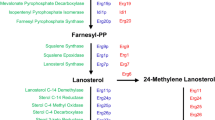
Abstract
Sterols are important components of biological membranes that determine the physicochemical properties of lipid bilayer and regulate the functioning of membrane proteins. Being insoluble in water, sterols cannot diffuse between the membrane compartments separated by an aqueous phase. For this reason, distribution of sterols across cellular membranes is rather uneven. Membrane-to-membrane transport of sterols occurs mainly in a non-vesicular fashion and is provided by Lam and Osh proteins. In this review, we discuss the consequences of impairments in sterol biosynthesis and transport mostly relying on the studies performed on the model organism Saccharomyces cerevisiae. Despite the fact that molecular mechanisms underlying the functioning of Lam and Osh proteins are well established, the biological roles of these proteins are still unclear, because deletions of corresponding genes do not affect yeast phenotype. At the same time, disruptions in the biosynthesis of ergosterol, the major sterol of S. cerevisiae, lead to either cell death or reduced stress resistance. However, under certain conditions (e.g., mild salt or thermal stresses), a decrease in the ergosterol levels causes an increase in cell resistance. This suggests that the cells possess a mechanism facilitating rapid adjustment of the plasma membrane sterol content. We argue that the biological role of Lam proteins is, in particular, fast optimization of sterol composition of cell membranes.
Similar content being viewed by others
Abbreviations
- ER:
-
endoplasmic reticulum
- Lam proteins:
-
lipid transfer proteins anchored at the membrane contact sites
- MCS:
-
membrane contact site
- MDR:
-
multidrug resistance
- ORD:
-
OSBP-related domain (400-a.a.-long lipid-bindingdomain)
- OSBP:
-
oxysterolbinding protein
- Osh proteins:
-
oxysterolbinding protein homologs
- PI4P:
-
phosphatidylinositol 4 phosphate
- PM:
-
plasma membrane
- StART:
-
steroidogenic acute regulatory transfer
References
Desmond, E., and Gribaldo, S. (2009) Phylogenomics of sterol synthesis: insights into the origin, evolution, and diversity of a key eukaryotic feature, Genome Biol. Evol., 1, 364–381, doi: https://doi.org/10.1093/gbe/evp036.
Weete, J. D., Abril, M., and Blackwell, M. (2010) Phylogenetic distribution of fungal sterols, PLoS One, 5, e10899, doi: https://doi.org/10.1371/journal.pone.0010899.
Souza, C. M., and Pichler, H. (2007) Lipid requirements for endocytosis in yeast, Biochim. Biophys. Acta, 1771, 442–454, doi: https://doi.org/10.1016/j.bbalip.2006.08.006.
Gimpl, G., and Fahrenholz, F. (2002) Cholesterol as stabilizer of the oxytocin receptor, Biochim. Biophys. Acta, 1564, 384–392.
Mayor, S., Sabharanjak, S., and Maxfield, F. R. (1998) Cholesterol–dependent retention of GPI–anchored proteins in endosomes, EMBO J., 17, 4626–4638, doi: https://doi.org/10.1093/emboj/17.16.4626.
Umebayashi, K., and Nakano, A. (2003) Ergosterol is required for targeting of tryptophan permease to the yeast plasma membrane, J. Cell. Biol., 161, 1117–1131, doi: https://doi.org/10.1083/jcb.200303088.
Lucero, H. A., and Robbins, P. W. (2004) Lipid rafts–protein association and the regulation of protein activity, Arch. Biochem. Biophys., 426, 208–224, doi: https://doi.org/10.1016/j.abb.2004.03.020.
Mullner, H., and Daum, G. (2004) Dynamics of neutral lipid storage in yeast, Acta Biochim. Polonica, 51, 323–347, doi: org/035001323.
Gimpl, G., Burger, K., and Fahrenholz, F. (1997) Cholesterol as modulator of receptor function, Biochemistry, 36, 10959–10974, doi: https://doi.org/10.1021/bi963138w.
