-
PDF
- Split View
-
Views
-
Cite
Cite
Patricia Álvarez-Campos, Gonzalo Giribet, Guillermo San Martín, Greg W. Rouse, Ana Riesgo, Straightening the striped chaos: systematics and evolution of Trypanosyllis and the case of its pseudocryptic type species Trypanosyllis krohnii (Annelida, Syllidae), Zoological Journal of the Linnean Society, Volume 179, Issue 3, 1 March 2017, Pages 492–540, https://doi.org/10.1111/zoj.12443
Close - Share Icon Share
Abstract
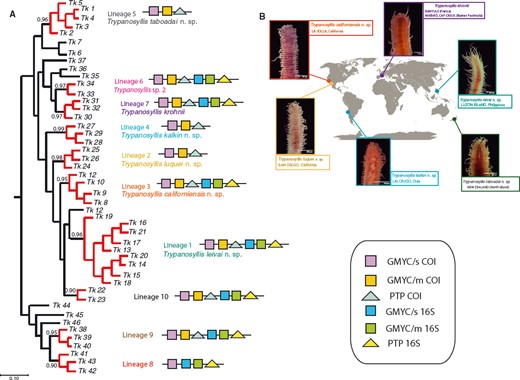
All members of the syllid genus Trypanosyllis show distinctive flattened, ribbon-like bodies and a pharynx armed with a trepan; however, the phylogenetic relationships within this genus remain unsettled, especially with respect to the genera Eurysyllis and Xenosyllis (morphologically similar). To resolve this systematic uncertainty we analysed the phylogenetic relationships of a worldwide sampling of specimens of Trypanosyllis and three related genera using multiple molecular markers. We show that Trypanosyllis as presently construed is paraphyletic, and identify a clade of striped species that were previously all considered to be Trypanosyllis zebra (Grube, 1860). We outline the case to consider Trypanosyllis krohnii Claparède, 1864 as the type species of the genus, instead of Trypanosyllis zebra. Trypanosyllis krohnii (interpreted as Trypanosyllis zebra by recent authors) was previously believed to be cosmopolitan, but we show that it includes at least seven cryptic and pseudocryptic species, five of which are described herein: Trypanosyllis kalkin sp. nov., Trypanosyllis californiensis sp. nov., Trypanosyllis luquei sp. nov., Trypanosyllis leivai sp. nov., and Trypanosyllis taboadai sp. nov. In addition, Trypanedenta gemmipara (Johnson, 1901) comb. nov. and Trypanedenta gigantea (McIntosh, 1885) comb. nov., previously included in Trypanosyllis, are here transferred to Trypanedenta Imajima & Hartman, 1964, and Pseudosyllis brevipennis Grube, 1863 [previously named Trypanosyllis coeliaca (Claparède, 1868)] is transferred to the resurrected genus Pseudosyllis Grube, 1863. Overall our results show a complex scenario of speciation, with cases of pseudocryptic species that correspond to geographically restricted lineages.
Introduction
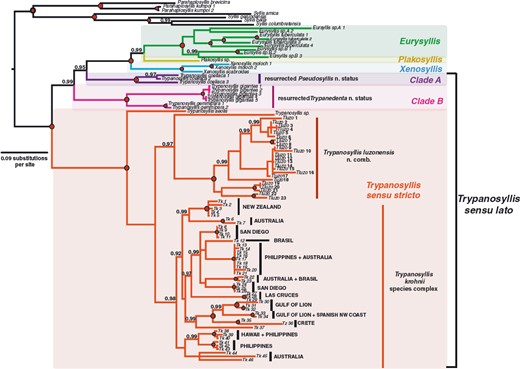
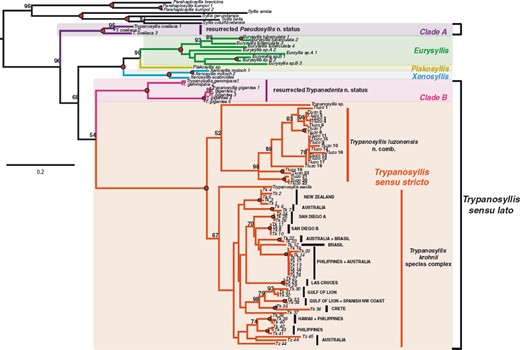
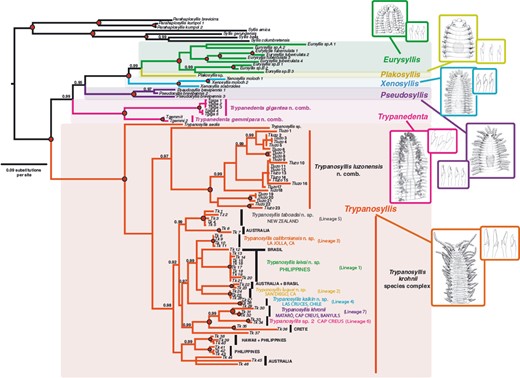
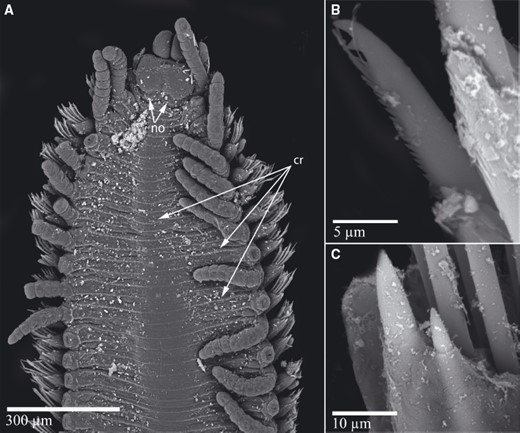
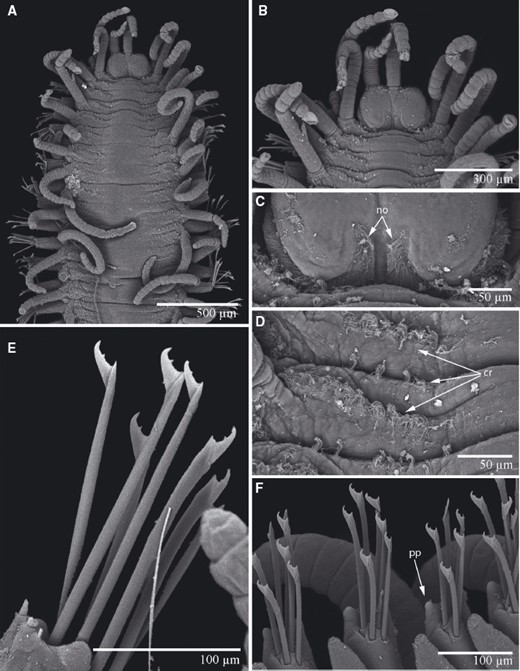
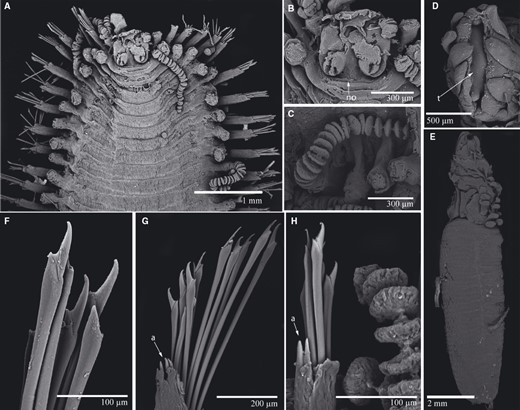
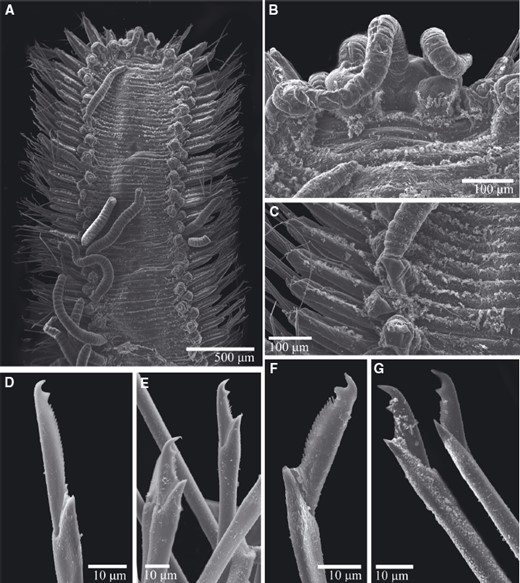
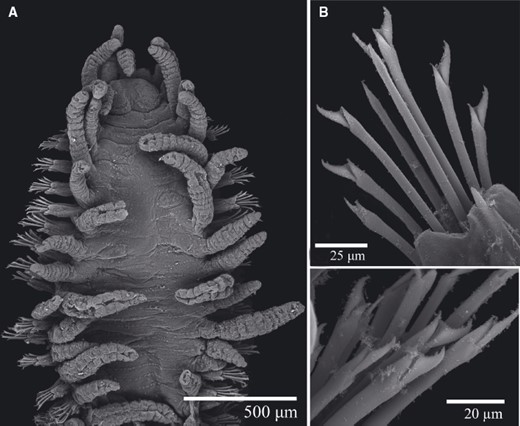
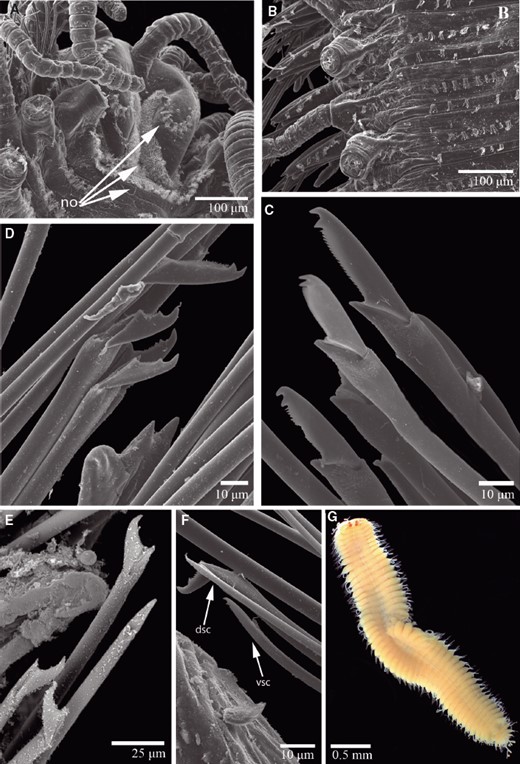
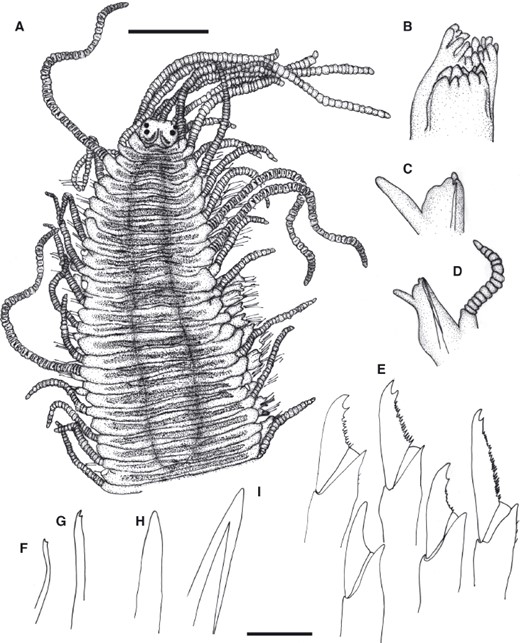
TrypanosyllisClaparède, 1864 is a genus within Syllidae, the members of which are widely distributed in all oceans, mainly inhabiting algal mats and coral rubble (San Martín, 2003). A flattened, ribbon-like body and the presence of a muscular axial pharynx armed with a trepan—a chitinous inner lining forming a complete ring—characterize members of this taxon (San Martín, 2003). Even though the morphological features of Trypanosyllis have been previously studied, its phylogenetic position remains a matter of debate, and the handful of available studies have used molecular sequence data from just a few specimens to disentangle its relationships. Although the first study recovered the genus as monophyletic (Aguado, Nygren & Siddall, 2007), more recent analyses by (Aguado, San Martín & Siddall 2012), (Aguado et al. 2015) presented a tree in which Trypanosyllis appeared paraphyletic, although only two Trypanosyllis species, Trypanosyllis coeliaca Claparède, 1868 and Trypanosyllis zebra (Grube, 1860), were included in these studies. Currently, Trypanosyllis comprises 32 species (WoRMS Editorial Board, 2015), some with limited distributions (e.g. Verrill, 1882; Potts, 1911; Day, 1960; Hartmann-Schröder, 1965; Nogueira & Fukuda, 2008), whereas others are apparently widely distributed across oceans (e.g. Çinar & Ergen, 2003; Imajima, 2003; San Martín, 2003; Çinar, 2007). Trypanosyllis krohniiClaparède, 1864 has long been considered a synonym of the proposed type species, Trypanosyllis zebra. Trypanosyllis krohnii is characterized by having a colour pattern with two transverse violet to brown bands on each segment (Claparède, 1864) and falcigerous bidentate chaetae with dorsoventral gradation along the length of the blades. These features, similar to those of Trypanosyllis zebra, provided the grounds for the synonymy of both species (Langerhans, 1879), and since then all authors have followed this synonymy and have treated Trypanosyllis zebra as the type species of the genus. We show in this paper that this synonymy was an error and that Trypanosyllis krohnii is the correct type species. As Trypanosyllis krohnii is the valid type species of the genus (see the morphological remarks of the species), the complex will be considered hereafter to belong within Trypanosyllis krohnii instead of Trypanosyllis zebra.
