Abstract—
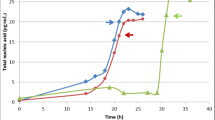
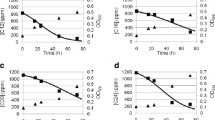
The role of heat shock proteins in ability of Rhodococcus pyridinivorans 5Ap to degrade hydrocarbons at different temperatures was studied. The presence of the Сpn60.1–Сpn10 chaperons and of the Hrc regulatory protein was found to be required for hexadecane degradation at 42°C. When genetic determinants responsible for synthesis of these proteins were inactivated, the efficiency of hexadecane degradation decreased 1.7 and 2.7 times, respectively. Mutations in the cpn and hrcA genes did not affect the viability of R. pyridinivorans 5Ар: the original strain and the mutants exhibited the same growth rates at all temperatures in the minimal medium with succinate and in full-strength medium. In the absence of the Сpn60.1–Сpn10 heat shock proteins, the growth rate at 42°C decreased in the case of minimal agar media with kerosene, diesel fuel, acetone, naphthalene, 2-methylnaphthalene, or phenanthrene.
Similar content being viewed by others
REFERENCES
Barreiro, C., González-Lavado, E., Pátek, M., and Martín, J.F., Transcriptional analysis of the groES-groEL1, groEL2, and dnaK genes in Corynebacterium glutamicum: characterization of heat shock-induced promoters, J. Bacteriol., 2004, vol. 186, pp. 4813–4817.
Bullock, W.O., Fernandez, J.M., and Short, J.M., XL1-Blue–a high-efficiency plasmid transforming recA Escherichia coli strain with β-galactosidase selection, Biotechniques, 1987, vol. 5, pp. 376–379.
Charniauskaya, M.I., Bukliarevich, A.A., Akhremchuk, A.E., Valentovich, L.N., and Titok, M.A., Primary analysis of the genome of oil-degrading bacteria Rhodococcus pyridinivorans 5Ap, Tr. Belorus. Gos. Univ., 2016, vol. 11, no. 1, pp. 219–223.
Charniauskaya, M.I., Bukliarevich, A.A., Delegan, Ya.A., Akhremchuk, A.E., Filonov, A.E., and Titok, M.A., Biodiversity of hydrocarbon-oxidizing soil bacteria from various climatic zones, Microbiology (Moscow), 2018, vol. 87, pp. 699–711.
Chaturongakul, S., Raengpradub, S., Palmer, M.E., Bergholz, T.M., Orsi, R.H., Hu, Y., Ollinger, J., Wiedmann, M., and Boor, K.J., Transcriptomic and phenotypic analyses identify coregulated, overlapping regulons among PrfA, CtsR, HrcA, and the alternative sigma factors sigmaB, sigmaC, sigmaH, and sigmaL in Listeria monocytogenes, Appl. Environ. Microbiol., 2011, no. 77, pp. 187–200.
Crombie, A.T., Khawand, M.E., Rhodius, V.A., Fengler, K.A., Miller, M.C., Whited, G.M., McGenity, T.J., and Murrell, J.C., Regulation of plasmid-encoded isoprene metabolism in Rhodococcus, a representative of an important link in the global isoprene cycle, Environ. Microbiol., 2015, vol. 17, pp. 3314–3329.
Furuya, T., Hayashi, M., Semba, H., and Kino, K., The mycobacterial binuclear iron monooxygenases require a specific chaperonin-like protein for functional expression in a heterologous host, FEBS J., 2013, vol. 280, pp. 817–826.
Goyal, K., Qamra, R., and Mande, S.C., Multiple gene duplication and rapid evolution in the groEL gene: functional implications, J. Mol. Evol., 2006, vol. 63, pp. 781–787.
Grandvalet, C., Rapoport, G., and Mazodier, P., hrcA, encoding the repressor of the groEL genes in Streptomyces albus G, is associated with a second dnaJ gene, J. Bacteriol., 1998, vol. 180, pp. 5129–5134.