Blackwell, M. (2017) Made for each other: ascomycete yeasts and insects, Microbiol. Spectr., 5, doi: https://doi.org/10.1128/microbiolspec.FUNK-0081-2016.
Clark, A. J., and Block, K. (1959) The absence of sterol synthesis in insects, J. Biol. Chem., 234, 2578–2582.
Rietveld, A., Neutz, S., Simons, K., and Eaton, S. (1999) Association of sterol–and glycosylphosphatidylinositol–linked proteins with Drosophila raft lipid microdomains, J. Biol. Chem., 274, 12049–12054.
C. elegans Sequencing Consortium (1998) Genome sequence of the nematode C. elegans: a platform for investigating biology, Science, 282, 2012–2008.
Kurzchalia, T. V., and Ward, S. (2003) Why do worms need cholesterol? Nat. Cell Biol., 5, 684–688, doi: https://doi.org/10.1038/ncb0803-684.
Huang, J., and Feigenson, G. W. (1999) A microscopic interaction model of maximum solubility of cholesterol in lipid bilayers, Biophys. J., 76, 2142–2157, doi: https://doi.org/10.1016/S0006-3495(99)77369-8.
Simons, K., and Ehehalt, R. (2002) Cholesterol, lipid rafts, and disease, J. Clin. Invest., 110, 597–603, doi: https://doi.org/10.1172/JCI16390.
Mitsui, K., Hatakeyama, K., Matsushita, M., and Kanazawa, H. (2009) Saccharomyces cerevisiae Na+/H+ antiporter Nha1p associates with lipid rafts and requires sphingolipid for stable localization to the plasma membrane, J. Biochem., 145, 709–720, doi: https://doi.org/10.1093/jb/mvp032.
Bagnat, M., Chang, A., and Simons, K. (2001) Plasma membrane proton ATPase Pma1p requires raft association for surface delivery in yeast, Mol. Biol. Cell, 12, 4129–4138, doi: https://doi.org/10.1091/mbc.12.12.4129.
Ejsing, C. S., Sampaio, J. L., Surendranath, V., Duchoslav, E., Ekroos, K., Klemm, R. W., and Shevchenko, A. (2009) Global analysis of the yeast lipidome by quantitative shotgun mass spectrometry, Proc. Natl. Acad. Sci. USA, 106, 2136–2141, doi: https://doi.org/10.1073/pnas.0811700106.
Sampaio, J. L., Gerl, M. J., Klose, C., Ejsing, C. S., Beug, H., Simons, K., and Shevchenko, A. (2011) Membrane lipidome of an epithelial cell line, Proc. Natl. Acad. Sci. USA, 108, 1903–1907, doi: https://doi.org/10.1073/pnas.1019267108.
Kachroo, A. H., Laurent, J. M., Yellman, C. M., Meyer, A. G., Wilke, C. O., and Marcotte, E. M. (2015) Evolution. Systematic humanization of yeast genes reveals conserved functions and genetic modularity, Science, 348, 921–925, doi: https://doi.org/10.1126/science.aaa0769.
Daum, G., Lees, N. D., Bard, M., and Dickson, R. (1998) Biochemistry, cell biology and molecular biology of lipids of Saccharomyces cerevisiae, Yeast, 14, 1471–1510.
Kristan, K., and Rizner, T. L. (2012) Steroid–transforming enzymes in fungi, J. Steroid Biochem. Mol. Biol., 129, 79–91, doi: https://doi.org/10.1016/j.jsbmb.2011.08.012.
Klug, L., and Daum, G. (2014) Yeast lipid metabolism at a glance, FEMS Yeast Res., 14, 369–388, doi: https://doi.org/10.1111/1567-1364.12141.
Hu, Z., He, B., Ma, L., Sun, Y., Niu, Y., and Zeng, B. (2017) Recent advances in ergosterol biosynthesis and regulation mechanisms in Saccharomyces cerevisiae, Indian J. Microbiol., 57, 270–277, doi: https://doi.org/10.1007/s12088-017-0657-1.