For many years, Trypanosyllis krohnii has been reported from a variety of substrates and depths, with a worldwide distribution, including the Mediterranean and Red seas, and the Atlantic, Indian, and Pacific basins (San Martín, 1991; Núñez, San Martín & Brito, 1992; San Martín, Hutchings & Aguado, 2008). Species with such a wide distribution are considered cosmopolitan (sensuSpellerberg & Sawyer, 1999). Cosmopolitan species are often more common among marine organisms with long-lived planktotrophic larvae, as they may have a higher degree of connectivity among geographically distant populations (Sanford & Kelly, 2011) because of an apparent lack of dispersal barriers (Grosberg & Cunningham, 2001); however, many marine taxa that are considered to be cosmopolitan have been shown to be two or more morphologically indistinguishable cryptic species (e.g. Knowlton, 1993, 2000; Bickford et al., 2007). This apparent lack of defining morphological features occurs because speciation is not always accompanied by morphological changes. In addition, in some cases, even if there are slight morphological differences between two given populations, sometimes they are not distinguishable because of the state of sample preservation. In this sense, molecular tools are extremely helpful to test species boundaries and enable us to better quantify the real biodiversity and species distribution ranges in poorly known marine ecosystems (e.g. Knowlton, 2000; Witt, Threloff & Hebert, 2006; Bucklin, Steinke & Blanco-Bercial, 2011; Carr et al., 2011; Ahrens et al., 2013).
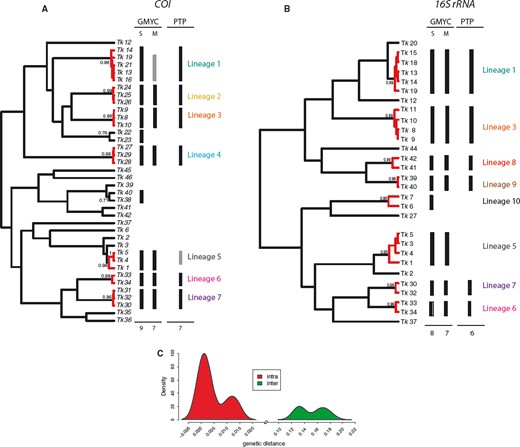
Cryptic speciation has been detected using molecular tools in many different marine groups, including sponges (e.g. Klautau et al., 1999; Xavier et al., 2010), molluscs (e.g. Calvo et al., 2009; Wilson, Schrödl & Halanych, 2009; Kawauchi & Giribet, 2011), tunicates (e.g. Pérez-Portela et al., 2013), echinoderms (e.g. Hemery et al., 2012), and crustaceans (e.g. Held, 2003). In particular, among annelids, studies confirming large geographic ranges for putative cosmopolitan species are scarce (Staton & Rice, 1999; Westheide et al., 2003; Meyer et al., 2008; Kawauchi & Giribet, 2010, 2014; Schüller & Hutchings, 2012), and in turn, strong geographical population structure is common (e.g. Barroso et al., 2010; Carr et al., 2011; Nygren & Pleijel, 2011; Borda et al., 2013; Glasby, Wei & Gibb, 2013; Stiller et al., 2013; Nygren, 2014). Surprisingly, even though syllids are a diverse and abundant annelid clade, studies documenting cryptic speciation and/or cosmopolitanism are scarce (Westheide & Haβ-Cordes, 2001). Surprisingly, among syllids, the distinctive striped coloration of Trypanosyllis krohnii around the world has made it prone to be considered a cosmopolitan species (San Martín, 1991; Núñez et al., 1992; San Martín et al., 2008), although many authors have noted that they may include more than one species (e.g., Núñez et al., 1992; San Martín, 2003; Nogueira & Fukuda, 2008; San Martín et al., 2008). In this sense, the case of Trypanosyllis krohnii appears as an ideal example to study species boundaries in this diverse and abundant group of annelids.
In our study we aim to assess the monophyly of Trypanosyllis, including the analysis of the closely related genera Eurysyllis, Plakosyllis, and Xenosyllis, with a multi-locus data set (two mitochondrial and two nuclear markers) for 101 specimens, including 92 new specimens collected around the world. In addition, we investigate the problem of the striped colouring in the Trypanosyllis krohnii species complex and provide morphological evidence for the differentiation of possible cryptic species.
Material and Methods
Sampling and morphological examination
Specimens were collected in biological surveys between 2010 and 2014, by hand, snorkelling, or SCUBA diving, in algae and rocks from intertidal and subtidal zones in each location (Table 1). Catalogue numbers, locality information, coordinates, substrates, and collecting dates are provided in the taxonomic section and in Table 1. Prior to fixation, selected specimens were anaesthetized with 7% magnesium chloride in fresh water and photographed under a microscope. All specimens included in the study were later fixed and preserved in 96% ethanol, both for morphological and molecular analyses. Further examination and identification was completed under a Nikon Optiphot light microscope with a differential interference contrast system (Nomarsky) at the Universidad Autónoma de Madrid (UAM). Using light microscopy, drawings were made to scale with a camera lucida attached to a Nikon Optiphot microscope. The width of specimens was measured at the level of the proventricle, excluding the parapodia.
Localities, substrates, date of sampling, coordinates, catalogue numbers, and GenBank accession numbers for all specimens sequenced
| Species . | Code . | N . | Locality . | Substrate . | Collection date . | Coordinates (decimal degrees) . | Museum numbers . | 28S . | 18S . | 16S . | COI . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Eurysyllis tuberculata | Eurysyllis tuberculata 1 | 1 | Cap Falcata uberculataegr | Red algae | 16 September 2011 | 42.433333, 3.174722 | MCZ 25312 | KX084805 | KX084852 | – | KX084931 |
| Eurysyllis tuberculata 2 | 1 | San Vicente do Mar, Galicia, Spain | Algae, 1 m | 11 March 2008 | 42.48, −8.901389 | MCZ 25287 | KX084807 | KX084851 | – | KX084934 | |
| Eurysyllis tuberculata 3 | 1 | Shark Bay, WA, Australia | – | – | −25.5, 113.5 | – | – | JF903594 | – | JF903787 | |
| Eurysyllis tuberculata 4 | 1 | Balayan Bay, Luzon Island, Philippines | Coral rubble with hydrozoans, 2–4 m | 4 December 2010 | 13.740556, 120.892778 | MNCN 16.01/16033, MNCN/ADN 85706 | KX084806 | KX084853 | KX084925 | KX084932 | |
| Eurysyllis sp. A | Eurysyllis sp. A1 | 1 | Balayan Bay, Luzon Island, Philippines | Coral rubble, 2–4 m | 7 December 2010 | 13.740556, 120.892778 | MNCN 16.01/16030, MNCN/ADN 85707 | KX084804 | – | – | KX084930 |
| Eurysyllis sp. A2 | 1 | Raja Ampat 2013, Indonesia | Reef flat just off beach, 1 m | 13 October 2013 | −0.551389, 130.695833 | SIO A6141 | – | KX084854 | – | KX084945 | |
| Eurysyllis sp. B | Eurysyllis sp. B1 | 1 | Sombrero Island, Balayan Bay, Luzon Island, Philippines | Coral rubble, 20 m | 9 December 2010 | −0.551389, 130.695833 | MNCN 16.01/16031, MNCN/ADN 85639 | – | KX084850 | KX084862 | KX084943 |
| Eurysyllis sp. B2 | 1 | Sombrero Island, Balayan Bay, Luzon Island, Philippines | Coral rubble, 17 m | 6 December 2010 | −0.551389, 130.695833 | MNCN 16.01/16063, MNCN/ADN 85640 | – | KX084849 | – | KX084960 | |
| Eurysyllis sp. B3 | 1 | Sombrero Island, Balayan Bay, Luzon Island, Philippines | Coral rubble, 2 m | 6 December 2010 | −0.551389, 130.695833 | MNCN 16.01/16032, MNCN/ADN 85641 | – | KX084848 | – | KX084959 | |
| Parahaplosyllis brevicirra | – | 1 | Port Jackson, NSW, Australia | – | – | −35.858333, 151.233333 | – | – | JF903679 | JF903706 | JF903784 |
| Parahaplosyllis kumpol | Parahaplosyllis kumpol 1 | 1 | Cavalli Islands, New Zealand | Unidentified sponge, 15 m | 1 February 2012 | −34.984444, 173.944167 | MNCN 16.01/16036, MNCN/ADN 85642 | – | – | KX084922 | KX084966 |
| Parahaplosyllis kumpol 2 | 1 | Cavalli Islands, New Zealand | Unidentified sponge, 15 m | 1 February 2012 | −34.984444, 173.944167 | MNCN 16.01/16037, MNCN/ADN 85643 | – | – | KX084923 | KX084967 | |
| Plakosyllis sp. | – | 1 | El Nido, Palawan Island, Philippines | Unidentified sponge, 12 m | 18 December 2012 | 11.197222, 119.317222 | MNCN 16.01/16038, MNCN/ADN 85644 | KX084808 | KX084855 | – | KX084933 |
| Pseudosyllis brevipennis | Pseudosyllis brevipennis 1 | 1 | Port de la Selva, Girona, Spain | Posidonia oceanica, 10 m | 21 September 2004 | 42.3375, 3.203333 | MNCN/ADN 9622 | – | EF123878 | EF123816 | EF123785 |
| Pseudosyllis brevipennis 2 | 1 | Alborpe Sea, Spain | Algae, 42–48 m | 24 September 2011 | 35.95 −2.966667 | MNCN 16.01/16040, MNCN/ADN 85645 | – | – | KX084917 | – | |
| Pseudosyllis brevipennis 3 | 1 | Matarpe Barcelona, Spain | Algae, 0 m | March 2014 | 41.5325, 2.453056 | MNCN 16.01/16041, MNCN/ADN 85646 | – | – | – | KX084969 | |
| Syllis amica | – | 1 | Puerto Colera, Girona, Spain | Lithophyllum tortuosum, 0 m | 14 September 2011 | 42.403333, 3.165278 | MCZ 25188 MNCN/ADN 85711 | KX084858 | – | KX084924 | KX084927 |
| Syllis bella | – | 1 | ‘Sepok Point’, between Balayan Bay and Batangas Bay, Luzon Island, Philippines | Coral rubble, 6 m | 10 December 2010 | 13.683889, 120.895833 | MCZ 25190 MNCN/ADN 85712 | – | KX084846 | KX084859 | KX084926 |
| Syllis colum bretensis | – | 1 | Cap de Creus, Girona, Spain | Paramuricea clavata, 40 m | 16 September 2011 | 42.320278, 3.320556 | MCZ 25191 MNCN/ADN 85713 | – | KX084845 | KX084860 | KX084928 |
| Syllis gerundensis | – | 1 | El Toro Island, Mallorca, Spain | Cladocora cespitosa and Miriapora sp., 12 m | 18 June 2012 | 39.4891, 2.4809 | MCZ 25194 MNCN/ADN 85714 | – | KX084847 | KX084861 | KX084929 |
| Trypanosyllis aeolis | – | 1 | El Toro Island, Mallorca, Spain | Cladocora cespitosa and Miriapora sp., 12 m | 18 June 2012 | 39.4891, 2.4809 | MNCN 16.01/16039, MNCN/ADN 85647 | – | KX084817 | KX084913 | KX084968 |
| Trypanedenta gemmipara comb. nov. | Tgemmi 1 | 1 | Cavalli Islands, New Zealand | Rainbow Warrior wreck, 25 m | 1 February 2012 | −36.834167, 174.771389 | MNCN 16.01/16042, MNCN/ADN 85648 | – | – | KX084920 | – |
| Tgemmi 2 | 1 | Cavalli Islands, New Zealand | Rainbow Warrior wreck, 25 m | 1 February 2012 | −36.834167, 174.771389 | MNCN 16.01/16043 (SEM), MNCN/ADN 85649 | – | – | KX084921 | – | |
| Trypanedenta giganteacomb. nov. | Tgiga 1 | 1 | Elephant Island, Antarctica | Blake Trawl, 223–242 m | 22 October 2011 | −61.163333, −54.997222 | SIO A3211 | – | KX084842 | KX084918 | KX084946 |