Hu, Y., Henderson, B., Lund, P.A., Tormay, P., Ahmed, M.T., Gurcha, S.S., Besra, G.S., and Coates, A.R., A Mycobacterium tuberculosis mutant lacking the groEL homologue cpn60.1 is viable but fails to induce an inflammatory response in animal models of infection, Infect. Immun., 2008, vol. 76, pp. 1535–1546.
Katalog shtammov regional’noi profilirovannoi kollektsii alkanotrofnykh mikroorganizmov (Strain Catalog of the Regional Profiled Collection of Alkanotrophic Microorganisms), Ivshina, I.B., Ed., 1994.
Khosravinia, S., Mahdavi, M.A., Gheshlaghi, R., and Dehghani, H., Characterization of truncated dsz operon responsible for dibenzothiophene biodesulfurization in Rhodococcus sp. FUM94, Appl. Biochem. Biotechnol., 2018, vol. 184, pp. 885–896.
Kim, A.I., Ghosh, P., Aaron, M.A., Bibb, L.A., Jain, S.,and Hatfull, G.F., Mycobacteriophage Bxb1 integrates into the Mycobacterium smegmatis groEL1 gene, Mol. Microbiol., 2003, vol. 50, pp. 463–473.
Kolaj, O., Spada, S., Robin, S., and Wall, J.G., Use of folding modulators to improve heterologous protein production in Escherichia coli [Electronic resource], Microbial Cell Factories, 2009, vol. 8, no. 9. Date of access: 14.10.2018.https://doi.org/10.1186/1475-2859-8-9
Lin, Z., Madan, D., and Rye, H.S., GroEL stimulates protein folding through forced unfolding, Nat. Struct. Mol. Biol., 2008, vol. 15, pp. 303–311.
Lund, P.A., Multiple chaperonins in bacteria–why so many?, FEMS Microbiol. Rev., 2009, vol. 334, pp. 785–800.
Maniatis, T., Fritsh, E.E., and Sambrook, J., Molecular Cloning. A Laboratory Manual, Cold Spring Harbor Laboratory, 1982.
Manual of Methods for General Bacteriology, Gerhardt, P., Murray, R.G.E., Costilow, R.N., Nester, E.W., Wood, W.A., Krieg, N.R., and Phillips, G.B., Eds., Washington: Amer. Soc. Microbiol., 1981.
McCarl, V., Somerville, M.V., Ly, M.A., Henry, R., Liew, E.F., Wilson, N.L., Holmes, A.J., and Coleman, N.V., Heterologous expression of Mycobacterium alkene monooxygenases in gram-positive and gram-negative bacterial hosts [Electronic resource], Appl. Environ Microbiol., 2018, vol. 84, no. 15. Date of access: 14.10.2018.https://doi.org/10.1128/AEM.00397-18
Metcalf, W.W., Jiang, W., and Wanner, B.L., Use of the rep technique for allele replacement to construct new Escherichia coli hosts for maintenance of R6Kgamma origin plasmids at different copy numbers, Gene, 1994, vol. 138, pp. 1–7.
Miller, J.H., Experiments in Molecular Genetics, Cold Spring Harbor: Cold Spring Harbor Lab., 1972.
Nicolaou, S.A., Gaida, S.M., and Papoutsakis, E.T., A comparative view of metabolic and substrate stress and tolerance in microbial bioprocessing: from biofuels and chemicals, to biocatalysis and bioremediation, Metabolic Engineering., 2010, vol. 12, pp. 307–331.
Ojha, A., Anand, M., Bhatt, A., Kremer, L., Jacobs, W.R., Jr., and Hatfull, G.F., GroEL1: a dedicated chaperone involved in mycolic acid biosynthesis during biofilm formation in mycobacteria, Cell, 2005, vol. 123, pp. 861–873.
