Abstract
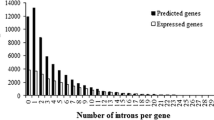
As determined by computer sequence analysis, the average exon length in Arabidopsis thaliana, Oryza sativa, Caenorhabditis elegans, and Homo sapiens genes decreases with an increasing number of introns. In A. thaliana and O. sativa, variations in intron and exon lengths with an increasing number of introns are highly correlated. Linear correlation is observed between the total exon length and the number of introns, while the gene length increases in proportion to the number of introns. In human, the average intron and gene lengths depended on the gene density in DNA.
Similar content being viewed by others
References
Wheeler D.L., Barrett T., Benson D.A., et al. 2007. The molecular biology database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 35, D5–D12.
Naora H., Deacon N.J. 1982. Relationship between the total size of exons and introns in protein-coding genes of higher eukaryotes. Proc. Natl. Acad. Sci. USA. 79. 6196–6200.
Hawkins J.D. 1988. A survey on intron and exon lengths. Nucleic Acids Res. 16. 9893–9908.
Smith M.W. 1988. Structure of vertebrate genes: A statistical analysis implicating selection. J. Mol. Evol. 27, 45–55.
Goffeau A., Barrell B.G., Bussey H., et al. 1996. Life with 6000 genes. Science. 274, 546–547.
Spingola M., Grate L., Haussler D.A. 1999. Genomewide bioinformatic and molecular analysis of intron in S. cerevisiae. RNA. 5, 221–234.
The Arabidopsis Genome Initiative. 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 408, 796–813.
The Caenorhabditis elegans Sequencing Consortium. 1998. Genome sequence of the nematode Caenorhabditis elegans: A platform for investigating biology. Science. 282, 2012–2018.
Adams M.D., Celiker S.E., Holit R.A., et al., 2000. The genome sequence of Drosophila melanogaster. Science. 287, 2185–2195.
Venter J.C., Adams M.D., Myers E.W., et al., 2001. The sequence of the human genome. Science. 291, 1304–1351.
Deutsch M., Long M. 1999. Intron-exon structures of eukaryotic model organisms. Nucleic Acids Res. 27, 3219–3228.
Sakharkar M.K., Chow V.T.K., Kangueane P. 2004. Distributions of exons and introns in the human genome. In Silico Biol. 4, 0032.
Schisler N.J., Palmer J.D. 2000. The IDB and IEDB: Intron sequence and evolution databases. Nucleic Acids Res. 28, 181–184.
Bultrini E., Pizzi E., Giudice P., Frontani C. 2003. Pentamer vocabularies characterizing introns and intron-like intergenic tracts from Caenorhabditis elegans and Drosophila melanogaster. Gene. 304, 183–192.
Comeron J.M., Kreitman M. 2000. The correlation between intron length and recombination in Drosophila: Dynamic equilibrium between mutational and selective forces. Genetics. 156, 1175–1190.
Cho S., Jin S-W., Cohen A., Ellis R.E. 2004. A phylogeny of Caenorhabditis reveals frequent loss of introns during nematode evolution. Genet. Res. 14, 1207–1220.
Coghlan A., Wolfe K.H. 2004. Origins of recently gained introns in Caenorhabditis. Proc. Natl. Acad. Sci. USA. 101, 11 362–11 367.
Wendel J.F., Cronn R.C., Alvarez I., Liu B., Small R.L., Senchina D.S. 2002. Intron size and genome size in plants. Mol. Biol. Evol. 19, 2346–2352.
Katinka M.D., Duprat S., Cornillot E., et al. 2001. Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature. 414, 450–453.
Loftus B.J., Fung E., Roncaglia P., et al. 2005. The genome of the basidiomycetous yeast and human pathogen Cryptococcus neoformans. Science. 307, 1321–1324.
Zhang M.Q. 1998. Statistical features of human exons and their flanking regions. Human Mol. Genet. 7, 919–932.
Duret L., Mouchiroud D., Gautier C. 1995. Statistical analysis of vertebrate sequences reveals that long genes are scarce in GC-rich isochores. J. Mol. Evol., 40, 308–317.
Bernardi G. 2001. Misunderstandings about isochores. Part 1. Gene. 276, 3–13.
Carels N., Bernardi G. 2000. Two classes of genes in plants. Genetics. 154, 1819–1825.
Oliver J.L., Marin A. 1996. A relationship between GC content and coding-sequence length. J. Mol. Evol. 43, 216–223.