| Tgiga 2 | 1 | Elephant Island, Antarctica | Blake Trawl, 223–242 m | 22 October 2011 | −61.163333, −54.997222 | SIO A2935 | – | – | KX084919 | KX084984 | |
| Tgiga 3 | 1 | Bransfield Strait, Antarctica | Blake Trawl, 150–247 m | 24 October 2011 | −63.080556, −59.156389 | SIO A2948 | – | – | – | KX084992 | |
| Tgiga 4 | 1 | Burdwood Bank East, Antarctica | Blake Trawl, 90–92 m | 24 April 2013 | −54.558889, −56.828889 | SIO A3565 | – | KX084843 | – | KX084947 | |
| Tgiga 5 | 1 | Burdwood Bank East, Antarctica | Blake Trawl, 122–123 m | 24 April 2013 | −54.540833, −56.626944 | SIO A3515 | – | KX084844 | – | KX084948 | |
| Trypanosyllis sp. 1 | – | 1 | ‘Sepok Point’, between Balayan Bay and Batangas Bay, Luzon Island, Philippines | Coral rubble, 6 m | 10 December 2010 | 13.683889, 120.895833 | MNCN 16.01/16056, MNCN/ADN 85650 | KX084809 | KX084818 | KX084914 | KX084935 |
| Trypanosyllis luzonensiscomb. nov. | Tluzo 1 | 1 | Hong Kong University of Science and Technology, Hong Kong, China | Aquarium system | 9 May 2014 | 22.338056, 114.2675 | SIO A6142 | – | KX084838 | KX084892 | KX084963 |
| Tluzo 2 | 1 | ‘Twin Rocks’, El Nido, Palawan Island, Philippines | Unidentified sponges, 6 m | 17 December 2010 | 11.297222, 119.318333 | MNCN 16.01/16053, MNCN/ADN 85652 | – | KX084839 | KX084893 | – | |
| Tluzo 3 | 1 | ‘Mainif Point’, between Balayan Bay and Batangas Bay, Luzon Island, Philippines | Coral rubble, 2 m | 8 December 2010 | 13.68, 120.855556 | MNCN 16.01/16054, MNCN/ADN 85653 | – | – | KX084895 | KX084978 | |
| Tluzo 4 | 1 | Sombrero Island, Balayan Bay, Luzon Island, Philippines | Coral rubble, 20 m | 9 December 2010 | 13.697778, 120.829722 | MNCN 16.01/16050, MNCN/ADN 85654 | – | – | KX084896 | KX084979 | |
| Tluzo 5 | 1 | Sombrero Island, Balayan Bay, Luzon Island, Philippines | Unidentified sponges, 2 m | 6 December 2010 | 13.697778, 120.829722 | MNCN 16.01/16055, MNCN/ADN 85655 | – | – | KX084894 | – | |
| Tluzo 6 | 1 | Montgomery Reef, WA, Australia | Acropora sp., 0 m | 23 October 2009 | −16.020556, 14.159167 | AM W.42432, MNCN/ADN 85656 | – | – | KX084897 | – | |
| Tluzo 7 | 1 | Sombrero Island, Balayan Bay, Luzon Island, Philippines | Coral rubble, 20 m | 9 December 2010 | 13.697778, 120.829722 | MNCN 16.01/16052, MNCN/ADN 85657 | – | – | KX084904 | KX084980 | |
| Tluzo 8 | 1 | Sombrero Island, Balayan Bay, Luzon Island, Philippines | Coral rubble, 20 m | 9 December 2010 | 13.697778, 120.829722 | MNCN 16.01/16044, MNCN/ADN 85658 | – | – | – | KX084986 | |
| Tluzo 9 | 1 | Port Jackson, NSW, Australia | Encrusting sponges and dead shells on subtidal rock ledge, 9 m | 6 October 2005 | −35.858333, 151.233333 | AM W.47275 MNCN/ADN 85710 | – | – | KX084898 | KX084977 | |
| Tluzo 10 | 1 | Port Jackson, NSW, Australia | Encrusting sponges and dead shells on subtidal rock ledge, 9 m | 6 October 2005 | −35.858333, 151.233333 | AM W.47276, MNCN/ADN 85659 | – | – | KX084907 | KX084981 | |
| Tluzo 11 | 1 | Adele Island, WA, Australia | Sublittoral fore-reef slope, 12.5 m | 18 October 2009 | −15.557778, 123.133889 | AM W.41723, MNCN/ADN 85660 | – | – | KX084899 | – | |
| Tluzo 12 | 1 | Ningaloo Reef, WA, Australia | Coarse coral rubble, 7 m | 20 May 2009 | −22.623611, 113.641111 | AM W.41647, MNCN/ADN 85661 | – | – | KX084900 | – | |
| Tluzo 13 | 1 | Ningaloo Reef, WA, Australia | Brown algae and coral rubble, 24 m | 17 May 2009 | −22.623611, 113.641111 | AM W.41648, MNCN/ADN 85662 | – | – | KX084901 | – | |
| Tluzo 14 | 1 | Ningaloo Reef, WA, Australia | Coarse coral rubble, 7 m | 20 May 2009 | −22.623611, 113.641111 | AM W.41646 (SEM), MNCN/ADN 85663 | – | – | KX084902 | – | |
| Tluzo 15 | 1 | Ningaloo Reef, WA, Australia | Coarse coral rubble, 7 m | 20 May 2009 | −22.623611, 113.641111 | AM W.41645, MNCN/ADN 85664 | – | – | KX084903 | – | |
| Tluzo 16 | 1 | Long Reef, WA, Australia | Sublitoral reef platform, 4 m | 21 October 2010 | −13.856667, 125.825 | AM W.41639 (SEM), MNCN/ADN 85665 | – | – | KX084905 | – | |
| Tluzo 17 | 1 | Prince of Wales Island, QLD, Australia | – | – | −10.681944, 142.188611 | – | – | – | JF903748 | – | |
| Tluzo 18 | 1 | Sombrero Island, Balayan Bay, Luzon Island, Philippines | Coral rubble, 20 m | 9 December 2010 | 13.697778, 120.829722 | MNCN 16.01/16046, MNCN/ADN 85666 | – | – | KX084906 | – | |
| Tluzo 19 | 1 | ‘Sepok Point’, between Balayan Bay and Batangas Bay, Luzon Island, Philippines | Coral rubble, 6 m | 10 December 2010 | 13.683889, 120.895833 | MNCN 16.01/16047, MNCN/ADN 85667 | – | KX084840 | KX084908 | KX084964 | |
| Tluzo 20 | 1 | ‘Koala Point’, Balayan Bay, Luzon Island, Philippines | Coral rubble, 16 m | 5 December 2010 | 13.798889, 120.869444 | MNCN 16.01/16045, MNCN/ADN 85668 | – | – | KX084909 | KX084982 | |
| Tluzo 21 | 1 | ‘Koala Point’, Balayan Bay, Luzon Island, Philippines | Thalyssias sp. and Acanthella sp. sponges, 16 m | 5 December 2010 | 13.798889, 120.869444 | MNCN 16.01/16048, MNCN/ADN 85669 | – | KX084841 | KX084910 | KX084965 | |
| Tluzo 22 | 1 | ‘Sepok Point’, between Balayan Bay and Batangas Bay, Luzon Island, Philippines | Coral rubble, 6 m | 10 December 2010 | 13.683889, 120.895833 | MNCN 16.01/16051, MNCN/ADN 85670 | – | – | KX084911 | KX084983 | |
| Tluzo 23 | 1 | Adele Island, edge of Frazer Inlet, WA, Australia | Sublittoral channel slope 0 m | 22 October 2009 | −15.444722, 123.170833 | AM W.41649, MNCN/ADN 85671 | – | – | KX084912 | – | |
| Trypanosyllis taboadaisp. nov. (lineage 5) | Tk 1 | Holotype 1 | Cavalli Islands, New Zealand | Calcareous algae, 17 m | 2 February 2012 | −34.984444, 173.944167 | MNCN 16.01/16087, MNCN/ADN 85672 | KX084810 | KX084819 | KX084866 | KX084936 |
| Tk 2 | 1 | Maitai Bay, Karikari Peninsule, New Zealand | Corallina sp. and unidentified brown algae, 3 m | 31 January 2012 | −34.831111, 173.409444 | MNCN 16.01/16086, MNCN/ADN 85673 | KX084811 | KX084832 | KX084868 | KX084937 | |
| Tk 3 | 1 | Cavalli Islands, New Zealand | Calcareous algae, 17 m | 2 February 2012 | −34.984444, 173.944167 | MNCN 16.01/16080, MNCN/ADN 85674 | – | KX084828 | KX084865 | KX084949 | |
| Tk 4 | 1 | Cavalli Islands, New Zealand | Unidentified sponges, algae 15 m | 31 January 2012 | −34.984444, 173.944167 | MNCN 16.01/16079, MNCN/ADN 85675 | – | KX084824 | KX084863 | KX084951 | |
| Tk5 | 1 | Maitai Bay, Karikari Peninsule, New Zealand | Kelp, 3 m | 31 January 2012 | −34.831111, 173.409444 | MNCN 16.01/16078, MNCN/ADN 85677 | – | KX084825 | KX084864 | KX084952 | |
| Trypanosyllis cf. krohnii (Australia) | Tk 6 | 1 | Port Jackson, NSW, Australia | Balanoid barnacles and sponges, 1 m | 19 February 2006 | −35.858333, 151.233333 | AM W.47278, MNCN/ADN 85677 | – | KX084831 | KX084878 | KX084954 |
| Tk 7 | 1 | Port Jackson, Vauclusse, Bottle&Glass Rock, Sydney, NSW, Australia | On Ecklonia sp. holdfast and under rocks, 8 m | 8 April 2010 | −35.858333, 151.233333 | AM W.42428, MNCN/ADN 85708 | – | – | KX084879 | – | |
| Trypanosyllis californiensissp. nov. (lineage 3) | Tk 8 | 1 | La Jolla, San Diego, California | Holdfast, 5 m | 18 April 2014 | 32.866944, −117.255833 | SIO A5007 (SEM) | KX084812 | KX084821 | KX084869 | KX084938 |
| Tk 9 | 1 | La Jolla, San Diego, California | Algae, 0 m | 18 April 2014 | 32.866944, −117.255833 | SIO A5009 | KX084813 | – | KX084870 | KX084939 | |
| Tk 10 | Holotype 1 | La Jolla, San Diego, California | Holdfast, 5 m | 18 April 2014 | 32.866944, −117.255833 | SIO A5008 | – | KX084820 | KX084871 | KX084950 | |
| Tk 11 | 1 | La Jolla, San Diego, California | Algae, 0 m | 18 April 2014 | 32.866944, −117.255833 | SIO A5006 | – | – | KX084872 | – | |
| Trypanosyllis cf. krohnii | Tk 12 | 1 | Itapua Beach, Bahgo, California&Glil | Halimeda opuntia, 0 m | June 2014 | −12.956944, −38.36 | MNCN/ADN 85680 | KX084814 | KX084823 | KX084881 | KX084940 |
| Trypanosyllis leivaisp. nov. (lineage 1) | Tk 13 | 1 | ‘Mainif Point’, between Balayan Bay and Batangas Bay, Luzon Island, Philippines | Coral rubble, 2 m | 8 December 2010 | 13.68, 120.855556 | MNCN 16.01/16081, MNCN/ADN 85679 | KX084816 | – | KX084874 | KX084942 |
| Tk 14 | 1 | ‘Beatrice Point’, Sombrero Island, Balayan Bay, Luzon Island, Philippines | Unidentified sponge, 2 m | 9 December 2010 | 13.697778, 120.829722 | MNCN/ADN 85680 | – | KX084827 | KX084876 | KX084953 | |
| Tk 15 | 1 | Sombrero Island, Balayan Bay, Luzon Island, Philippines | Unidentified sponges, 2 m | 6 December 2010 | 13.697778, 120.829722 | MNCN 16.01/16075, MNCN/ADN 85681 | – | KX084829 | KX084873 | – | |
| Tk 16 | 1 | ‘Koala Point’, Balayan Bay, Luzon Island, Philippines | Dead coral, 2 m | 5 December 2010 | 13.798889, 120.869444 | MNCN 16.01/16074, MNCN/ADN 85682 | – | KX084830 | – | KX084955 | |
| Tk 17 | 1 | El Nido, Palawan Island, Philippines | Unidentified sponge, 12 m | 18 December 2012 | 11.197222, 119.317222 | MNCN/ADN 85683 | – | KX084826 | – | KX084956 | |
| Tk 18 | Holotype 1 | ‘Mainif Point’, between Balayan Bay and Batangas Bay, Luzon Island, Philippines | Coral rubble, 2 m | 8 December 2010 | 13.68, 120.855556 | MNCN 16.01/16082, MNCN/ADN 85684 | – | KX084822 | KX084877 | – | |
| Tk 19 | 1 | ‘Mainif Point’, between Balayan Bay and Batangas Bay, Luzon Island, Philippines | Coral rubble, 2 m | 8 December 2010 | 13.68, 120.855556 | MNCN 16.01/16062, MNCN/ADN 85685 | – | – | KX084875 | KX084972 | |
| Tk 20 | 1 | Shark Bay, WA, Australia | – | – | −25.5, 113.5 | – | – | JF903677 | JF903751 | – | |
| Tk 21 | 1 | ‘Polpollcan’, El Nido, Palawan Island, Philippines | Coral rubble, 3 m | 15 December 2010 | 11.197222, 119.285 | MNCN 16.01/16061, MNCN/ADN 85686 | – | – | – | KX084987 | |
| Trypanosyllis cf. krohnii (lineage 10) | Tk 22 | 1 | Bahea de Aras 61, Philippinesangas Paulo, Brazil | Sponges, ascidians, 0 m | 10 May 2013 | 11.197222, 119.285 | SIO A6143 (SEM), MNCN/ADN 85687 | – | – | – | KX084985 |
| Tk 23 | 1 | Port Jackson, NSW, Australia | – | – | −35.858333, 151.233333 | – | – | JF903678 | JF903752 | JF903790 | |
