohan K., Ueda G., Kim A.R., Jude K.M., Fallas J.A., Guo Y., Hafer M., Miao Y., Saxton R.A., Piehler J., Sankaran V.G., Baker D., Garcia K.C. 2019. Topological control of cytokine receptor signaling induces differential effects in hematopoiesis. Science. 364, eaav7532. https://doi.org/10.1126/science.aav7532
Plückthun A. 2015. Designed ankyrin repeat proteins (DARPins): Binding proteins for research, diagnostics, and therapy. Annu. Rev. Pharmacol. Toxicol. 55, 489–511. https://doi.org/10.1146/annurev-pharmtox-010611-134654
Boersma Y.L. 2018. Advances in the application of designed ankyrin repeat proteins (DARPins) as research tools and protein therapeutics. Methods Mol. Biol. 1798, 307–327. https://doi.org/10.1007/978-1-4939-7893-9_23
Schilling J., Schöppe J., Plückthun A. 2014. From DARPins to LoopDARPins: Novel LoopDARPin design allows the selection of low picomolar binders in a single round of ribosome display. J. Mol. Biol. 426, 691–721. https://doi.org/10.1016/j.jmb.2013.10.026
Stumpp M.T., Amstutz P. 2007. DARPins: A true alternative to antibodies. Curr. Opin. Drug Discov. Dev. 10, 153–159. PMID:17436550
Schweizer A., Rusert P., Berlinger L., Ruprecht C.R., Mann A., Corthésy S., Turville S.G., Aravantinou M., Fischer M., Robbiani M., Amstutz P., Trkola A. 2008. CD4-specific designed ankyrin repeat proteins are novel potent HIV entry inhibitors with unique characteristics. PLoS Pathog. 4, e1000109. https://doi.org/10.1371/journal.ppat.1000109
Zahnd C., Kawe M., Stumpp M.T., de Pasquale C., Tamaskovic R., Nagy-Davidescu G., Dreier B., Schibli R., Binz H.K., Waibel R., Plückthun A. 2010. Efficient tumor targeting with high-affinity designed ankyrin repeat proteins: Effects of affinity and molecular size. Cancer Res. 70, 1595–1605. https://doi.org/10.1158/0008-5472.CAN-09-2724
Reichen C., Madhurantakam C., Plückthun A., Mittl P.R.E. 2014. Crystal structures of designed armadillo repeat proteins: Implications of construct design and crystallization conditions on overall structure. Protein Sci. 23, 1572–1583. https://doi.org/10.1002/pro.2535
Madhurantakam C., Varadamsetty G., Grütter M.G., Plückthun A., Mittl P.R.E. 2012. Structure-based optimization of designed Armadillo-repeat proteins. Protein Sci. 21, 1015–1028. https://doi.org/10.1002/pro.2085
Reichen C., Madhurantakam C., Hansen S., Grütter M.G., Plückthun A., Mittl P.R.E. 2016. Structures of designed armadillo-repeat proteins show propagation of inter-repeat interface effects. Acta Crystallogr. D: Struct. Biol. 72, 168–175. https://doi.org/10.1107/S2059798315023116
Ernst P., Honegger A., van der Valk F., Ewald C., Mittl P.R.E., Plückthun A. 2019. Rigid fusions of designed helical repeat binding proteins efficiently protect a binding surface from crystal contacts. Sci. Rep. 9, 16162. https://doi.org/10.1038/s41598-019-52121-9
Park K., Shen B.W., Parmeggiani F., Huang P.-S., Stoddard B.L., Baker D. 2015. Control of repeat-protein curvature by computational protein design. Nat. Struct. Mol. Biol. 22, 167–174. https://doi.org/10.1038/nsmb.2938
Rämisch S., Weininger U., Martinsson J., Akke M., André I. 2014. Computational design of a leucine-rich repeat protein with a predefined geometry. Proc. Natl. Acad. Sci. U. S. A. 111, 17875–17880. https://doi.org/10.1073/pnas.1413638111
Stumpp M.T., Forrer P., Binz H.K., Plückthun A. 2003. Designing repeat proteins: Modular leucine-rich repeat protein libraries based on the mammalian ribonuclease inhibitor family. J. Mol. Biol. 332, 471–487. https://doi.org/10.1016/s0022-2836(03)00897-0
Ernst P., Plückthun A. 2017. Advances in the design and engineering of peptide-binding repeat proteins. Biol. Chem. 398, 23–29. https://doi.org/10.1515/hsz-2016-0233
Reichen C., Hansen S., Plückthun A. 2014. Modular peptide binding: From a comparison of natural binders to designed armadillo repeat proteins. J. Struct. Biol. 185, 147–162. https://doi.org/10.1016/j.jsb.2013.07.012
Schlehuber S., Skerra A. 2002. Tuning ligand affinity, specificity, and folding stability of an engineered lipocalin variant—a so-called “anticalin”—using a molecular random approach. Biophys. Chem. 96, 213–228. https://doi.org/10.1016/s0301-4622(02)00026-1