Orro, A., Cappelletti, M., D’Ursi, P., Milanesi, L., Di Canito, A., Zampolli, J., Collina, E., Decorosi, F., Viti, C., Fedi, S., Presentato, A., Zannoni, D., and Di Gennaro, P., Genome and phenotype microarray analyses of Rhodococcus sp. BCP1 and Rhodococcus opacus R7: genetic determinants and metabolic abilities with environmental relevance [Electronic resource], PloS One, 2015, vol. 10, no. 10. Date of access: 14.10.2018.https://doi.org/10.1371/journal.pone.0139467
RF Patent no. 2617941, 2017.
Romanenko, V.I. and Kuznetsov, S.I., Ekologiya mikroorganizmov presnykh vod: Laboratornoe rukovodstvo (Ecology of Freshwater Microorganisms: A Laboratory Manual), Moscow: Nauka, 1974.
Roncarati, D. and Scarlato, V., Regulation of heat shock genes in bacteria: from signal sensing to gene expression output, FEMS Microbiol. Rev., 2017, vol. 41, pp. 549–574.
Schäfer, A., Tauch, A., Jäger, W., Kalinowski, J., Thierbach, G., and Pühler, A., Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum, Gene, 1994, vol. 145, pp. 69–73.
Servant, P., Thompson, C.J., and Mazodier, P., Post-transcriptional regulation of the groEL1 gene of Streptomyces albus, Mol. Microbiol., 1994, vol. 12, pp. 423–432.
Stewart, G.R., Wernisch, L., Stabler, R., Mangan, J.A., Hinds, J., Laing, K.G., Young, D.B, and Butcher, P.D., Dissection of the heatshock response in Mycobacterium tuberculosis using mutants and microarrays, Microbiology (UK), 2002, vol. 148, pp. 3129–3138.
Takihara, H., Ogihara, J., Yoshida, T., Okuda, S., Nakajima, M., Iwabuchi, N., and Sunairi, M. Enhanced translocation and growth of Rhodococcus erythropolis PR4 in the alkane phase of aqueous-alkane two phase cultures were mediated by GroEL2 overexpression, Microbes Environ., 2014, vol. 29, pp. 346–352.
te Riele, H., Michel, B., and Ehrlich, S.D., Single-stranded plasmid DNA in Bacillus subtilis and Staphylococcus aureus, Proc. Natl. Acad. Sci. U. S. A., 1986, vol. 83, pp. 2541–2545.
Tomas, C.A., Welker, N.E., and Papoutsakis, E.T., Overexpression of groESL in Clostridium acetobutylicum results in increased solvent production and tolerance, prolonged metabolism, and changes in the cell’s transcriptional program, Appl. Environ. Microbiol., 2003, vol. 69, pp. 4951–4965.
van der Geize, R., Hessels, G.I., van Gerwen, R., van der Meijden, P., and Dijkhuizen, L. Unmarked gene deletion mutagenesis of kstD, encoding 3-ketosteroid Delta1-dehydrogenase, in Rhodococcus erythropolis SQ1 using sacB as counter-selectable marker, FEMS Microbiol. Lett., 2001, vol. 205, pp. 197–202.
Funding
The study was supported by the Belarusian Republican Foundation for -Fundamental Research (project no. B18-070).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interests. The authors declare that they have no conflict of interest.
Statement on the welfare of animals. This article does not contain any studies involving animals performed by any of the authors.
Additional information
Translated by D. Timchenko
Rights and permissions
About this article
Cite this article
Bukliarevich, H.A., Charniauskaya, M.I., Akhremchuk, A.E. et al. Effect of the Structural and Regulatory Heat Shock Proteins on Hydrocarbon Degradation by Rhodococcus pyridinivorans 5Ap. Microbiology 88, 573–579 (2019). https://doi.org/10.1134/S0026261719050023
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0026261719050023