| Trypanosyllis luqueisp. nov. (lineage 2) | Tk 24 | 1 | Anza Cove, San Diego, California | Bryozoans, 0 m | October 2013 | 32.795278, −117.212778 | SIO A5003 | – | – | – | KX084991 |
| Tk 25 | 1 | Anza Cove, San Diego, California | Bryozoans, 0 m | October 2013 | 32.795278, −117.212778 | SIO A5004 (SEM) | – | – | – | KX084994 | |
| Tk 26 | Holotype 1 | Anza Cove, San Diego, California | Bryozoans, 0 m | October 2013 | 32.795278, −117.212778 | SIO A5005 | – | – | – | KX084993 | |
| Trypanosyllis kalkinsp. nov. (lineage 4) | Tk 27 | 1 | Las Cruces, Valparan, Californiaas P | Unidentified sponge, 18 m | 16 January 2013 | −33.847778, −72.0575 | MNCN 16.01/16060, MNCN/ADN 85688 | – | – | KX084880 | – |
| Tk 28 | 1 | Las Cruces, Valpara, Californiaas P | Dendrymenia skottsbergii, 0 m | 15 January 2013 | −33.847778, −72.0575 | MNCN 16.01/16057, MNCN/ADN 85689 | – | – | – | KX084970 | |
| Tk 29 | Holotype 1 | Las Cruces, Valparaergiiliforniaas P | Dendrymenia skottsbergii, 0 m | 15 January 2013 | −33.847778, −72.0575 | MNCN 16.01/16058, MNCN/ADN 85690 | – | – | – | KX084990 | |
| Trypanosyllis krohnii (lineage 7) | Tk 30 | 1 | Matarge Barcelona Spain | Algae, 0 m | March 2014 | 41.5325, 2.453056 | MNCN 16.01/16187, MNCN/ADN 85691 | – | – | KX084867 | KX084971 |
| Tz 31 | 1 | Cap de Creus, Girona, Spain | Petrosia sp., 16 m | 16 September 2011 | 41.5325, 2.453056 | MNCN 16.01/16066, MNCN/ADN 85692 | – | – | KX084887 | KX084973 | |
| Tk 32 | Neotype 1 | Banyuls-sur-Mer, France | Shallow water | 19 April 2001 | 42.483333, 3.133333 | MNCN/ADN 9623 | – | JF903676 | EF123817 | EF123786 | |
| Trypanosyllis sp. 2 (lineage 6) | Tk 33 | Holotype 1 | Cap de Creus, Girona, Spain | Paramuricea clavata, 40 m | 16 September 2011 | 42.320278, 3.320556 | MNCN 16.01/16065, MNCN/ADN 85693 | – | – | KX084888 | KX084974 |
| Tk 34 | 1 | Cap de Creus, Girona, Spain | Calcareous algae, 36 m | 16 September 2011 | 42.320278, 3.320556 | MNCN 16.01/16064, MNCN/ADN 85694 | – | – | KX084889 | KX084975 | |
| Trypanosyllis cf. krohnii (Crete clade) | Tk35 | 1 | Alykes, Crete | Cystoseira barbata | 20 June 2008 | 35.415833, 24.9875 | MNCN 16.01/16069, MNCN/ADN 85695 | – | – | – | KX084988 |
| Tk 36 | 1 | Elounda, Crete | Calcareous red algae, 1 m | 20 June 2008 | 35.251667, 25.758333 | MNCN 16.01/16190, MNCN/ADN 85696 | – | – | – | KX084989 | |
| Trypanosyllis cf. krohnii | Tk 37 | 1 | Port Philip, Vic, Australia | Rock covered with epibionts, 1 m | 9 February 2010 | −38.103056, 144.4375 | AM W.42429, MNCN/ADN 85697 | – | – | KX084890 | KX084976 |
| Trypanosyllis cf. krohnii (lineage 9) | Tk 38 | 1 | Maunalaya Bay Beach Park, South shore of Oahu, Hawaii | Unidentified algae, 0 m | 25 September 2013 | 21.300556, −157.992778 | MNCN 16.01/16089 (SEM) | KX084815 | – | – | KX084941 |
| Tk 39 | 1 | ‘Mainif Point’, between Balayan Bay and Batangas Bay, Luzon Island, Philippines | Coral rubble, 2 m | 8 December 2010 | 13.68, 120.855556 | MNCN 16.01/16083, MNCN/ADN 85698 | – | KX084836 | KX084885 | KX084961 | |
| Tk 40 | 1 | ‘Mainif Point’, between Balayan Bay and Batangas Bay, Luzon Island, Philippines | Coral rubble, 2 m | 8 December 2010 | 13.68, 120.855556 | MNCN 16.01/16076, MNCN/ADN 85699 | – | KX084837 | KX084886 | KX084962 | |
| Trypanosyllis cf. krohnii (lineage 8) | Tk 41 | 1 | ‘Beatrice Point’, Sombrero Island, Balayan Bay, Luzon Island, Philippines | Sedimet, 3 m | 9 December 2010 | 13.68, 120.855556 | MNCN 16.01/16085, MNCN/ADN 85700 | – | KX084833 | KX084884 | KX084957 |
| Tk 42 | 1 | Balayan Bay, Luzon Island, Philippines | Coral rubble, 2–4 m | 7 December 2010 | 13.740556, 120.892778 | MNCN 16.01/16084, MNCN/ADN 85701 | – | KX084834 | KX084882 | KX084958 | |
| Tk 43 | 1 | Sombrero Island, Balayan Bay, Luzon Island, Philippines | Unidentified sponges, 2 m | 6 December 2010 | 13.697778, 120.829722 | MNCN 16.01/16088, MNCN/ADN 85702 | – | KX084835 | KX084883 | – | |
| Trypanosyllis cf. krohnii | Tk 44 | 1 | Port Jackson, Vauclusse, Bottle&Glass Rock, Sydney, NSW, Australia | On Ecklonia sp. holdfast and under rocks, 4 m | 8 April 2010 | −35.858333, 151.233333 | AM W.42431, MNCN/ADN 85709 | – | – | KX084891 | – |
| Tk 45 | 1 | Lizard island, QLD, Australia | – | – | −14.668889, 145.459444 | – | – | – | JF903750 | JF903793 | |
| Tk 46 | 1 | Port Jackson, NSW, Australia | Balanoid barnacles and sponges, 1 m | 19 February 2006 | −35.858333, 151.233333 | AM W.47277, MNCN/ADN 85703 | – | – | – | KX084995 | |
| Xenosyllis moloch | Xenosyllis moloch 1 | 1 | ‘Sepok Point’, between Balayan Bay and Batangas Bay, Luzon Island, Philippines | Coral rubble, 6 m | 10 December 2010 | 13.683889, 120.895833 | MCZ 251314, MNCN/ADN 85704 | – | KX084856 | KX084915 | – |
| Xenosyllis moloch 2 | 1 | ‘Koala Point’, Balayan Bay, Luzon Island, Philippines | Thalyssias sp. and Acanthella sp. sponges, 16 m | 5 December 2010 | 13.798889, 120.869444 | MCZ 251315, MNCN/ADN 85705 | – | KX084857 | KX084916 | KX084944 | |
| Xenosyllis scabroides | Xenosyllis scabroides | 1 | Lizard Island, QLD, Australia | – | – | −14.668889, 145.459444 | – | – | JF913974 | JF903753 | – |
| Species . | Code . | N . | Locality . | Substrate . | Collection date . | Coordinates (decimal degrees) . | Museum numbers . | 28S . | 18S . | 16S . | COI . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Eurysyllis tuberculata | Eurysyllis tuberculata 1 | 1 | Cap Falcata uberculataegr | Red algae | 16 September 2011 | 42.433333, 3.174722 | MCZ 25312 | KX084805 | KX084852 | – | KX084931 |
| Eurysyllis tuberculata 2 | 1 | San Vicente do Mar, Galicia, Spain | Algae, 1 m | 11 March 2008 | 42.48, −8.901389 | MCZ 25287 | KX084807 | KX084851 | – | KX084934 | |
| Eurysyllis tuberculata 3 | 1 | Shark Bay, WA, Australia | – | – | −25.5, 113.5 | – | – | JF903594 | – | JF903787 | |
| Eurysyllis tuberculata 4 | 1 | Balayan Bay, Luzon Island, Philippines | Coral rubble with hydrozoans, 2–4 m | 4 December 2010 | 13.740556, 120.892778 | MNCN 16.01/16033, MNCN/ADN 85706 | KX084806 | KX084853 | KX084925 | KX084932 | |
| Eurysyllis sp. A | Eurysyllis sp. A1 | 1 | Balayan Bay, Luzon Island, Philippines | Coral rubble, 2–4 m | 7 December 2010 | 13.740556, 120.892778 | MNCN 16.01/16030, MNCN/ADN 85707 | KX084804 | – | – | KX084930 |
| Eurysyllis sp. A2 | 1 | Raja Ampat 2013, Indonesia | Reef flat just off beach, 1 m | 13 October 2013 | −0.551389, 130.695833 | SIO A6141 | – | KX084854 | – | KX084945 | |
| Eurysyllis sp. B | Eurysyllis sp. B1 | 1 | Sombrero Island, Balayan Bay, Luzon Island, Philippines | Coral rubble, 20 m | 9 December 2010 | −0.551389, 130.695833 | MNCN 16.01/16031, MNCN/ADN 85639 | – | KX084850 | KX084862 | KX084943 |
| Eurysyllis sp. B2 | 1 | Sombrero Island, Balayan Bay, Luzon Island, Philippines | Coral rubble, 17 m | 6 December 2010 | −0.551389, 130.695833 | MNCN 16.01/16063, MNCN/ADN 85640 | – | KX084849 | – | KX084960 | |
| Eurysyllis sp. B3 | 1 | Sombrero Island, Balayan Bay, Luzon Island, Philippines | Coral rubble, 2 m | 6 December 2010 | −0.551389, 130.695833 | MNCN 16.01/16032, MNCN/ADN 85641 | – | KX084848 | – | KX084959 | |
| Parahaplosyllis brevicirra | – | 1 | Port Jackson, NSW, Australia | – | – | −35.858333, 151.233333 | – | – | JF903679 | JF903706 | JF903784 |
| Parahaplosyllis kumpol | Parahaplosyllis kumpol 1 | 1 | Cavalli Islands, New Zealand | Unidentified sponge, 15 m | 1 February 2012 | −34.984444, 173.944167 | MNCN 16.01/16036, MNCN/ADN 85642 | – | – | KX084922 | KX084966 |
| Parahaplosyllis kumpol 2 | 1 | Cavalli Islands, New Zealand | Unidentified sponge, 15 m | 1 February 2012 | −34.984444, 173.944167 | MNCN 16.01/16037, MNCN/ADN 85643 | – | – | KX084923 | KX084967 | |
| Plakosyllis sp. | – | 1 | El Nido, Palawan Island, Philippines | Unidentified sponge, 12 m | 18 December 2012 | 11.197222, 119.317222 | MNCN 16.01/16038, MNCN/ADN 85644 | KX084808 | KX084855 | – | KX084933 |
| Pseudosyllis brevipennis | Pseudosyllis brevipennis 1 | 1 | Port de la Selva, Girona, Spain | Posidonia oceanica, 10 m | 21 September 2004 | 42.3375, 3.203333 | MNCN/ADN 9622 | – | EF123878 | EF123816 | EF123785 |
| Pseudosyllis brevipennis 2 | 1 | Alborpe Sea, Spain | Algae, 42–48 m | 24 September 2011 | 35.95 −2.966667 | MNCN 16.01/16040, MNCN/ADN 85645 | – | – | KX084917 | – | |
| Pseudosyllis brevipennis 3 | 1 | Matarpe Barcelona, Spain | Algae, 0 m | March 2014 | 41.5325, 2.453056 | MNCN 16.01/16041, MNCN/ADN 85646 | – | – | – | KX084969 | |
| Syllis amica | – | 1 | Puerto Colera, Girona, Spain | Lithophyllum tortuosum, 0 m | 14 September 2011 | 42.403333, 3.165278 | MCZ 25188 MNCN/ADN 85711 | KX084858 | – | KX084924 | KX084927 |
| Syllis bella | – | 1 | ‘Sepok Point’, between Balayan Bay and Batangas Bay, Luzon Island, Philippines | Coral rubble, 6 m | 10 December 2010 | 13.683889, 120.895833 | MCZ 25190 MNCN/ADN 85712 | – | KX084846 | KX084859 | KX084926 |
| Syllis colum bretensis | – | 1 | Cap de Creus, Girona, Spain | Paramuricea clavata, 40 m | 16 September 2011 | 42.320278, 3.320556 | MCZ 25191 MNCN/ADN 85713 | – | KX084845 | KX084860 | KX084928 |
| Syllis gerundensis | – | 1 | El Toro Island, Mallorca, Spain | Cladocora cespitosa and Miriapora sp., 12 m | 18 June 2012 | 39.4891, 2.4809 | MCZ 25194 MNCN/ADN 85714 | – | KX084847 | KX084861 | KX084929 |
| Trypanosyllis aeolis | – | 1 | El Toro Island, Mallorca, Spain | Cladocora cespitosa and Miriapora sp., 12 m | 18 June 2012 | 39.4891, 2.4809 | MNCN 16.01/16039, MNCN/ADN 85647 | – | KX084817 | KX084913 | KX084968 |
| Trypanedenta gemmipara comb. nov. | Tgemmi 1 | 1 | Cavalli Islands, New Zealand | Rainbow Warrior wreck, 25 m | 1 February 2012 | −36.834167, 174.771389 | MNCN 16.01/16042, MNCN/ADN 85648 | – | – | KX084920 | – |
| Tgemmi 2 | 1 | Cavalli Islands, New Zealand | Rainbow Warrior wreck, 25 m | 1 February 2012 | −36.834167, 174.771389 | MNCN 16.01/16043 (SEM), MNCN/ADN 85649 | – | – | KX084921 | – | |