Horibe T., Kohno M., Haramoto M., Ohara K., Kawakami K. 2011. Designed hybrid TPR peptide targeting Hsp90 as a novel anticancer agent. J. Transl. Med. 9, 8. https://doi.org/10.1186/1479-5876-9-8
Cortajarena A.L., Yi F., Regan L. 2008. Designed TPR modules as novel anticancer agents. ACS Chem. Biol. 3, 161–166. https://doi.org/10.1021/cb700260z
Horibe T., Torisawa A., Kohno M., Kawakami K. 2012. Molecular mechanism of cytotoxicity induced by Hsp90-targeted Antp-TPR hybrid peptide in glioblastoma cells. Mol. Cancer. 11, 59. https://doi.org/10.1186/1476-4598-11-59
Mejias S.H., Aires A., Couleaud P., Cortajarena A.L. 2016. Designed repeat proteins as building blocks for nanofabrication. Adv. Exp. Med. Biol. 940, 61–81. https://doi.org/10.1007/978-3-319-39196-0_4
Grove T.Z., Regan L., Cortajarena A.L. 2013. Nanostructured functional films from engineered repeat proteins. J. R. Soc. Interface. 10, 20130051. https://doi.org/10.1098/rsif.2013.0051
Carter N.A., Grove T.Z. 2015. Repeat-proteins films exhibit hierarchical anisotropic mechanical properties. Biomacromolecules. 16, 706–714. https://doi.org/10.1021/bm501578j
Mejías S.H., López-Andarias J., Sakurai T., Yoneda S., Erazo K.P., Seki S., Atienza C., Martín N., Cortajarena A.L. 2016. Repeat protein scaffolds: Ordering photo- and electroactive molecules in solution and solid state. Chem. Sci. 7, 4842–4847. https://doi.org/10.1039/c6sc01306f
Couleaud P., Adan-Bermudez S., Aires A., Mejías S.H., Sot B., Somoza A., Cortajarena A.L. 2015. Designed modular proteins as scaffolds to stabilize fluorescent nanoclusters. Biomacromolecules. 16, 3836–3844. https://doi.org/10.1021/acs.biomac.5b01147
Masakari Y., Hara C., Araki Y., Gomi K., Ito K. 2020. Improvement in the thermal stability of Mucor prainii-derived FAD-dependent glucose dehydrogenase via protein chimerization. Enzyme Microb. Technol. 132, 109387. https://doi.org/10.1016/j.enzmictec.2019.109387
Crennell S.J., Garman E.F., Laver W.G., Vimr E.R., Taylor G.L. 1993. Crystal structure of a bacterial sialidase (from Salmonella typhimurium LT2) shows the same fold as an influenza virus neuraminidase. Proc. Natl. Acad. Sci. U. S. A. 90, 9852–9856. https://doi.org/10.1073/pnas.90.21.9852
Glanz V.Y., Myasoedova V.A., Grechko A.V., Ore-khov A.N. 2018. Inhibition of sialidase activity as a therapeutic approach. Drug Des. Devel. Ther. 12, 3431–3437. https://doi.org/10.2147/DDDT.S176220
Sacramento C.Q., Jordão A.K., Abrantes J.L., Alves C.M., Marttorelli A., Fintelman-Rodrigues N., de Freitas C.S., de Melo G.R., Cunha A.C., Ferreira V.F., Souza T.M.L. 2020. Neuraminidase from influenza A and B viruses is susceptible to the compound 4-(4-phenyl-1H-1,2,3-triazol-1-yl)-2,2,6,6-tetramethylpiperidine-1-oxyl. Curr. Top. Med. Chem. 20, 132–139. https://doi.org/10.2174/1568026620666191227142433
Voet A.R.D., Noguchi H., Addy C., Simoncini D., Terada D., Unzai S., Park S.-Y., Zhang K.Y.J., Tame J.R.H. 2014. Computational design of a self-assembling symmetrical β-propeller protein. Proc. Natl. Acad. Sci. U. S. A. 111, 15102–15107. https://doi.org/10.1073/pnas.1412768111
Noguchi H., Addy C., Simoncini D., Wouters S., My-lemans B., Van Meervelt L., Schiex T., Zhang K.Y.J., Tame J.R.H., Voet A.R.D. 2019. Computational design of symmetrical eight-bladed β-propeller proteins. IUCrJ. 6, 46–55. https://doi.org/10.1107/S205225251801480X
Mylemans B., Laier I., Kamata K., Akashi S., Noguchi H., Tame J.R.H., Voet A.R.D. 2021. Structural plasticity of a designer protein sheds light on β-propeller protein evolution. FEBS J. 288, 530–545. https://doi.org/10.1111/febs.15347
Urvoas A., Guellouz A., Valerio-Lepiniec M., Graille M., Durand D., Desravines D.C., van Tilbeurgh H., Desmadril M., Minard P. 2010. Design, production and molecular structure of a new family of artificial alpha-helicoidal repeat proteins (αRep) based on thermostable HEAT-like repeats. J. Mol. Biol. 404, 307–327. https://doi.org/10.1016/j.jmb.2010.09.048
