Abstract
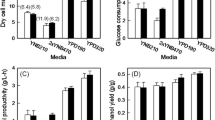
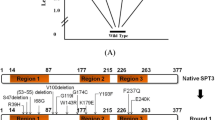
The SPT15 gene of Saccharomyces cerevisiae obtained using site-saturation mutagenesis was connected to the expression vector pYES2NTc. Then, the mutant library was constructed. Twelve single point mutant genes were obtained, and the recombinant plasmids were constructed with wild gene SPT15 and control gene. The S. cerevisiae strains were obtained by converting recombinant plasmids into the S. cerevisiae INVSC1 by lithium acetate method. Using glucose as substrate, the recombinant S. cerevisiae was inoculated and fermented to determine its ethanol yield. The ethanol yields of recombinant S. cerevisiae INVSC1-SPT15-K127M and S. cerevisiae INVSC1-SPT15-K127N were greatly increased by 43.0 ± 0.9 and 43.7 ± 0.2 g/L, i.e., 26.5 and 28.5%, respectively, compared with that of the control strain S. cerevisiae INVSC1-Neo. The site-saturation mutagenesis technique was used instead of random mutagenesis, and 2 mutant strains with phenotypic improvements were screened in the limited mutant library, which indicated that 127 amino acids had an important effect on the binding efficiency of Spt15 and TATA.




Similar content being viewed by others
REFERENCES
Kasavi, C., Eraslan, S., Arga, K. Y., Oner, E. T. and Kirdar, B., BMC Syst. Biol., 2014, vol. 8, p. 90.
Deparis, Q., Claes, A., Foulquie-Moreno, M. R. and Thevelein, J. M., FEMS Yeast Res., 2017, vol. 17, no. 4, pp. 1–17.
Alper, H. and Stephanopoulos, G., Metab Eng., 2007, vol. 9, no. 3, pp. 258–267.
Cadwell, R. C. and Joyce, G. F., PCR Methods Appl., 1992, vol. 2, no. 1, pp. 28–33.
Crameri, A., Raillard, S. A., Bermudez, E. and Stemmer, W. P., Nature, 1998, vol. 391, no. 6664, pp. 288–291.
Bhaumik, S. R. and Green, M. R., Mol. Cell Biol., 2002, vol. 22, no. 21, pp. 7365–7371.
Hallborn, J., Walfridsson, M., Airaksinen, U., Ojamo, H., Hahn-Hagerdal, B., Penttila, M. and Kerasnen, S., Biotechnology (N. Y.), 1991, vol. 9, no. 11, pp. 1090–1095.
Yang, J., Bae, J. Y., Lee, Y. M., Kwon, H., Moon, H. Y., Kang, H. A., et al., Biotechnol. Bioeng., 2011, vol. 108, no. 8, pp. 1776–1787.
El-Rotail, A., Zhang, L., Li, Y., Liu, S. P. and Shi, G. Y., AMB Express, 2017, vol. 7, no. 1, p. 111.
Tan, F., Wu, B., Dai, L., Qin, H., Shui, Z., Wang, J., et al., Microb. Cell Fact., 2016, vol. 15. p. 4. https://doi.org/10.1186/s12934-015-0398-y
Darst, R. P., Dasgupta, A., Zhu, C., Hsu, J. Y., Vroom, A., Muldrow, T. and Auble, D. T., J. Biol. Chem., 2003, vol. 278, no. 15, pp. 13216–13226.
Zheng, L., Baumann, U. and Reymond, J. L., Nucleic Acids Res., 2004, vol. 32, no. 14, p. e115.
Gietz, R. D., Schiestl, R. H., Willems, A. R. and Woods, R. A., Yeast, 1995, vol. 11, no. 4, pp. 355–360.
Gietz, R. D. and Woods R. A., Methods Mol. Biol., 2006, vol. 313, pp. 107–120.
Seong, Y. J., Park, H., Yang, J., Kim, S. J., Choi, W., Kim, K. H. and Park, Y. C., Appl. Microbiol. Biotechnol., 2017, vol. 101, no. 9, pp. 3567–3575.
Hou, L., Cao, X., Wang, C. and Lu, M., Lett. Appl. Microb-iol., 2009, vol. 49, no. 1, pp. 14–19.
Funding
This work was supported by Research Planning Project of Basic and Advanced Technology of Henan Province (nos. 20181847 and 202102310022), the Open Project Program of the State Key Laboratory of Motor Vehicle Biofuel Technology (nos. KFKT2018001, KFKT2018002) and Natural Science Foundation of Henan Educational Committee (nos. 18A180025).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
The authors declare that they have no conflict of interest. This article does not contain any studies involving animals or human participants performed by any of the authors.
Rights and permissions
About this article
Cite this article
Ke, T., Liu, J., Zhao, S. et al. Using Global Transcription Machinery Engineering (GTME) and Site-Saturation Mutagenesis Technique to Improve Ethanol Yield of Saccharomyces cerevisiae . Appl Biochem Microbiol 56, 563–568 (2020). https://doi.org/10.1134/S0003683820050087
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0003683820050087