| Trypanedenta giganteacomb. nov. | Tgiga 1 | 1 | Elephant Island, Antarctica | Blake Trawl, 223–242 m | 22 October 2011 | −61.163333, −54.997222 | SIO A3211 | – | KX084842 | KX084918 | KX084946 |
| Tgiga 2 | 1 | Elephant Island, Antarctica | Blake Trawl, 223–242 m | 22 October 2011 | −61.163333, −54.997222 | SIO A2935 | – | – | KX084919 | KX084984 | |
| Tgiga 3 | 1 | Bransfield Strait, Antarctica | Blake Trawl, 150–247 m | 24 October 2011 | −63.080556, −59.156389 | SIO A2948 | – | – | – | KX084992 | |
| Tgiga 4 | 1 | Burdwood Bank East, Antarctica | Blake Trawl, 90–92 m | 24 April 2013 | −54.558889, −56.828889 | SIO A3565 | – | KX084843 | – | KX084947 | |
| Tgiga 5 | 1 | Burdwood Bank East, Antarctica | Blake Trawl, 122–123 m | 24 April 2013 | −54.540833, −56.626944 | SIO A3515 | – | KX084844 | – | KX084948 | |
| Trypanosyllis sp. 1 | – | 1 | ‘Sepok Point’, between Balayan Bay and Batangas Bay, Luzon Island, Philippines | Coral rubble, 6 m | 10 December 2010 | 13.683889, 120.895833 | MNCN 16.01/16056, MNCN/ADN 85650 | KX084809 | KX084818 | KX084914 | KX084935 |
| Trypanosyllis luzonensiscomb. nov. | Tluzo 1 | 1 | Hong Kong University of Science and Technology, Hong Kong, China | Aquarium system | 9 May 2014 | 22.338056, 114.2675 | SIO A6142 | – | KX084838 | KX084892 | KX084963 |
| Tluzo 2 | 1 | ‘Twin Rocks’, El Nido, Palawan Island, Philippines | Unidentified sponges, 6 m | 17 December 2010 | 11.297222, 119.318333 | MNCN 16.01/16053, MNCN/ADN 85652 | – | KX084839 | KX084893 | – | |
| Tluzo 3 | 1 | ‘Mainif Point’, between Balayan Bay and Batangas Bay, Luzon Island, Philippines | Coral rubble, 2 m | 8 December 2010 | 13.68, 120.855556 | MNCN 16.01/16054, MNCN/ADN 85653 | – | – | KX084895 | KX084978 | |
| Tluzo 4 | 1 | Sombrero Island, Balayan Bay, Luzon Island, Philippines | Coral rubble, 20 m | 9 December 2010 | 13.697778, 120.829722 | MNCN 16.01/16050, MNCN/ADN 85654 | – | – | KX084896 | KX084979 | |
| Tluzo 5 | 1 | Sombrero Island, Balayan Bay, Luzon Island, Philippines | Unidentified sponges, 2 m | 6 December 2010 | 13.697778, 120.829722 | MNCN 16.01/16055, MNCN/ADN 85655 | – | – | KX084894 | – | |
| Tluzo 6 | 1 | Montgomery Reef, WA, Australia | Acropora sp., 0 m | 23 October 2009 | −16.020556, 14.159167 | AM W.42432, MNCN/ADN 85656 | – | – | KX084897 | – | |
| Tluzo 7 | 1 | Sombrero Island, Balayan Bay, Luzon Island, Philippines | Coral rubble, 20 m | 9 December 2010 | 13.697778, 120.829722 | MNCN 16.01/16052, MNCN/ADN 85657 | – | – | KX084904 | KX084980 | |
| Tluzo 8 | 1 | Sombrero Island, Balayan Bay, Luzon Island, Philippines | Coral rubble, 20 m | 9 December 2010 | 13.697778, 120.829722 | MNCN 16.01/16044, MNCN/ADN 85658 | – | – | – | KX084986 | |
| Tluzo 9 | 1 | Port Jackson, NSW, Australia | Encrusting sponges and dead shells on subtidal rock ledge, 9 m | 6 October 2005 | −35.858333, 151.233333 | AM W.47275 MNCN/ADN 85710 | – | – | KX084898 | KX084977 | |
| Tluzo 10 | 1 | Port Jackson, NSW, Australia | Encrusting sponges and dead shells on subtidal rock ledge, 9 m | 6 October 2005 | −35.858333, 151.233333 | AM W.47276, MNCN/ADN 85659 | – | – | KX084907 | KX084981 | |
| Tluzo 11 | 1 | Adele Island, WA, Australia | Sublittoral fore-reef slope, 12.5 m | 18 October 2009 | −15.557778, 123.133889 | AM W.41723, MNCN/ADN 85660 | – | – | KX084899 | – | |
| Tluzo 12 | 1 | Ningaloo Reef, WA, Australia | Coarse coral rubble, 7 m | 20 May 2009 | −22.623611, 113.641111 | AM W.41647, MNCN/ADN 85661 | – | – | KX084900 | – | |
| Tluzo 13 | 1 | Ningaloo Reef, WA, Australia | Brown algae and coral rubble, 24 m | 17 May 2009 | −22.623611, 113.641111 | AM W.41648, MNCN/ADN 85662 | – | – | KX084901 | – | |
| Tluzo 14 | 1 | Ningaloo Reef, WA, Australia | Coarse coral rubble, 7 m | 20 May 2009 | −22.623611, 113.641111 | AM W.41646 (SEM), MNCN/ADN 85663 | – | – | KX084902 | – | |
| Tluzo 15 | 1 | Ningaloo Reef, WA, Australia | Coarse coral rubble, 7 m | 20 May 2009 | −22.623611, 113.641111 | AM W.41645, MNCN/ADN 85664 | – | – | KX084903 | – | |
| Tluzo 16 | 1 | Long Reef, WA, Australia | Sublitoral reef platform, 4 m | 21 October 2010 | −13.856667, 125.825 | AM W.41639 (SEM), MNCN/ADN 85665 | – | – | KX084905 | – | |
| Tluzo 17 | 1 | Prince of Wales Island, QLD, Australia | – | – | −10.681944, 142.188611 | – | – | – | JF903748 | – | |
| Tluzo 18 | 1 | Sombrero Island, Balayan Bay, Luzon Island, Philippines | Coral rubble, 20 m | 9 December 2010 | 13.697778, 120.829722 | MNCN 16.01/16046, MNCN/ADN 85666 | – | – | KX084906 | – | |
| Tluzo 19 | 1 | ‘Sepok Point’, between Balayan Bay and Batangas Bay, Luzon Island, Philippines | Coral rubble, 6 m | 10 December 2010 | 13.683889, 120.895833 | MNCN 16.01/16047, MNCN/ADN 85667 | – | KX084840 | KX084908 | KX084964 | |
| Tluzo 20 | 1 | ‘Koala Point’, Balayan Bay, Luzon Island, Philippines | Coral rubble, 16 m | 5 December 2010 | 13.798889, 120.869444 | MNCN 16.01/16045, MNCN/ADN 85668 | – | – | KX084909 | KX084982 | |
| Tluzo 21 | 1 | ‘Koala Point’, Balayan Bay, Luzon Island, Philippines | Thalyssias sp. and Acanthella sp. sponges, 16 m | 5 December 2010 | 13.798889, 120.869444 | MNCN 16.01/16048, MNCN/ADN 85669 | – | KX084841 | KX084910 | KX084965 | |
| Tluzo 22 | 1 | ‘Sepok Point’, between Balayan Bay and Batangas Bay, Luzon Island, Philippines | Coral rubble, 6 m | 10 December 2010 | 13.683889, 120.895833 | MNCN 16.01/16051, MNCN/ADN 85670 | – | – | KX084911 | KX084983 | |
| Tluzo 23 | 1 | Adele Island, edge of Frazer Inlet, WA, Australia | Sublittoral channel slope 0 m | 22 October 2009 | −15.444722, 123.170833 | AM W.41649, MNCN/ADN 85671 | – | – | KX084912 | – | |
| Trypanosyllis taboadaisp. nov. (lineage 5) | Tk 1 | Holotype 1 | Cavalli Islands, New Zealand | Calcareous algae, 17 m | 2 February 2012 | −34.984444, 173.944167 | MNCN 16.01/16087, MNCN/ADN 85672 | KX084810 | KX084819 | KX084866 | KX084936 |
| Tk 2 | 1 | Maitai Bay, Karikari Peninsule, New Zealand | Corallina sp. and unidentified brown algae, 3 m | 31 January 2012 | −34.831111, 173.409444 | MNCN 16.01/16086, MNCN/ADN 85673 | KX084811 | KX084832 | KX084868 | KX084937 | |
| Tk 3 | 1 | Cavalli Islands, New Zealand | Calcareous algae, 17 m | 2 February 2012 | −34.984444, 173.944167 | MNCN 16.01/16080, MNCN/ADN 85674 | – | KX084828 | KX084865 | KX084949 | |
| Tk 4 | 1 | Cavalli Islands, New Zealand | Unidentified sponges, algae 15 m | 31 January 2012 | −34.984444, 173.944167 | MNCN 16.01/16079, MNCN/ADN 85675 | – | KX084824 | KX084863 | KX084951 | |
| Tk5 | 1 | Maitai Bay, Karikari Peninsule, New Zealand | Kelp, 3 m | 31 January 2012 | −34.831111, 173.409444 | MNCN 16.01/16078, MNCN/ADN 85677 | – | KX084825 | KX084864 | KX084952 | |
| Trypanosyllis cf. krohnii (Australia) | Tk 6 | 1 | Port Jackson, NSW, Australia | Balanoid barnacles and sponges, 1 m | 19 February 2006 | −35.858333, 151.233333 | AM W.47278, MNCN/ADN 85677 | – | KX084831 | KX084878 | KX084954 |
| Tk 7 | 1 | Port Jackson, Vauclusse, Bottle&Glass Rock, Sydney, NSW, Australia | On Ecklonia sp. holdfast and under rocks, 8 m | 8 April 2010 | −35.858333, 151.233333 | AM W.42428, MNCN/ADN 85708 | – | – | KX084879 | – | |
| Trypanosyllis californiensissp. nov. (lineage 3) | Tk 8 | 1 | La Jolla, San Diego, California | Holdfast, 5 m | 18 April 2014 | 32.866944, −117.255833 | SIO A5007 (SEM) | KX084812 | KX084821 | KX084869 | KX084938 |
| Tk 9 | 1 | La Jolla, San Diego, California | Algae, 0 m | 18 April 2014 | 32.866944, −117.255833 | SIO A5009 | KX084813 | – | KX084870 | KX084939 | |
| Tk 10 | Holotype 1 | La Jolla, San Diego, California | Holdfast, 5 m | 18 April 2014 | 32.866944, −117.255833 | SIO A5008 | – | KX084820 | KX084871 | KX084950 | |
| Tk 11 | 1 | La Jolla, San Diego, California | Algae, 0 m | 18 April 2014 | 32.866944, −117.255833 | SIO A5006 | – | – | KX084872 | – | |
| Trypanosyllis cf. krohnii | Tk 12 | 1 | Itapua Beach, Bahgo, California&Glil | Halimeda opuntia, 0 m | June 2014 | −12.956944, −38.36 | MNCN/ADN 85680 | KX084814 | KX084823 | KX084881 | KX084940 |
| Trypanosyllis leivaisp. nov. (lineage 1) | Tk 13 | 1 | ‘Mainif Point’, between Balayan Bay and Batangas Bay, Luzon Island, Philippines | Coral rubble, 2 m | 8 December 2010 | 13.68, 120.855556 | MNCN 16.01/16081, MNCN/ADN 85679 | KX084816 | – | KX084874 | KX084942 |
| Tk 14 | 1 | ‘Beatrice Point’, Sombrero Island, Balayan Bay, Luzon Island, Philippines | Unidentified sponge, 2 m | 9 December 2010 | 13.697778, 120.829722 | MNCN/ADN 85680 | – | KX084827 | KX084876 | KX084953 | |
| Tk 15 | 1 | Sombrero Island, Balayan Bay, Luzon Island, Philippines | Unidentified sponges, 2 m | 6 December 2010 | 13.697778, 120.829722 | MNCN 16.01/16075, MNCN/ADN 85681 | – | KX084829 | KX084873 | – | |
| Tk 16 | 1 | ‘Koala Point’, Balayan Bay, Luzon Island, Philippines | Dead coral, 2 m | 5 December 2010 | 13.798889, 120.869444 | MNCN 16.01/16074, MNCN/ADN 85682 | – | KX084830 | – | KX084955 | |
| Tk 17 | 1 | El Nido, Palawan Island, Philippines | Unidentified sponge, 12 m | 18 December 2012 | 11.197222, 119.317222 | MNCN/ADN 85683 | – | KX084826 | – | KX084956 | |
| Tk 18 | Holotype 1 | ‘Mainif Point’, between Balayan Bay and Batangas Bay, Luzon Island, Philippines | Coral rubble, 2 m | 8 December 2010 | 13.68, 120.855556 | MNCN 16.01/16082, MNCN/ADN 85684 | – | KX084822 | KX084877 | – | |
| Tk 19 | 1 | ‘Mainif Point’, between Balayan Bay and Batangas Bay, Luzon Island, Philippines | Coral rubble, 2 m | 8 December 2010 | 13.68, 120.855556 | MNCN 16.01/16062, MNCN/ADN 85685 | – | – | KX084875 | KX084972 | |
| Tk 20 | 1 | Shark Bay, WA, Australia | – | – | −25.5, 113.5 | – | – | JF903677 | JF903751 | – | |
| Tk 21 | 1 | ‘Polpollcan’, El Nido, Palawan Island, Philippines | Coral rubble, 3 m | 15 December 2010 | 11.197222, 119.285 | MNCN 16.01/16061, MNCN/ADN 85686 | – | – | – | KX084987 | |
| Trypanosyllis cf. krohnii (lineage 10) | Tk 22 | 1 | Bahea de Aras 61, Philippinesangas Paulo, Brazil | Sponges, ascidians, 0 m | 10 May 2013 | 11.197222, 119.285 | SIO A6143 (SEM), MNCN/ADN 85687 | – | – | – | KX084985 |