Zinser, E., Paltauf, F., and Daum, G. (1993) Sterol composition of yeast organelle membranes and subcellular distribution of enzymes involved in sterol metabolism, J. Bacteriol., 175, 2853–2858.
Lange, Y., Swaisgood, M. H., Ramos, B. V., and Steck, T. L. (1989) Plasma membranes contain half the phospholipid and 90% of the cholesterol and sphingomyelin in cultured human fibroblasts, J. Biol. Chem., 264, 3786–3793.
Schneiter, R., Brugger, B., Sandhoff, R., Zellnig, G., Leber, A., Lampl, M., Athenstaedt, K., Hrastnik, C., Eder, S., Daum, G., Paltauf, F., Wieland, F. T., and Kohlwein, S. D. (1999) Electrospray ionization tandem mass spectrometry (ESI–MS/MS) analysis of the lipid molecular species composition of yeast subcellular membranes reveals acyl chain–based sorting/remodeling of distinct molecular species en route to the plasma membrane, J. Cell Biol., 146, 741–754.
Brugger, B., Sandhoff, R., Wegehingel, S., Gorgas, K., Malsam, J., Helms, J. B., Lehmann, W. D., Nickel, W., and Wieland, F. T. (2000) Evidence for segregation of sphingomyelin and cholesterol during formation of COPI–coated vesicles, J. Cell Biol., 151, 507–518.
Klemm, R. W., Ejsing, C. S., Surma, M. A., Kaiser, H.–J., Gerl, M. J., Sampaio, J. L., Robillard, Q., Ferguson, C., Proszynski, T. J., Shevchenko, A., and Simons, K. (2009) Segregation of sphingolipids and sterols during formation of secretory vesicles at the trans–Golgi network, J. Cell Biol., 185, 601–612, doi: https://doi.org/10.1083/jcb.200901145.
Mesmin, B., and Maxfield, F. R. (2009) Intracellular sterol dynamics, Biochim. Biophys. Acta, 1791, 636–645, doi: https://doi.org/10.1016/j.bbalip.2009.03.002.
Baumann, N. A., Sullivan, D. P., Ohvo–Rekila, H., Simonot, C., Pottekat, A., Klaassen, Z., Beh, C. T., and Menon, A. K. (2005) Transport of newly synthesized sterol to the sterol–enriched plasma membrane occurs via non–vesicular equilibration, Biochemistry, 44, 5816–5826, doi: https://doi.org/10.1021/bi048296z.
Sullivan, D. P., Ohvo–Rekila, H., Baumann, N. A., Beh, C. T., and Menon, A. K. (2006) Sterol trafficking between the endoplasmic reticulum and plasma membrane in yeast, Biochem. Soc. Trans., 34, 356–358, doi: https://doi.org/10.1042/BST0340356.
Kentala, H., Weber–Boyvat, M., and Olkkonen, V. M. (2016) OSBP–related protein family: mediators of lipid transport and signaling at membrane contact sites, Intern. Rev. Cell. Mol. Biol., 321, 299–340, doi: https://doi.org/10.1016/bs.ircmb.2015.09.006.
Tong, J., Manik, M. K., Yang, H., and Im, Y. J. (2016) Structural insights into nonvesicular lipid transport by the oxysterol binding protein homologue family, Biochim. Biophys. Acta, 1861, 928–939, doi: https://doi.org/10.1016/j.bbalip.2016.01.008.
Raychaudhuri, S., Im, Y. J., Hurley, J. H., and Prinz, W. A. (2006) Nonvesicular sterol movement from plasma membrane to ER requires oxysterol–binding protein–related proteins and phosphoinositides, J. Cell Biol., 173, 107–119, doi: https://doi.org/10.1083/jcb.200510084.
De Saint–Jean, M., Delfosse, V., Douguet, D., Chicanne, G., Payrastre, B., Bourguet, W., Antonny, B., and Drin, G. (2011) Osh4p exchanges sterols for phosphatidylinositol 4–phosphate between lipid bilayers, J. Cell Biol., 195, 965–978, doi: https://doi.org/10.1083/jcb.201104062.
