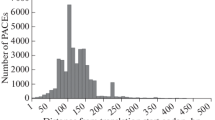
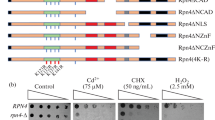
Abstract
Expression of Saccharomyces cerevisiae proteasomal genes is regulated in a coordinated manner by a system that includes the ScRpn4 transcription factor and its binding site known as PACE. Earlier we showed that, Rpn4-like proteins from the biotechnologically important yeast species Komagataellapfaffii (Pichiapastoris), Yarrowia lipolytica, and Debaryomyces hansenii are capable of complementing the RPN4 deletion in S. cerevisiae in spite of their low structural similarity to ScRpn4. The opportunistic yeast pathogen Candida glabrata has a gene coding for a Rpn4-like protein, which has not been characterized experimentally yet. The C. glabrata ortholog ScRpn4 was expressed heterologously and found to restore the stress resistance and expression of proteasomal genes in a mutant S. cerevisiae strain with a RPN4 deletion. This complementation required the unique N-terminal region of CgRpn4. The results indicate that CgRpn4 acts as a transcriptional activator of proteasomal genes. The S. cerevisiae model can be used for further structural and functional analyses of CgRpn4.




Similar content being viewed by others
REFERENCES
Gabaldon T., Carrete L. 2016. The birth of a deadly yeast: tracing the evolutionary emergence of virulence traits in Candida glabrata. FEMS Yeast Res. 16, fov110.
Mannhaupt G., Feldmann H. 2007. Genomic evolution of the proteasome system among hemiascomycetous yeasts. J. Mol. Evol. 65, 529–540.
Vermitsky J.-P., Earhart K.D., Smith W.L., Homayouni R., Edlind T.D., Rogers P.D. 2006. Pdr1 regulates multidrug resistance in Candida glabrata: Gene disruption and genome-wide expression studies. Mol. Microbiol. 61, 704–722.
Owsianik G., Balzil L., Ghislain M. 2002. Control of 26S proteasome expression by transcription factors regulating multidrug resistance in Saccharomyces cerevisiae. Mol. Microbiol. 43, 1295–1308.
Kapranov A.B., Kuryatova MV., Preobrazhenskaya O.V., Tutyaeva V.V., Stucka R., Feldmann H., Karpov V.L. 2001. Isolation and identification of PACE-binding protein Rpn4, a new transcriptional activator regulating 26S-proteasomal and other genes. Mol. Biol. (Moscow). 35 (3), 356–364.
Mannhaupt G., Schnall R., Karpov V., Vetter I., Feldmann H. 1999. Rpn4 acts as a transcription factor by binding to PACE, a nonamer box found upstream of 26S proteasomal and other genes in yeast. FEBS Lett. 450, 27–34.
Spasskaya D.S., Karpov D.S., Mironov A.S., Karpov V.L. 2014. Transcription factor Rpn4 promotes a complex antistress response in Saccharomyces cerevisiae cells exposed to methyl methanesulfonate. Mol. Biol. (Moscow). 48 (1), 141–149.
Grineva E.N., Leinsoo A.T., Spsskaya D.S., Karpov D.S., Karpov V.L. 2014. Functional analysis of Rpn4-like proteins from Komagataella (Pichia) pastoris and Yarrowia lipolytica in a heterologous Saccharomyces cerevisiae system. Biotekhnologiya. 6, 8–17.
Karpov D.S., Grineva E.N., Leinsoo A.T., Nadolinskaia N.I., Danilenko N.K., Tutyaeva V.V., Spasskaya D.S., Preobrazhenskaya O.V., Lysov Y.P., Karpov V.L. 2017. Functional analysis of Debaryomyces hansenii Rpn4 on a genetic background of Saccharomyces cerevisiae. FEMS Yeast Res. 17, fow098.
Enjalbert B., Smith D.A., Cornell M.J., Alam I., Nicholls S., Brown A.J.P., Quinn J. 2006. Role of the Hog1 stress-activated protein kinase in the global transcriptional response to stress in the fungal pathogen Candida albicans. Mol. Biol. Cell. 17, 1018‒1032.
Gietz R.D., Woods R.A. 2002. Transformation of yeast by the LiAc/ss carrier DNA/PEG method. Methods Enzymol. 350, 87–96.