| Tk 23 | 1 | Port Jackson, NSW, Australia | – | – | −35.858333, 151.233333 | – | – | JF903678 | JF903752 | JF903790 | |
| Trypanosyllis luqueisp. nov. (lineage 2) | Tk 24 | 1 | Anza Cove, San Diego, California | Bryozoans, 0 m | October 2013 | 32.795278, −117.212778 | SIO A5003 | – | – | – | KX084991 |
| Tk 25 | 1 | Anza Cove, San Diego, California | Bryozoans, 0 m | October 2013 | 32.795278, −117.212778 | SIO A5004 (SEM) | – | – | – | KX084994 | |
| Tk 26 | Holotype 1 | Anza Cove, San Diego, California | Bryozoans, 0 m | October 2013 | 32.795278, −117.212778 | SIO A5005 | – | – | – | KX084993 | |
| Trypanosyllis kalkinsp. nov. (lineage 4) | Tk 27 | 1 | Las Cruces, Valparan, Californiaas P | Unidentified sponge, 18 m | 16 January 2013 | −33.847778, −72.0575 | MNCN 16.01/16060, MNCN/ADN 85688 | – | – | KX084880 | – |
| Tk 28 | 1 | Las Cruces, Valpara, Californiaas P | Dendrymenia skottsbergii, 0 m | 15 January 2013 | −33.847778, −72.0575 | MNCN 16.01/16057, MNCN/ADN 85689 | – | – | – | KX084970 | |
| Tk 29 | Holotype 1 | Las Cruces, Valparaergiiliforniaas P | Dendrymenia skottsbergii, 0 m | 15 January 2013 | −33.847778, −72.0575 | MNCN 16.01/16058, MNCN/ADN 85690 | – | – | – | KX084990 | |
| Trypanosyllis krohnii (lineage 7) | Tk 30 | 1 | Matarge Barcelona Spain | Algae, 0 m | March 2014 | 41.5325, 2.453056 | MNCN 16.01/16187, MNCN/ADN 85691 | – | – | KX084867 | KX084971 |
| Tz 31 | 1 | Cap de Creus, Girona, Spain | Petrosia sp., 16 m | 16 September 2011 | 41.5325, 2.453056 | MNCN 16.01/16066, MNCN/ADN 85692 | – | – | KX084887 | KX084973 | |
| Tk 32 | Neotype 1 | Banyuls-sur-Mer, France | Shallow water | 19 April 2001 | 42.483333, 3.133333 | MNCN/ADN 9623 | – | JF903676 | EF123817 | EF123786 | |
| Trypanosyllis sp. 2 (lineage 6) | Tk 33 | Holotype 1 | Cap de Creus, Girona, Spain | Paramuricea clavata, 40 m | 16 September 2011 | 42.320278, 3.320556 | MNCN 16.01/16065, MNCN/ADN 85693 | – | – | KX084888 | KX084974 |
| Tk 34 | 1 | Cap de Creus, Girona, Spain | Calcareous algae, 36 m | 16 September 2011 | 42.320278, 3.320556 | MNCN 16.01/16064, MNCN/ADN 85694 | – | – | KX084889 | KX084975 | |
| Trypanosyllis cf. krohnii (Crete clade) | Tk35 | 1 | Alykes, Crete | Cystoseira barbata | 20 June 2008 | 35.415833, 24.9875 | MNCN 16.01/16069, MNCN/ADN 85695 | – | – | – | KX084988 |
| Tk 36 | 1 | Elounda, Crete | Calcareous red algae, 1 m | 20 June 2008 | 35.251667, 25.758333 | MNCN 16.01/16190, MNCN/ADN 85696 | – | – | – | KX084989 | |
| Trypanosyllis cf. krohnii | Tk 37 | 1 | Port Philip, Vic, Australia | Rock covered with epibionts, 1 m | 9 February 2010 | −38.103056, 144.4375 | AM W.42429, MNCN/ADN 85697 | – | – | KX084890 | KX084976 |
| Trypanosyllis cf. krohnii (lineage 9) | Tk 38 | 1 | Maunalaya Bay Beach Park, South shore of Oahu, Hawaii | Unidentified algae, 0 m | 25 September 2013 | 21.300556, −157.992778 | MNCN 16.01/16089 (SEM) | KX084815 | – | – | KX084941 |
| Tk 39 | 1 | ‘Mainif Point’, between Balayan Bay and Batangas Bay, Luzon Island, Philippines | Coral rubble, 2 m | 8 December 2010 | 13.68, 120.855556 | MNCN 16.01/16083, MNCN/ADN 85698 | – | KX084836 | KX084885 | KX084961 | |
| Tk 40 | 1 | ‘Mainif Point’, between Balayan Bay and Batangas Bay, Luzon Island, Philippines | Coral rubble, 2 m | 8 December 2010 | 13.68, 120.855556 | MNCN 16.01/16076, MNCN/ADN 85699 | – | KX084837 | KX084886 | KX084962 | |
| Trypanosyllis cf. krohnii (lineage 8) | Tk 41 | 1 | ‘Beatrice Point’, Sombrero Island, Balayan Bay, Luzon Island, Philippines | Sedimet, 3 m | 9 December 2010 | 13.68, 120.855556 | MNCN 16.01/16085, MNCN/ADN 85700 | – | KX084833 | KX084884 | KX084957 |
| Tk 42 | 1 | Balayan Bay, Luzon Island, Philippines | Coral rubble, 2–4 m | 7 December 2010 | 13.740556, 120.892778 | MNCN 16.01/16084, MNCN/ADN 85701 | – | KX084834 | KX084882 | KX084958 | |
| Tk 43 | 1 | Sombrero Island, Balayan Bay, Luzon Island, Philippines | Unidentified sponges, 2 m | 6 December 2010 | 13.697778, 120.829722 | MNCN 16.01/16088, MNCN/ADN 85702 | – | KX084835 | KX084883 | – | |
| Trypanosyllis cf. krohnii | Tk 44 | 1 | Port Jackson, Vauclusse, Bottle&Glass Rock, Sydney, NSW, Australia | On Ecklonia sp. holdfast and under rocks, 4 m | 8 April 2010 | −35.858333, 151.233333 | AM W.42431, MNCN/ADN 85709 | – | – | KX084891 | – |
| Tk 45 | 1 | Lizard island, QLD, Australia | – | – | −14.668889, 145.459444 | – | – | – | JF903750 | JF903793 | |
| Tk 46 | 1 | Port Jackson, NSW, Australia | Balanoid barnacles and sponges, 1 m | 19 February 2006 | −35.858333, 151.233333 | AM W.47277, MNCN/ADN 85703 | – | – | – | KX084995 | |
| Xenosyllis moloch | Xenosyllis moloch 1 | 1 | ‘Sepok Point’, between Balayan Bay and Batangas Bay, Luzon Island, Philippines | Coral rubble, 6 m | 10 December 2010 | 13.683889, 120.895833 | MCZ 251314, MNCN/ADN 85704 | – | KX084856 | KX084915 | – |
| Xenosyllis moloch 2 | 1 | ‘Koala Point’, Balayan Bay, Luzon Island, Philippines | Thalyssias sp. and Acanthella sp. sponges, 16 m | 5 December 2010 | 13.798889, 120.869444 | MCZ 251315, MNCN/ADN 85705 | – | KX084857 | KX084916 | KX084944 | |
| Xenosyllis scabroides | Xenosyllis scabroides | 1 | Lizard Island, QLD, Australia | – | – | −14.668889, 145.459444 | – | – | JF913974 | JF903753 | – |
Sequences generated for this study appear in bold.
Localities, substrates, date of sampling, coordinates, catalogue numbers, and GenBank accession numbers for all specimens sequenced
| Species . | Code . | N . | Locality . | Substrate . | Collection date . | Coordinates (decimal degrees) . | Museum numbers . | 28S . | 18S . | 16S . | COI . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Eurysyllis tuberculata | Eurysyllis tuberculata 1 | 1 | Cap Falcata uberculataegr | Red algae | 16 September 2011 | 42.433333, 3.174722 | MCZ 25312 | KX084805 | KX084852 | – | KX084931 |
| Eurysyllis tuberculata 2 | 1 | San Vicente do Mar, Galicia, Spain | Algae, 1 m | 11 March 2008 | 42.48, −8.901389 | MCZ 25287 | KX084807 | KX084851 | – | KX084934 | |
| Eurysyllis tuberculata 3 | 1 | Shark Bay, WA, Australia | – | – | −25.5, 113.5 | – | – | JF903594 | – | JF903787 | |
| Eurysyllis tuberculata 4 | 1 | Balayan Bay, Luzon Island, Philippines | Coral rubble with hydrozoans, 2–4 m | 4 December 2010 | 13.740556, 120.892778 | MNCN 16.01/16033, MNCN/ADN 85706 | KX084806 | KX084853 | KX084925 | KX084932 | |
| Eurysyllis sp. A | Eurysyllis sp. A1 | 1 | Balayan Bay, Luzon Island, Philippines | Coral rubble, 2–4 m | 7 December 2010 | 13.740556, 120.892778 | MNCN 16.01/16030, MNCN/ADN 85707 | KX084804 | – | – | KX084930 |
| Eurysyllis sp. A2 | 1 | Raja Ampat 2013, Indonesia | Reef flat just off beach, 1 m | 13 October 2013 | −0.551389, 130.695833 | SIO A6141 | – | KX084854 | – | KX084945 | |
| Eurysyllis sp. B | Eurysyllis sp. B1 | 1 | Sombrero Island, Balayan Bay, Luzon Island, Philippines | Coral rubble, 20 m | 9 December 2010 | −0.551389, 130.695833 | MNCN 16.01/16031, MNCN/ADN 85639 | – | KX084850 | KX084862 | KX084943 |
| Eurysyllis sp. B2 | 1 | Sombrero Island, Balayan Bay, Luzon Island, Philippines | Coral rubble, 17 m | 6 December 2010 | −0.551389, 130.695833 | MNCN 16.01/16063, MNCN/ADN 85640 | – | KX084849 | – | KX084960 | |
| Eurysyllis sp. B3 | 1 | Sombrero Island, Balayan Bay, Luzon Island, Philippines | Coral rubble, 2 m | 6 December 2010 | −0.551389, 130.695833 | MNCN 16.01/16032, MNCN/ADN 85641 | – | KX084848 | – | KX084959 | |
| Parahaplosyllis brevicirra | – | 1 | Port Jackson, NSW, Australia | – | – | −35.858333, 151.233333 | – | – | JF903679 | JF903706 | JF903784 |
| Parahaplosyllis kumpol | Parahaplosyllis kumpol 1 | 1 | Cavalli Islands, New Zealand | Unidentified sponge, 15 m | 1 February 2012 | −34.984444, 173.944167 | MNCN 16.01/16036, MNCN/ADN 85642 | – | – | KX084922 | KX084966 |
| Parahaplosyllis kumpol 2 | 1 | Cavalli Islands, New Zealand | Unidentified sponge, 15 m | 1 February 2012 | −34.984444, 173.944167 | MNCN 16.01/16037, MNCN/ADN 85643 | – | – | KX084923 | KX084967 | |
| Plakosyllis sp. | – | 1 | El Nido, Palawan Island, Philippines | Unidentified sponge, 12 m | 18 December 2012 | 11.197222, 119.317222 | MNCN 16.01/16038, MNCN/ADN 85644 | KX084808 | KX084855 | – | KX084933 |
| Pseudosyllis brevipennis | Pseudosyllis brevipennis 1 | 1 | Port de la Selva, Girona, Spain | Posidonia oceanica, 10 m | 21 September 2004 | 42.3375, 3.203333 | MNCN/ADN 9622 | – | EF123878 | EF123816 | EF123785 |
| Pseudosyllis brevipennis 2 | 1 | Alborpe Sea, Spain | Algae, 42–48 m | 24 September 2011 | 35.95 −2.966667 | MNCN 16.01/16040, MNCN/ADN 85645 | – | – | KX084917 | – | |
| Pseudosyllis brevipennis 3 | 1 | Matarpe Barcelona, Spain | Algae, 0 m | March 2014 | 41.5325, 2.453056 | MNCN 16.01/16041, MNCN/ADN 85646 | – | – | – | KX084969 | |
| Syllis amica | – | 1 | Puerto Colera, Girona, Spain | Lithophyllum tortuosum, 0 m | 14 September 2011 | 42.403333, 3.165278 | MCZ 25188 MNCN/ADN 85711 | KX084858 | – | KX084924 | KX084927 |
| Syllis bella | – | 1 | ‘Sepok Point’, between Balayan Bay and Batangas Bay, Luzon Island, Philippines | Coral rubble, 6 m | 10 December 2010 | 13.683889, 120.895833 | MCZ 25190 MNCN/ADN 85712 | – | KX084846 | KX084859 | KX084926 |
| Syllis colum bretensis | – | 1 | Cap de Creus, Girona, Spain | Paramuricea clavata, 40 m | 16 September 2011 | 42.320278, 3.320556 | MCZ 25191 MNCN/ADN 85713 | – | KX084845 | KX084860 | KX084928 |
| Syllis gerundensis | – | 1 | El Toro Island, Mallorca, Spain | Cladocora cespitosa and Miriapora sp., 12 m | 18 June 2012 | 39.4891, 2.4809 | MCZ 25194 MNCN/ADN 85714 | – | KX084847 | KX084861 | KX084929 |
| Trypanosyllis aeolis | – | 1 | El Toro Island, Mallorca, Spain | Cladocora cespitosa and Miriapora sp., 12 m | 18 June 2012 | 39.4891, 2.4809 | MNCN 16.01/16039, MNCN/ADN 85647 | – | KX084817 | KX084913 | KX084968 |
| Trypanedenta gemmipara comb. nov. | Tgemmi 1 | 1 | Cavalli Islands, New Zealand | Rainbow Warrior wreck, 25 m | 1 February 2012 | −36.834167, 174.771389 | MNCN 16.01/16042, MNCN/ADN 85648 | – | – | KX084920 | – |