Loewen, C. J. R., Roy, A., and Levine, T. P. (2003) A conserved ER targeting motif in three families of lipid binding proteins and in Opi1p binds VAP, EMBO J., 22, 2025–2035, doi: https://doi.org/10.1093/emboj/cdg201.
Levine, T. P., and Munro, S. (2002) Targeting of Golgi–specific pleckstrin homology domains involves both PtdIns 4–kinase–dependent and–independent components, Curr. Biol., 12, 695–704.
Raychaudhuri, S., and Prinz, W. A. (2010) The diverse functions of oxysterol–binding proteins, Ann. Rev. Cell. Dev. Biol., 26, 157–177, doi: https://doi.org/10.1146/annurev.cell-bio.042308.113334.
Maeda, K., Anand, K., Chiapparino, A., Kumar, A., Poletto, M., Kaksonen, M., and Gavin, A. C. (2013) Interactome map uncovers phosphatidylserine transport by oxysterol–binding proteins, Nature, 501, 257–261, doi: https://doi.org/10.1038/nature12430.
Moser von Filseck, J., Copic, A., Delfosse, V., Vanni, S., Jackson, C. L., Bourguet, W., and Drin, G. (2015) Intracellular transport. Phosphatidylserine transport by ORP/Osh proteins is driven by phosphatidylinositol 4–phosphate, Science, 349, 432–436, doi: https://doi.org/10.1126/sci-ence.aab1346.
Tian, S., Ohta, A., Horiuchi, H., and Fukuda, R. (2018) Oxysterol–binding protein homologs mediate sterol transport from the endoplasmic reticulum to mitochondria in yeast, J. Biol. Chem., 293, 5636–5648, doi: https://doi.org/10.1074/jbc.RA117.000596.
Beh, C. T., Cool, L., Phillips, J., and Rine, J. (2001) Overlapping functions of the yeast oxysterol–binding protein homologues, Genetics, 157, 1117–1140.
Alberts, A. W., Chen, J., Kuron, G., Hunt, V., Huff, J., Hoffman, C., Rothrock, J., Lopez, M., Joshua, H., Harris, E., Patchett, A., Monaghan, R., Currie, S., Stapley, E., Albers–Schonberg, G., Hensens, O., Hirshfield, J., Hoogsteen, K., Liesch, J., and Springer, J. (1980) Mevinolin: a highly potent competitive inhibitor of hydroxymethylglutaryl–coenzyme A reductase and a cholesterol–lowering agent, Proc. Natl. Acad. Sci. USA, 77, 3957–3961.
Woods, R. A. (1971) Nystatin–resistant mutants of yeast: alterations in sterol content, J. Bacteriol., 108, 69–73.
Gatta, A. T., Wong, L. H., Sere, Y. Y., Calderon–Norena, D. M., Cockcroft, S., Menon, A. K., and Levine, T. P. (2015) A new family of StART domain proteins at membrane contact sites has a role in ER–PM sterol transport, eLife, 4, doi: https://doi.org/10.7554/eLife.07253.
Horenkamp, F. A., Valverde, D. P., Nunnari, J., and Reinisch, K. M. (2018) Molecular basis for sterol transport by StART–like lipid transfer domains, EMBO J., 37, doi: https://doi.org/10.15252/embj.201798002.
Murley, A., Sarsam, R. D., Toulmay, A., Yamada, J., Prinz, W. A., and Nunnari, J. (2015) Ltc1 is an ER–localized sterol transporter and a component of ER–mitochondria and ER–vacuole contacts, J. Cell Biol., 209, 539–548, doi: https://doi.org/10.1083/jcb.201502033.
Wong, L. H., and Levine, T. P. (2016) Lipid transfer proteins do their thing anchored at membrane contact sites… but what is their thing? Biochem. Soc. Trans., 44, 517–527, doi: https://doi.org/10.1042/BST20150275.