Schwarzmuller T., Ma B., Hiller E., Istel F., Tscherner M., Brunke S., Ames L., Firon A., Green B., Cabral V., Marcet-Houben M., Jacobsen I.D., Quintin J., Seider K., Frohner I., et al. 2014. Systematic phenotyping of a large-scale Candida glabrata deletion collection reveals novel antifungal tolerance genes. PLoS Pathog. 10, e1004211.
Amberg D.C., Burke D.J., Strathern J.N. 2006. Isolation of yeast genomic DNA for southern blot analysis. CSH Protoc. 2006.
Karpov D.S., Spasskaya D.S., Tutyaeva V.V., Mironov A.S., Karpov V.L. 2013. Proteasome inhibition enhances resistance to DNA damage via upregulation of Rpn4-dependent DNA repair genes. FEBS Lett. 587, 3108–3114.
Spasskaya D.S., Karpov D.S., Karpov V.L. 2011. Escherichia coli Dam-methylase as a molecular tool for mapping binding sites of the yeast transcription factor Rpn4. Mol. Biol. (Moscow). 45 (4), 591–599.
Schmitt M.E., Brown T.A., Trumpower B.L. 1990. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 18, 3091–3092.
Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31, 3406–3415.
Ha S.W., Ju D., Xie Y. 2012. The N-terminal domain of Rpn4 serves as a portable ubiquitin-independent degron and is recognized by specific 19S RP subunits. Biochem. Biophys. Res. Commun. 419, 226–231.
Karpov D.S., Tutyaeva V.V., Karpov V.L. 2008. Mapping of yeast Rpn4p transactivation domains. FEBS Lett. 582, 3459–3464.
McWilliam H., Li W., Uludag M., Squizzato S., Park Y.M., Buso N., Cowley A.P., Lopez R. 2013. Analysis tool web services from the EMBL-EBI. Nucleic Acids Res. 41, W597–W600.
Persikov A., Singh M. 2014. De novo prediction of DNA-binding specificities for Cys2His2 zinc finger proteins. Nucleic Acids Res. 42, 97–108.
Wolfe S.A., Nekludova L., Pabo C.O. 2000. DNA recognition by Cys2His2 zinc finger proteins. Annu. Rev. Biophys. Biomol. Struct. 29, 183–212.
Dreier B., Segal D.J., Barbas C.F. 2000. Insights into the molecular recognition of the 5′-GNN-3′ family of DNA sequences by zinc finger domains. J. Mol. Biol. 303, 489– 502.
Crooks G.E., Hon G., Chandonia J.M., Brenner S.E. 2004. WebLogo: A sequence logo generator. Genome Res. 14, 1188–1190.
Brennan R.J., Schiestl R.H. 1996. Cadmium is an inducer of oxidative stress in yeast. Mutat. Res. 356, 171–178.
Haugen A.C., Kelley R., Collins J.B., Tucker C.J., Deng C., Afshari C.A., Brown J.M., Ideker T., Van Houten B. 2004. Integrating phenotypic and expression profiles to map arsenic-response networks. Genome Biol. 5, R95.
London M.K., Keck B.I., Ramos P.C., R. Dohmen R.J. 2004. Regulatory mechanisms controlling biogenesis of ubiquitin and the proteasome. FEBS Lett. 567, 259–264.
Lee J., Godon C., Lagniel G., Spector D., Garin J., Labarre J., Toledano M.B. 1999. Yap1 and Skn7 control two specialized oxidative stress response regulons in yeast. J. Biol. Chem. 274, 16040–16046.
Mitchell P.J., Tjian R. 1989. Transcriptional regulation in mammalian cells by sequence specific DNA binding proteins. Science. 245, 371–378.
Author information
Authors and Affiliations
Corresponding author
Additional information
Translated by T. Tkacheva
Abbreviations: PACE, proteasome-associated control element; MMS, methyl methane sulfonate; 4-NQO, 4-nitroquinoline-1-oxide; NAD, N-terminal acidic domain; CAD, C-terminal acidic domain; NTAD, N-terminal transactivation domain; N-ZnF, N-terminal zinc finger domain; C-ZnF, C-terminal zinc finger domain; QRR, glutamine-rich region.
Rights and permissions
About this article
Cite this article
Karpov, D.S., Grineva, E.N., Kiseleva, S.V. et al. Candida glabrata Rpn4-like Protein Complements the RPN4 Deletion in Saccharomyces cerevisiae. Mol Biol 53, 242–248 (2019). https://doi.org/10.1134/S0026893319020067
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0026893319020067