| Tgemmi 2 | 1 | Cavalli Islands, New Zealand | Rainbow Warrior wreck, 25 m | 1 February 2012 | −36.834167, 174.771389 | MNCN 16.01/16043 (SEM), MNCN/ADN 85649 | – | – | KX084921 | – | |
| Trypanedenta giganteacomb. nov. | Tgiga 1 | 1 | Elephant Island, Antarctica | Blake Trawl, 223–242 m | 22 October 2011 | −61.163333, −54.997222 | SIO A3211 | – | KX084842 | KX084918 | KX084946 |
| Tgiga 2 | 1 | Elephant Island, Antarctica | Blake Trawl, 223–242 m | 22 October 2011 | −61.163333, −54.997222 | SIO A2935 | – | – | KX084919 | KX084984 | |
| Tgiga 3 | 1 | Bransfield Strait, Antarctica | Blake Trawl, 150–247 m | 24 October 2011 | −63.080556, −59.156389 | SIO A2948 | – | – | – | KX084992 | |
| Tgiga 4 | 1 | Burdwood Bank East, Antarctica | Blake Trawl, 90–92 m | 24 April 2013 | −54.558889, −56.828889 | SIO A3565 | – | KX084843 | – | KX084947 | |
| Tgiga 5 | 1 | Burdwood Bank East, Antarctica | Blake Trawl, 122–123 m | 24 April 2013 | −54.540833, −56.626944 | SIO A3515 | – | KX084844 | – | KX084948 | |
| Trypanosyllis sp. 1 | – | 1 | ‘Sepok Point’, between Balayan Bay and Batangas Bay, Luzon Island, Philippines | Coral rubble, 6 m | 10 December 2010 | 13.683889, 120.895833 | MNCN 16.01/16056, MNCN/ADN 85650 | KX084809 | KX084818 | KX084914 | KX084935 |
| Trypanosyllis luzonensiscomb. nov. | Tluzo 1 | 1 | Hong Kong University of Science and Technology, Hong Kong, China | Aquarium system | 9 May 2014 | 22.338056, 114.2675 | SIO A6142 | – | KX084838 | KX084892 | KX084963 |
| Tluzo 2 | 1 | ‘Twin Rocks’, El Nido, Palawan Island, Philippines | Unidentified sponges, 6 m | 17 December 2010 | 11.297222, 119.318333 | MNCN 16.01/16053, MNCN/ADN 85652 | – | KX084839 | KX084893 | – | |
| Tluzo 3 | 1 | ‘Mainif Point’, between Balayan Bay and Batangas Bay, Luzon Island, Philippines | Coral rubble, 2 m | 8 December 2010 | 13.68, 120.855556 | MNCN 16.01/16054, MNCN/ADN 85653 | – | – | KX084895 | KX084978 | |
| Tluzo 4 | 1 | Sombrero Island, Balayan Bay, Luzon Island, Philippines | Coral rubble, 20 m | 9 December 2010 | 13.697778, 120.829722 | MNCN 16.01/16050, MNCN/ADN 85654 | – | – | KX084896 | KX084979 | |
| Tluzo 5 | 1 | Sombrero Island, Balayan Bay, Luzon Island, Philippines | Unidentified sponges, 2 m | 6 December 2010 | 13.697778, 120.829722 | MNCN 16.01/16055, MNCN/ADN 85655 | – | – | KX084894 | – | |
| Tluzo 6 | 1 | Montgomery Reef, WA, Australia | Acropora sp., 0 m | 23 October 2009 | −16.020556, 14.159167 | AM W.42432, MNCN/ADN 85656 | – | – | KX084897 | – | |
| Tluzo 7 | 1 | Sombrero Island, Balayan Bay, Luzon Island, Philippines | Coral rubble, 20 m | 9 December 2010 | 13.697778, 120.829722 | MNCN 16.01/16052, MNCN/ADN 85657 | – | – | KX084904 | KX084980 | |
| Tluzo 8 | 1 | Sombrero Island, Balayan Bay, Luzon Island, Philippines | Coral rubble, 20 m | 9 December 2010 | 13.697778, 120.829722 | MNCN 16.01/16044, MNCN/ADN 85658 | – | – | – | KX084986 | |
| Tluzo 9 | 1 | Port Jackson, NSW, Australia | Encrusting sponges and dead shells on subtidal rock ledge, 9 m | 6 October 2005 | −35.858333, 151.233333 | AM W.47275 MNCN/ADN 85710 | – | – | KX084898 | KX084977 | |
| Tluzo 10 | 1 | Port Jackson, NSW, Australia | Encrusting sponges and dead shells on subtidal rock ledge, 9 m | 6 October 2005 | −35.858333, 151.233333 | AM W.47276, MNCN/ADN 85659 | – | – | KX084907 | KX084981 | |
| Tluzo 11 | 1 | Adele Island, WA, Australia | Sublittoral fore-reef slope, 12.5 m | 18 October 2009 | −15.557778, 123.133889 | AM W.41723, MNCN/ADN 85660 | – | – | KX084899 | – | |
| Tluzo 12 | 1 | Ningaloo Reef, WA, Australia | Coarse coral rubble, 7 m | 20 May 2009 | −22.623611, 113.641111 | AM W.41647, MNCN/ADN 85661 | – | – | KX084900 | – | |
| Tluzo 13 | 1 | Ningaloo Reef, WA, Australia | Brown algae and coral rubble, 24 m | 17 May 2009 | −22.623611, 113.641111 | AM W.41648, MNCN/ADN 85662 | – | – | KX084901 | – | |
| Tluzo 14 | 1 | Ningaloo Reef, WA, Australia | Coarse coral rubble, 7 m | 20 May 2009 | −22.623611, 113.641111 | AM W.41646 (SEM), MNCN/ADN 85663 | – | – | KX084902 | – | |
| Tluzo 15 | 1 | Ningaloo Reef, WA, Australia | Coarse coral rubble, 7 m | 20 May 2009 | −22.623611, 113.641111 | AM W.41645, MNCN/ADN 85664 | – | – | KX084903 | – | |
| Tluzo 16 | 1 | Long Reef, WA, Australia | Sublitoral reef platform, 4 m | 21 October 2010 | −13.856667, 125.825 | AM W.41639 (SEM), MNCN/ADN 85665 | – | – | KX084905 | – | |
| Tluzo 17 | 1 | Prince of Wales Island, QLD, Australia | – | – | −10.681944, 142.188611 | – | – | – | JF903748 | – | |
| Tluzo 18 | 1 | Sombrero Island, Balayan Bay, Luzon Island, Philippines | Coral rubble, 20 m | 9 December 2010 | 13.697778, 120.829722 | MNCN 16.01/16046, MNCN/ADN 85666 | – | – | KX084906 | – | |
| Tluzo 19 | 1 | ‘Sepok Point’, between Balayan Bay and Batangas Bay, Luzon Island, Philippines | Coral rubble, 6 m | 10 December 2010 | 13.683889, 120.895833 | MNCN 16.01/16047, MNCN/ADN 85667 | – | KX084840 | KX084908 | KX084964 | |
| Tluzo 20 | 1 | ‘Koala Point’, Balayan Bay, Luzon Island, Philippines | Coral rubble, 16 m | 5 December 2010 | 13.798889, 120.869444 | MNCN 16.01/16045, MNCN/ADN 85668 | – | – | KX084909 | KX084982 | |
| Tluzo 21 | 1 | ‘Koala Point’, Balayan Bay, Luzon Island, Philippines | Thalyssias sp. and Acanthella sp. sponges, 16 m | 5 December 2010 | 13.798889, 120.869444 | MNCN 16.01/16048, MNCN/ADN 85669 | – | KX084841 | KX084910 | KX084965 | |
| Tluzo 22 | 1 | ‘Sepok Point’, between Balayan Bay and Batangas Bay, Luzon Island, Philippines | Coral rubble, 6 m | 10 December 2010 | 13.683889, 120.895833 | MNCN 16.01/16051, MNCN/ADN 85670 | – | – | KX084911 | KX084983 | |
| Tluzo 23 | 1 | Adele Island, edge of Frazer Inlet, WA, Australia | Sublittoral channel slope 0 m | 22 October 2009 | −15.444722, 123.170833 | AM W.41649, MNCN/ADN 85671 | – | – | KX084912 | – | |
| Trypanosyllis taboadaisp. nov. (lineage 5) | Tk 1 | Holotype 1 | Cavalli Islands, New Zealand | Calcareous algae, 17 m | 2 February 2012 | −34.984444, 173.944167 | MNCN 16.01/16087, MNCN/ADN 85672 | KX084810 | KX084819 | KX084866 | KX084936 |
| Tk 2 | 1 | Maitai Bay, Karikari Peninsule, New Zealand | Corallina sp. and unidentified brown algae, 3 m | 31 January 2012 | −34.831111, 173.409444 | MNCN 16.01/16086, MNCN/ADN 85673 | KX084811 | KX084832 | KX084868 | KX084937 | |
| Tk 3 | 1 | Cavalli Islands, New Zealand | Calcareous algae, 17 m | 2 February 2012 | −34.984444, 173.944167 | MNCN 16.01/16080, MNCN/ADN 85674 | – | KX084828 | KX084865 | KX084949 | |
| Tk 4 | 1 | Cavalli Islands, New Zealand | Unidentified sponges, algae 15 m | 31 January 2012 | −34.984444, 173.944167 | MNCN 16.01/16079, MNCN/ADN 85675 | – | KX084824 | KX084863 | KX084951 | |
| Tk5 | 1 | Maitai Bay, Karikari Peninsule, New Zealand | Kelp, 3 m | 31 January 2012 | −34.831111, 173.409444 | MNCN 16.01/16078, MNCN/ADN 85677 | – | KX084825 | KX084864 | KX084952 | |
| Trypanosyllis cf. krohnii (Australia) | Tk 6 | 1 | Port Jackson, NSW, Australia | Balanoid barnacles and sponges, 1 m | 19 February 2006 | −35.858333, 151.233333 | AM W.47278, MNCN/ADN 85677 | – | KX084831 | KX084878 | KX084954 |
| Tk 7 | 1 | Port Jackson, Vauclusse, Bottle&Glass Rock, Sydney, NSW, Australia | On Ecklonia sp. holdfast and under rocks, 8 m | 8 April 2010 | −35.858333, 151.233333 | AM W.42428, MNCN/ADN 85708 | – | – | KX084879 | – | |
| Trypanosyllis californiensissp. nov. (lineage 3) | Tk 8 | 1 | La Jolla, San Diego, California | Holdfast, 5 m | 18 April 2014 | 32.866944, −117.255833 | SIO A5007 (SEM) | KX084812 | KX084821 | KX084869 | KX084938 |
| Tk 9 | 1 | La Jolla, San Diego, California | Algae, 0 m | 18 April 2014 | 32.866944, −117.255833 | SIO A5009 | KX084813 | – | KX084870 | KX084939 | |
| Tk 10 | Holotype 1 | La Jolla, San Diego, California | Holdfast, 5 m | 18 April 2014 | 32.866944, −117.255833 | SIO A5008 | – | KX084820 | KX084871 | KX084950 | |
| Tk 11 | 1 | La Jolla, San Diego, California | Algae, 0 m | 18 April 2014 | 32.866944, −117.255833 | SIO A5006 | – | – | KX084872 | – | |
| Trypanosyllis cf. krohnii | Tk 12 | 1 | Itapua Beach, Bahgo, California&Glil | Halimeda opuntia, 0 m | June 2014 | −12.956944, −38.36 | MNCN/ADN 85680 | KX084814 | KX084823 | KX084881 | KX084940 |
| Trypanosyllis leivaisp. nov. (lineage 1) | Tk 13 | 1 | ‘Mainif Point’, between Balayan Bay and Batangas Bay, Luzon Island, Philippines | Coral rubble, 2 m | 8 December 2010 | 13.68, 120.855556 | MNCN 16.01/16081, MNCN/ADN 85679 | KX084816 | – | KX084874 | KX084942 |
| Tk 14 | 1 | ‘Beatrice Point’, Sombrero Island, Balayan Bay, Luzon Island, Philippines | Unidentified sponge, 2 m | 9 December 2010 | 13.697778, 120.829722 | MNCN/ADN 85680 | – | KX084827 | KX084876 | KX084953 | |
| Tk 15 | 1 | Sombrero Island, Balayan Bay, Luzon Island, Philippines | Unidentified sponges, 2 m | 6 December 2010 | 13.697778, 120.829722 | MNCN 16.01/16075, MNCN/ADN 85681 | – | KX084829 | KX084873 | – | |
| Tk 16 | 1 | ‘Koala Point’, Balayan Bay, Luzon Island, Philippines | Dead coral, 2 m | 5 December 2010 | 13.798889, 120.869444 | MNCN 16.01/16074, MNCN/ADN 85682 | – | KX084830 | – | KX084955 | |
| Tk 17 | 1 | El Nido, Palawan Island, Philippines | Unidentified sponge, 12 m | 18 December 2012 | 11.197222, 119.317222 | MNCN/ADN 85683 | – | KX084826 | – | KX084956 | |
| Tk 18 | Holotype 1 | ‘Mainif Point’, between Balayan Bay and Batangas Bay, Luzon Island, Philippines | Coral rubble, 2 m | 8 December 2010 | 13.68, 120.855556 | MNCN 16.01/16082, MNCN/ADN 85684 | – | KX084822 | KX084877 | – | |
| Tk 19 | 1 | ‘Mainif Point’, between Balayan Bay and Batangas Bay, Luzon Island, Philippines | Coral rubble, 2 m | 8 December 2010 | 13.68, 120.855556 | MNCN 16.01/16062, MNCN/ADN 85685 | – | – | KX084875 | KX084972 | |
| Tk 20 | 1 | Shark Bay, WA, Australia | – | – | −25.5, 113.5 | – | – | JF903677 | JF903751 | – | |