Finn, R. D., Bateman, A., Clements, J., Coggill, P., Eberhardt, R. Y., Eddy, S. R., Heger, A., Hetherington, K., Holm, L., Mistry, J., Sonnhammer, E. L., Tate, J., and Punta, M. (2014) Pfam: the protein families database, Nucleic Acids Res., 42, 222–230, doi: https://doi.org/10.1093/nar/gkt1223.
Knorre, D. A., Ojovan, S. M., Saprunova, V. B., Sokolov, S. S., Bakeeva, L. E., and Severin, F. F. (2008) Mitochondrial matrix fragmentation as a protection mechanism of yeast Saccharomyces cerevisiae, Biochemistry (Moscow), 73, 1254–1259.
Altmann, K., and Westermann, B. (2005) Role of essential genes in mitochondrial morphogenesis in Saccharomyces cerevisiae, Mol. Biol. Cell, 16, 5410–5417, doi: https://doi.org/10.1091/mbc.e05-07-0678.
Pozniakovsky, A. I., Knorre, D. A., Markova, O. V., Hyman, A. A., Skulachev, V. P., and Severin, F. F. (2005) Role of mitochondria in the pheromone–and amiodarone–induced programmed death of yeast, J. Cell. Biol., 168, 257–269, doi: https://doi.org/10.1083/jcb.200408145.
Sokolov, S., Knorre, D., Smirnova, E., Markova, O., Pozniakovsky, A., Skulachev, V., and Severin, F. (2006) Ysp2 mediates death of yeast induced by amiodarone or intracellular acidification, Biochim. Biophys. Acta, 1757, 1366–1370, doi: https://doi.org/10.1016/j.bbabio.2006.07.005.
Francois, I. E. J. A., Bink, A., Vandercappellen, J., Ayscough, K. R., Toulmay, A., Schneiter, R. (2009) Membrane rafts are involved in intracellular miconazole accumulation in yeast cells, J. Biol. Chem., 284, 32680–32685, doi: https://doi.org/10.1074/jbc.M109.014571.
Dos Santos, S. C., and Sa–Correia, I. (2011) A genome–wide screen identifies yeast genes required for protection against or enhanced cytotoxicity of the antimalarial drug quinine, Mol. Gen. Genom., 286, 333–346, doi: https://doi.org/10.1007/s00438-011-0649-5.
Murley, A., Yamada, J., Niles, B. J., Toulmay, A., Prinz, W. A., Powers, T., and Nunnari, J. (2017) Sterol transporters at membrane contact sites regulate TORC1 and TORC2 signaling, J. Cell Biol., 216, 2679–2689, doi: https://doi.org/10.1083/jcb.201610032.
Lees, N. D., Skaggs, B., Kirsch, D. R., and Bard, M. (1995) Cloning of the late genes in the ergosterol biosynthetic pathway of Saccharomyces cerevisiae–a review, Lipids, 30, 221–226.
Munn, A. L., Heese–Peck, A., Stevenson, B. J., Pichler, H., and Riezman, H. (1999) Specific sterols required for the internalization step of endocytosis in yeast, Mol. Biol. Cell, 10, 3943–3957, doi: https://doi.org/10.1091/mbc.10.11.3943.
Giaever, G., Chu, A. M., Ni, L., Connelly, C., Riles, L., Veronneau, S., et al. (2002) Functional profiling of the Saccharomyces cerevisiae genome, Nature, 418, 387–391, doi: https://doi.org/10.1038/nature00935.
Jakubkova, M., Dzugasova, V., Truban, D., Abelovska, L., Bhatia–Kissova, I., Valachovic, M., Klobucnikova, V., Zeiselova, L., Griac, P., Nosek, J., and Tomaska, L. (2016) Identification of yeast mutants exhibiting altered sensitivity to valinomycin and nigericin demonstrate pleiotropic effects of ionophores on cellular processes, PLoS One, 11, e0164175, doi: https://doi.org/10.1371/journal.pone.0164175.
Liu, G., Chen, Y., Faergeman, N. J., and Nielsen, J. (2017) Elimination of the last reactions in ergosterol biosynthesis alters the resistance of Saccharomyces cerevisiae to multiple stresses, FEMS Yeast Res., 17, doi: https://doi.org/10.1093/femsyr/fox063.