Castillo-Davis C.I., Mekhedov S.L., Hart D.L., Koonin E.V., Kondrashov F.A. 2002. Selection for short introns in highly expressed genes. Nature Genet. 31, 415–418.
Clark F., Thanaraj T.A. 2002. Categorization and characterization of transcript-confirmed constitutively and alternatively spliced introns and exons from human. Hum. Mol. Genet. 11, 451–464.
Bell M.V., Cowper A.E., Lefrane V.-P., Bell J.I., Sereaton G.R. 1998. Influence of intron length on alternative splicing of CD44. Mol. Cell. Biol., 18, 5930–5941.
Comeron J.M. 2004. Selective and mutational patterns associated with gene expression in humans: Influences on synonymous composition and intron presence. Genetics. 167, 1293–1304.
Nott A., Meislin S.H., Moore M.J. 2003. A quantitative analysis of intron effect on mammalian gene expression. RNA. 9, 607–617.
Das D., Clark T.A., Schweitzer A., et al. 2007. A correlation with exon expression approach to identify cis-regulatory elements for tissue-specific alternative splicing. Nucleic Acids Res. 35, 4845–4857.
Ren X.-Y., Vorst O., Fiers M., Stiekema W.J., Nap J.-P. 2006. In plants, highly expressed genes are the least compact. Trends Genet. 22, 528–532.
Chung B.Y.W., Simons C., Furth A.E., et al. 2006. Effect of 5′UTR introns on gene expression in Arabidopsis thaliana. BMS Genomics. 7, 120–133.
Coulombe-Huntington J., Majewski J. 2007. Characterization of intron loss events in mammals. Genome Res. 17, 23–32.
Liu M., Walch H., Wu S., Grigoriev A. 2005. Significant expansion of exon-bordering protein domains during animal proteome evolution. Nucleic Acids Res. 33, 95–105.
Bon E., Casaregola S., Blandin G., et al. 2003. Molecular evolution of eukaryotic genomes: Haemiascomycetous yeast spliceosomal introns. Nucleic Acids Res. 31, 1121–1135.
Carmel L., Wolf Y.I., Rogozin I.B., Koonin E.V. 2007. Three distinct modes of intron dynamics in the evolution of eukaryotes. Genome Res. 17, 1034–1044.
Roy S.W., Penny D. 2007. Intron length distributions and gene prediction. Nucleic Acids Res. 35, 4737–4742.
Stamm S., Ben-Ari S., Rafalska I., Tang Y., Zang Z., Toiber D., Thanaraj T.A., Soreq H. 2005. Function of alternative splicing. Gene. 344, 1–20.
Malko D.B., Makeev V.J., Mironov A.A., Gelfand M.S. 2006. Evolution of exon-intron structure and alternative splicing in fruit flies and malarial mosquito genomes. Genome Res. 16, 505–509.
Marcucci R., Baralle F.E., Romano M. 2007. Complex splicing control of the human thrombopoietin gene by intronic G runs. Nucleic Acids Res. 35, 132–142.
Kim E., Magen A., Ast G. 2007. Different levels of alternative splicing among eukaryotes. Nucleic Acids Res. 35, 125–131.
Sheth N., Roca X., Hastings M.L., Roeder T., Krainer A.R., Sachidanandam R. 2006. Comprehensive splice-state analysis using comparative genomics. Nucleic Acids Res. 34, 3955–3967.
Singh N.N., Singh R.N., Androphy E.J. 2007. Iodulating role of RNA structure in alternative splicing of a critical exon in the spinal muscular atrophy genes. Nucleic Acids Res. 35, 371–389.
Lakin G.F. 1990. Biometriya (Biometrics). Moscow: Vysshaya Shkola.
Author information
Authors and Affiliations
Corresponding author
Additional information
Original Russian Text © Sh.A. Atambayeva, V.A. Khailenko, A.T. Ivashchenko, 2008, published in Molekulyarnaya Biologiya, 2008, Vol. 42, No. 2, pp. 352–361.
An erratum to this article can be found online at http://dx.doi.org/10.1134/S0026893308040237.
Rights and permissions
About this article
Cite this article
Atambayeva, S.A., Khailenko, V.A. & Ivashchenko, A.T. Intron and exon length variation in Arabidopsis, rice, nematode, and human. Mol Biol 42, 312–320 (2008). https://doi.org/10.1134/S0026893308020180
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0026893308020180