Guellouz A., Valerio-Lepiniec M., Urvoas A., Chevrel A., Graille M., Fourati-Kammoun Z., Desmadril M., van Tilbeurgh H., Minard P. 2013. Selection of specific protein binders for pre-defined targets from an optimized library of artificial helicoidal repeat proteins (alphaRep). PLoS One. 8, e71512. https://doi.org/10.1371/journal.pone.0071512
Valerio-Lepiniec M., Urvoas A., Chevrel A., Guellouz A., Ferrandez Y., Mesneau A., de la Sierra-Gallay I.L., Aumont-Nicaise M., Desmadril M., van Tilbeurgh H., Minard P. 2015. The αRep artificial repeat protein scaffold: A new tool for crystallization and live cell applications. Biochem. Soc. Trans. 43, 819–824. https://doi.org/10.1042/BST20150075
Deng D., Yan C., Pan X., Mahfouz M., Wang J., Zhu J.-K., Shi Y., Yan N. 2012. Structural basis for sequence-specific recognition of DNA by TAL effectors. Science. 335, 720–723. https://doi.org/10.1126/science.1215670
Mak A.N.-S., Bradley P., Cernadas R.A., Bogda-nove A.J., Stoddard B.L. 2012. The crystal structure of TAL effector PthXo1 bound to its DNA target. Science. 335, 716–719. https://doi.org/10.1126/science.1216211
Flechsig H. 2014. TALEs from a spring–superelasticity of Tal effector protein structures. PLoS One. 9, e109919. https://doi.org/10.1371/journal.pone.0109919
Bogdanove A.J., Voytas D.F. 2011. TAL effectors: Customizable proteins for DNA targeting. Science. 333, 1843–1846. https://doi.org/10.1126/science.1204094
Scholze H., Boch J. 2011. TAL effectors are remote controls for gene activation. Curr. Opin. Microbiol. 14, 47–53. https://doi.org/10.1016/j.mib.2010.12.001
Schaper E., Anisimova M. 2015. The evolution and function of protein tandem repeats in plants. New Phytol. 206, 397–410. https://doi.org/10.1111/nph.13184
Moore A.D., Björklund A.K., Ekman D., Bornberg-Bauer E., Elofsson A. 2008. Arrangements in the modular evolution of proteins. Trends Biochem. Sci. 33, 444–451. https://doi.org/10.1016/j.tibs.2008.05.008
Verstrepen K.J., Jansen A., Lewitter F., Fink G.R. 2005. Intragenic tandem repeats generate functional variability. Nat. Genet. 37, 986–990. https://doi.org/10.1038/ng1618
Chevanne D., Saupe S.J., Clavé C., Paoletti M. 2010. WD-repeat instability and diversification of the Podospora anserina hnwd non-self recognition gene family. BMC Evol. Biol. 10, 134. https://doi.org/10.1186/1471-2148-10-134
Schüler A., Bornberg-Bauer E. 2016. Evolution of protein domain repeats in metazoa. Mol. Biol. Evol. 33, 3170–3182. https://doi.org/10.1093/molbev/msw194
McElhinny A.S., Kazmierski S.T., Labeit S., Gregorio C.C. 2003. Nebulin: The nebulous, multifunctional giant of striated muscle. Trends Cardiovasc. Med. 13, 195–201. https://doi.org/10.1016/s1050-1738(03)00076-8
Chaudhuri I., Söding J., Lupas A.N. 2008. Evolution of the beta-propeller fold. Proteins. 71, 795–803. https://doi.org/10.1002/prot.21764
Kopec K.O., Lupas A.N. 2013. β-Propeller blades as ancestral peptides in protein evolution. PLoS One. 8, e77074. https://doi.org/10.1371/journal.pone.0077074
Tompa P. 2003. Intrinsically unstructured proteins evolve by repeat expansion. Bioessays. 25, 847–855. https://doi.org/10.1002/bies.10324
Funding
This work was supported by the Russian Foundation for Basic Research (project no. 20-14-50211).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interests. The authors declare that they have no conflict of interest.
This work does not contain any studies involving animals or human subjects performed by the authors.
Additional information
Translated by T. Tkacheva
Abbreviations: TPR, tetratricopeptide repeat; LRR, leucine-rich repeat; AR, ankyrin repeat; ArmR, armadillo repeat; HEAT (Huntingtin-EF3-PP2A-TOR1), huntingtin, elongation factor 3, protein phosphatase 2A, kinase TOR1; TIM, triose phosphate isomerase; WD40, β-transducin; AFP, antifreeze protein; HMM, hidden Markov model; DARPin, designed ankyrin repeat protein; dArmRP, designed armadillo repeat protein; CTPR, consensus tetratricopeptide repeat protein.
Rights and permissions
About this article
Cite this article
Deryusheva, E.I., Machulin, A.V. & Galzitskaya, O.V. Structural, Functional, and Evolutionary Characteristics of Proteins with Repeats. Mol Biol 55, 683–704 (2021). https://doi.org/10.1134/S0026893321040038
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0026893321040038