Contamine, V., and Picard, M. (2000) Maintenance and integrity of the mitochondrial genome: a plethora of nuclear genes in the budding yeast, Microbiol. Mol. Biol. Rev., 64, 281–315.
Dupont, S., Lemetais, G., Ferreira, T., Cayot, P., Gervais, P., and Beney, L. (2012) Ergosterol biosynthesis: a fungal pathway for life on land? Evolution, 66, 2961–2968, doi: https://doi.org/10.1111/j.1558-5646.2012.01667.x.
Guan, X. L., Souza, C. M., Pichler, H., Dewhurst, G., Schaad, O., Kajiwara, K., Wakabayashi, H., Ivanova, T., Castillon, G. A., Piccolis, M., Abe, F., Loewith, R., Funato, K., Wenk, M. R., and Riezman, H. (2009) Functional interactions between sphingolipids and sterols in biological membranes regulating cell physiology, Mol. Biol. Cell, 20, 2083–2095, doi: https://doi.org/10.1091/mbc.E08-11-1126.
Abe, F., and Hiraki, T. (2009) Mechanistic role of ergosterol in membrane rigidity and cycloheximide resistance in Saccharomyces cerevisiae, Biochim. Biophys. Acta, 1788, 743–752, doi: https://doi.org/10.1016/j.bbamem.2008.12.002.
Leppert, G., McDevitt, R., Falco, S. C. (1990) Cloning by gene amplification of two loci conferring multiple drug resistance in Saccharomyces, Genetics, 125, 13–20.
Kaur, R., and Bachhawat, A. K. (1999) The yeast multidrug resistance pump, Pdr5p, confers reduced drug resistance in erg mutants of Saccharomyces cerevisiae, Microbiology, 145, 809–818, doi: https://doi.org/10.1099/13500872-145-4-809.
Desmoucelles, C., Pinson, B., Saint–Marc, C., and Daignan–Fornier, B. (2002) Screening the yeast “disruptome” for mutants affecting resistance to the immunosuppressive drug, mycophenolic acid, J. Biol. Chem., 277, 27036–27044, doi: https://doi.org/10.1074/jbc.M111433200.
Fleming, J. A., Lightcap, E. S., Sadis, S., Thoroddsen, V., Bulawa, C. E., and Blackman, R. K. (2002) Complementary whole–genome technologies reveal the cellular response to proteasome inhibition by PS–341, Proc. Natl. Acad. Sci. USA, 99, 1461–1466, doi: https://doi.org/10.1073/pnas.032516399.
Viladevall, L., Serrano, R., Ruiz, A., Domenech, G., Giraldo, J., Barcelo, A., and Arino, J. (2004) Characterization of the calcium–mediated response to alkaline stress in Saccharomyces cerevisiae, J. Biol. Chem., 279, 43614–43624, doi: https://doi.org/10.1074/jbc.M403606200.
Branco, M. R., Marinho, H. S., Cyrne, L., and Antunes, F. (2004) Decrease of H2O2 plasma membrane permeability during adaptation to H2O2 in Saccharomyces cerevisiae, J. Biol. Chem., 279, 6501–6506.
Montanes, F. M., Pascual–Ahuir, A., and Proft, M. (2011) Repression of ergosterol biosynthesis is essential for stress resistance and is mediated by the Hog1MAP kinase and the Mot3 and Rox1 transcription factors, Mol. Microbiol., 79, 1008–1023, doi: https://doi.org/10.1111/j.1365-2958.2010.07502.x.
Gazdag, Z., Mate, G., Certik, M., Turmer, K., Virag, E., Pocsi, I., and Pesti, M. (2014) Butyl hydroperoxide–induced differing plasma membrane and oxidative stress processes in yeast strains BY4741 and erg5Δ, J. Basic Microbiol., 54, 50–62, doi: https://doi.org/10.1002/jobm.201300925.
Bard, M., Lees, N. D., Burrows, L. S., and Kleinhans, F. W. (1978) Differences in crystal violet uptake and cation–induced death among yeast sterol mutants, J. Bacteriol., 135, 1146–1148.
Welihinda, A. A., Beavis, A. D., and Trumbly, R. J. (1994) Mutations in LIS1 (ERG6) gene confer increased sodium and lithium uptake in Saccharomyces cerevisiae, Biochim. Biophys. Acta, 1193, 107–117, doi: https://doi.org/10.1016/0005-2736(94)90339-5.
Kodedova, M., and Sychrova, H. (2015) Changes in the sterol composition of the plasma membrane affect membrane potential, salt tolerance and the activity of multidrug resistance pumps in Saccharomyces cerevisiae, PLoS One, 10, e0139306, doi: https://doi.org/10.1371/journal.pone.0139306.
Espenshade, P. J., and Hughes, A. L. (2007) Regulation of sterol synthesis in eukaryotes, Ann. Rev. Genet., 41, 401–427, doi: https://doi.org/10.1146/annurev.genet.41.110306.130315.
Aguilera, F., Peinado, R. A., Millan, C., Ortega, J. M., and Mauricio, J. C. (2006) Relationship between ethanol tolerance, H+–ATPase activity and the lipid composition of the plasma membrane in different wine yeast strains, Intern. J. Food Microbiol., 110, 34–42, doi: https://doi.org/10.1016/j.ijfoodmi-cro.2006.02.002.
Yang, H., Tong, J., Lee, C. W., Ha, S., Eom, S. H., and Im, Y. J. (2015) Structural mechanism of ergosterol regulation by fungal sterol transcription factor Upc2, Nat. Commun., 6, 6129, doi: https://doi.org/10.1038/ncomms7129.
Zavrel, M., Hoot, S. J., and White, T. C. (2013) Comparison of sterol import under aerobic and anaerobic conditions in three fungal species, Candida albicans, Candida glabrata, and Saccharomyces cerevisiae, Eukaryot. Cell, 12, 725–738, doi: https://doi.org/10.1128/EC.00345-12.
Tiwari, R., Koffel, R., and Schneiter, R. (2007) An acetylation/deacetylation cycle controls the export of sterols and steroids from S. cerevisiae, EMBO J., 26, 5109–5119, doi: https://doi.org/10.1038/sj.emboj.7601924.
Choudhary, V., Darwiche, R., Gfeller, D., Zoete, V., Michielin, O., and Schneiter, R. (2014) The caveolin–bind–ing motif of the pathogen–related yeast protein Pry1, a member of the CAP protein superfamily, is required for in vivo export of cholesteryl acetate, J. Lipid Res., 55, 883–894, doi: https://doi.org/10.1194/jlr.M047126.
Korber, M., Klein, I., and Daum, G. (2017) Steryl ester synthesis, storage and hydrolysis: a contribution to sterol homeostasis, Biochim. Biophys. Acta Mol. Cell Biol. Lipids, 1862, 1534–1545, doi: https://doi.org/10.1016/j.bbalip.2017.09.002.
Yang, H., Bard, M., Bruner, D. A., Gleeson, A., Deckelbaum, R. J., Aljinovic, G., Pohl, T. M., Rothstein, R., and Sturley, S. L. (1996) Sterol esterification in yeast: a two–gene process, Science, 272, 1353–1356.
Yu, C., Kennedy, N. J., Chang, C. C., and Rothblatt, J. A. (1996) Molecular cloning and characterization of two iso–forms of Saccharomyces cerevisiae acyl–CoA:sterol acyl–transferase, J. Biol. Chem., 271, 24157–24163.
Zweytick, D., Leitner, E., Kohlwein, S. D., Yu, C., Rothblatt, J., and Daum, G. (2000) Contribution of Are1p and Are2p to steryl ester synthesis in the yeast Saccharomyces cerevisiae, Eur. J. Biochem., 267, 1075–1082.
Koffel, R., and Schneiter, R. (2006) Yeh1 constitutes the major steryl ester hydrolase under heme–deficient conditions in Saccharomyces cerevisiae, Eukaryot. Cell, 5, 1018–1025, doi: https://doi.org/10.1128/EC.00002-06.
Georgiev, A. G., Sullivan, D. P., Kersting, M. C., Dittman, J. S., Beh, C. T., and Menon, A. K. (2011) Osh proteins regulate membrane sterol organization but are not required for sterol movement between the ER and PM, Traffic, 12, 1341–1355, doi: https://doi.org/10.1111/j.1600-0854.2011.01234.x.
Mullner, H., Deutsch, G., Leitner, E., Ingolic, E., and Daum, G. (2005) YEH2/YLR020c encodes a novel steryl ester hydrolase of the yeast Saccharomyces cerevisiae, J. Biol. Chem., 280, 13321–13328, doi: https://doi.org/10.1074/jbc.M409914200.
Jandrositz, A., Petschnigg, J., Zimmermann, R., Natter, K., Scholze, H., Hermetter, A., Kohlwein, S. D., and Leber, R. (2005) The lipid droplet enzyme Tgl1p hydrolyzes both steryl esters and triglycerides in the yeast, Saccharomyces cerevisiae, Biochim. Biophys. Acta, 1735, 50–58, doi: https://doi.org/10.1016/j.bbalip.2005.04.005.
Walther, T. C., and Farese, R. V., Jr. (2012) Lipid droplets and cellular lipid metabolism, Ann. Rev. Biochem., 81, 687–714, doi: https://doi.org/10.1146/annurev-biochem-061009-102430.
Wang, C.–W. (2015) Lipid droplet dynamics in budding yeast, Cell. Mol. Life Sci., 72, 2677–2695, doi: https://doi.org/10.1007/s00018-015-1903-5.
Ueno, K., Nagano, M., Shimizu, S., Toshima, J. Y., and Toshima, J. (2016) Lipid droplet proteins, Lds1p, Lds2p, and Rrt8p, are implicated in membrane protein transport associated with ergosterol, Biochem. Biophys. Res. Commun., 475, 315–321, doi: https://doi.org/10.1016/j.bbrc.2016.05.099.
Tinkelenberg, A. H., Liu, Y., Alcantara, F., Khan, S., Guo, Z., Bard, M., and Sturley, S. L. (2000) Mutations in yeast ARV1 alter intracellular sterol distribution and are complemented by human ARV1, J. Biol. Chem., 275, 40667–40670, doi: https://doi.org/10.1074/jbc.C000710200.
Tong, F., Billheimer, J., Shechtman, C. F., Liu, Y., Crooke, R., Graham, M., Cohen, D. E., Sturley, S. L., and Rader, D. J. (2010) Decreased expression of ARV1 results in cholesterol retention in the endoplasmic reticulum and abnormal bile acid metabolism, J. Biol. Chem., 285, 33632–33641, doi: https://doi.org/10.1074/jbc.M110.165761.
Georgiev, A. G., Johansen, J., Ramanathan, V. D., Sere, Y. Y., Beh, C. T., and Menon, A. K. (2013) Arv1 regulates PM and ER membrane structure and homeostasis but is dispensable for intracellular sterol transport, Traffic, 14, 912–921, doi: https://doi.org/10.1111/tra.12082.
Heuck, A. P., Moe, P. C., and Johnson, B. B. (2010) The cholesterol–dependent cytolysin family of gram–positive bacterial toxins, Sub–Cell. Biochem., 51, 551–577, doi: https://doi.org/10.1007/978-90-481-8622-8_20.
Author information
Authors and Affiliations
Corresponding author
Additional information
Published in Russian in Biokhimiya, 2019, Vol. 84, No. 4, pp. 481–493.
Rights and permissions
About this article
Cite this article
Sokolov, S.S., Trushina, N.I., Severin, F.F. et al. Ergosterol Turnover in Yeast: An Interplay between Biosynthesis and Transport. Biochemistry Moscow 84, 346–357 (2019). https://doi.org/10.1134/S0006297919040023
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0006297919040023