| Tk 21 | 1 | ‘Polpollcan’, El Nido, Palawan Island, Philippines | Coral rubble, 3 m | 15 December 2010 | 11.197222, 119.285 | MNCN 16.01/16061, MNCN/ADN 85686 | – | – | – | KX084987 | |
| Trypanosyllis cf. krohnii (lineage 10) | Tk 22 | 1 | Bahea de Aras 61, Philippinesangas Paulo, Brazil | Sponges, ascidians, 0 m | 10 May 2013 | 11.197222, 119.285 | SIO A6143 (SEM), MNCN/ADN 85687 | – | – | – | KX084985 |
| Tk 23 | 1 | Port Jackson, NSW, Australia | – | – | −35.858333, 151.233333 | – | – | JF903678 | JF903752 | JF903790 | |
| Trypanosyllis luqueisp. nov. (lineage 2) | Tk 24 | 1 | Anza Cove, San Diego, California | Bryozoans, 0 m | October 2013 | 32.795278, −117.212778 | SIO A5003 | – | – | – | KX084991 |
| Tk 25 | 1 | Anza Cove, San Diego, California | Bryozoans, 0 m | October 2013 | 32.795278, −117.212778 | SIO A5004 (SEM) | – | – | – | KX084994 | |
| Tk 26 | Holotype 1 | Anza Cove, San Diego, California | Bryozoans, 0 m | October 2013 | 32.795278, −117.212778 | SIO A5005 | – | – | – | KX084993 | |
| Trypanosyllis kalkinsp. nov. (lineage 4) | Tk 27 | 1 | Las Cruces, Valparan, Californiaas P | Unidentified sponge, 18 m | 16 January 2013 | −33.847778, −72.0575 | MNCN 16.01/16060, MNCN/ADN 85688 | – | – | KX084880 | – |
| Tk 28 | 1 | Las Cruces, Valpara, Californiaas P | Dendrymenia skottsbergii, 0 m | 15 January 2013 | −33.847778, −72.0575 | MNCN 16.01/16057, MNCN/ADN 85689 | – | – | – | KX084970 | |
| Tk 29 | Holotype 1 | Las Cruces, Valparaergiiliforniaas P | Dendrymenia skottsbergii, 0 m | 15 January 2013 | −33.847778, −72.0575 | MNCN 16.01/16058, MNCN/ADN 85690 | – | – | – | KX084990 | |
| Trypanosyllis krohnii (lineage 7) | Tk 30 | 1 | Matarge Barcelona Spain | Algae, 0 m | March 2014 | 41.5325, 2.453056 | MNCN 16.01/16187, MNCN/ADN 85691 | – | – | KX084867 | KX084971 |
| Tz 31 | 1 | Cap de Creus, Girona, Spain | Petrosia sp., 16 m | 16 September 2011 | 41.5325, 2.453056 | MNCN 16.01/16066, MNCN/ADN 85692 | – | – | KX084887 | KX084973 | |
| Tk 32 | Neotype 1 | Banyuls-sur-Mer, France | Shallow water | 19 April 2001 | 42.483333, 3.133333 | MNCN/ADN 9623 | – | JF903676 | EF123817 | EF123786 | |
| Trypanosyllis sp. 2 (lineage 6) | Tk 33 | Holotype 1 | Cap de Creus, Girona, Spain | Paramuricea clavata, 40 m | 16 September 2011 | 42.320278, 3.320556 | MNCN 16.01/16065, MNCN/ADN 85693 | – | – | KX084888 | KX084974 |
| Tk 34 | 1 | Cap de Creus, Girona, Spain | Calcareous algae, 36 m | 16 September 2011 | 42.320278, 3.320556 | MNCN 16.01/16064, MNCN/ADN 85694 | – | – | KX084889 | KX084975 | |
| Trypanosyllis cf. krohnii (Crete clade) | Tk35 | 1 | Alykes, Crete | Cystoseira barbata | 20 June 2008 | 35.415833, 24.9875 | MNCN 16.01/16069, MNCN/ADN 85695 | – | – | – | KX084988 |
| Tk 36 | 1 | Elounda, Crete | Calcareous red algae, 1 m | 20 June 2008 | 35.251667, 25.758333 | MNCN 16.01/16190, MNCN/ADN 85696 | – | – | – | KX084989 | |
| Trypanosyllis cf. krohnii | Tk 37 | 1 | Port Philip, Vic, Australia | Rock covered with epibionts, 1 m | 9 February 2010 | −38.103056, 144.4375 | AM W.42429, MNCN/ADN 85697 | – | – | KX084890 | KX084976 |
| Trypanosyllis cf. krohnii (lineage 9) | Tk 38 | 1 | Maunalaya Bay Beach Park, South shore of Oahu, Hawaii | Unidentified algae, 0 m | 25 September 2013 | 21.300556, −157.992778 | MNCN 16.01/16089 (SEM) | KX084815 | – | – | KX084941 |
| Tk 39 | 1 | ‘Mainif Point’, between Balayan Bay and Batangas Bay, Luzon Island, Philippines | Coral rubble, 2 m | 8 December 2010 | 13.68, 120.855556 | MNCN 16.01/16083, MNCN/ADN 85698 | – | KX084836 | KX084885 | KX084961 | |
| Tk 40 | 1 | ‘Mainif Point’, between Balayan Bay and Batangas Bay, Luzon Island, Philippines | Coral rubble, 2 m | 8 December 2010 | 13.68, 120.855556 | MNCN 16.01/16076, MNCN/ADN 85699 | – | KX084837 | KX084886 | KX084962 | |
| Trypanosyllis cf. krohnii (lineage 8) | Tk 41 | 1 | ‘Beatrice Point’, Sombrero Island, Balayan Bay, Luzon Island, Philippines | Sedimet, 3 m | 9 December 2010 | 13.68, 120.855556 | MNCN 16.01/16085, MNCN/ADN 85700 | – | KX084833 | KX084884 | KX084957 |
| Tk 42 | 1 | Balayan Bay, Luzon Island, Philippines | Coral rubble, 2–4 m | 7 December 2010 | 13.740556, 120.892778 | MNCN 16.01/16084, MNCN/ADN 85701 | – | KX084834 | KX084882 | KX084958 | |
| Tk 43 | 1 | Sombrero Island, Balayan Bay, Luzon Island, Philippines | Unidentified sponges, 2 m | 6 December 2010 | 13.697778, 120.829722 | MNCN 16.01/16088, MNCN/ADN 85702 | – | KX084835 | KX084883 | – | |
| Trypanosyllis cf. krohnii | Tk 44 | 1 | Port Jackson, Vauclusse, Bottle&Glass Rock, Sydney, NSW, Australia | On Ecklonia sp. holdfast and under rocks, 4 m | 8 April 2010 | −35.858333, 151.233333 | AM W.42431, MNCN/ADN 85709 | – | – | KX084891 | – |
| Tk 45 | 1 | Lizard island, QLD, Australia | – | – | −14.668889, 145.459444 | – | – | – | JF903750 | JF903793 | |
| Tk 46 | 1 | Port Jackson, NSW, Australia | Balanoid barnacles and sponges, 1 m | 19 February 2006 | −35.858333, 151.233333 | AM W.47277, MNCN/ADN 85703 | – | – | – | KX084995 | |
| Xenosyllis moloch | Xenosyllis moloch 1 | 1 | ‘Sepok Point’, between Balayan Bay and Batangas Bay, Luzon Island, Philippines | Coral rubble, 6 m | 10 December 2010 | 13.683889, 120.895833 | MCZ 251314, MNCN/ADN 85704 | – | KX084856 | KX084915 | – |
| Xenosyllis moloch 2 | 1 | ‘Koala Point’, Balayan Bay, Luzon Island, Philippines | Thalyssias sp. and Acanthella sp. sponges, 16 m | 5 December 2010 | 13.798889, 120.869444 | MCZ 251315, MNCN/ADN 85705 | – | KX084857 | KX084916 | KX084944 | |
| Xenosyllis scabroides | Xenosyllis scabroides | 1 | Lizard Island, QLD, Australia | – | – | −14.668889, 145.459444 | – | – | JF913974 | JF903753 | – |
| Species . | Code . | N . | Locality . | Substrate . | Collection date . | Coordinates (decimal degrees) . | Museum numbers . | 28S . | 18S . | 16S . | COI . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Eurysyllis tuberculata | Eurysyllis tuberculata 1 | 1 | Cap Falcata uberculataegr | Red algae | 16 September 2011 | 42.433333, 3.174722 | MCZ 25312 | KX084805 | KX084852 | – | KX084931 |
| Eurysyllis tuberculata 2 | 1 | San Vicente do Mar, Galicia, Spain | Algae, 1 m | 11 March 2008 | 42.48, −8.901389 | MCZ 25287 | KX084807 | KX084851 | – | KX084934 | |
| Eurysyllis tuberculata 3 | 1 | Shark Bay, WA, Australia | – | – | −25.5, 113.5 | – | – | JF903594 | – | JF903787 | |
| Eurysyllis tuberculata 4 | 1 | Balayan Bay, Luzon Island, Philippines | Coral rubble with hydrozoans, 2–4 m | 4 December 2010 | 13.740556, 120.892778 | MNCN 16.01/16033, MNCN/ADN 85706 | KX084806 | KX084853 | KX084925 | KX084932 | |
| Eurysyllis sp. A | Eurysyllis sp. A1 | 1 | Balayan Bay, Luzon Island, Philippines | Coral rubble, 2–4 m | 7 December 2010 | 13.740556, 120.892778 | MNCN 16.01/16030, MNCN/ADN 85707 | KX084804 | – | – | KX084930 |
| Eurysyllis sp. A2 | 1 | Raja Ampat 2013, Indonesia | Reef flat just off beach, 1 m | 13 October 2013 | −0.551389, 130.695833 | SIO A6141 | – | KX084854 | – | KX084945 | |
| Eurysyllis sp. B | Eurysyllis sp. B1 | 1 | Sombrero Island, Balayan Bay, Luzon Island, Philippines | Coral rubble, 20 m | 9 December 2010 | −0.551389, 130.695833 | MNCN 16.01/16031, MNCN/ADN 85639 | – | KX084850 | KX084862 | KX084943 |
| Eurysyllis sp. B2 | 1 | Sombrero Island, Balayan Bay, Luzon Island, Philippines | Coral rubble, 17 m | 6 December 2010 | −0.551389, 130.695833 | MNCN 16.01/16063, MNCN/ADN 85640 | – | KX084849 | – | KX084960 | |
| Eurysyllis sp. B3 | 1 | Sombrero Island, Balayan Bay, Luzon Island, Philippines | Coral rubble, 2 m | 6 December 2010 | −0.551389, 130.695833 | MNCN 16.01/16032, MNCN/ADN 85641 | – | KX084848 | – | KX084959 | |
| Parahaplosyllis brevicirra | – | 1 | Port Jackson, NSW, Australia | – | – | −35.858333, 151.233333 | – | – | JF903679 | JF903706 | JF903784 |
| Parahaplosyllis kumpol | Parahaplosyllis kumpol 1 | 1 | Cavalli Islands, New Zealand | Unidentified sponge, 15 m | 1 February 2012 | −34.984444, 173.944167 | MNCN 16.01/16036, MNCN/ADN 85642 | – | – | KX084922 | KX084966 |
| Parahaplosyllis kumpol 2 | 1 | Cavalli Islands, New Zealand | Unidentified sponge, 15 m | 1 February 2012 | −34.984444, 173.944167 | MNCN 16.01/16037, MNCN/ADN 85643 | – | – | KX084923 | KX084967 | |
| Plakosyllis sp. | – | 1 | El Nido, Palawan Island, Philippines | Unidentified sponge, 12 m | 18 December 2012 | 11.197222, 119.317222 | MNCN 16.01/16038, MNCN/ADN 85644 | KX084808 | KX084855 | – | KX084933 |
| Pseudosyllis brevipennis | Pseudosyllis brevipennis 1 | 1 | Port de la Selva, Girona, Spain | Posidonia oceanica, 10 m | 21 September 2004 | 42.3375, 3.203333 | MNCN/ADN 9622 | – | EF123878 | EF123816 | EF123785 |
| Pseudosyllis brevipennis 2 | 1 | Alborpe Sea, Spain | Algae, 42–48 m | 24 September 2011 | 35.95 −2.966667 | MNCN 16.01/16040, MNCN/ADN 85645 | – | – | KX084917 | – | |
| Pseudosyllis brevipennis 3 | 1 | Matarpe Barcelona, Spain | Algae, 0 m | March 2014 | 41.5325, 2.453056 | MNCN 16.01/16041, MNCN/ADN 85646 | – | – | – | KX084969 | |
| Syllis amica | – | 1 | Puerto Colera, Girona, Spain | Lithophyllum tortuosum, 0 m | 14 September 2011 | 42.403333, 3.165278 | MCZ 25188 MNCN/ADN 85711 | KX084858 | – | KX084924 | KX084927 |
| Syllis bella | – | 1 | ‘Sepok Point’, between Balayan Bay and Batangas Bay, Luzon Island, Philippines | Coral rubble, 6 m | 10 December 2010 | 13.683889, 120.895833 | MCZ 25190 MNCN/ADN 85712 | – | KX084846 | KX084859 | KX084926 |
| Syllis colum bretensis | – | 1 | Cap de Creus, Girona, Spain | Paramuricea clavata, 40 m | 16 September 2011 | 42.320278, 3.320556 | MCZ 25191 MNCN/ADN 85713 | – | KX084845 | KX084860 | KX084928 |
| Syllis gerundensis | – | 1 | El Toro Island, Mallorca, Spain | Cladocora cespitosa and Miriapora sp., 12 m | 18 June 2012 | 39.4891, 2.4809 | MCZ 25194 MNCN/ADN 85714 | – | KX084847 | KX084861 | KX084929 |
| Trypanosyllis aeolis | – | 1 | El Toro Island, Mallorca, Spain | Cladocora cespitosa and Miriapora sp., 12 m | 18 June 2012 | 39.4891, 2.4809 | MNCN 16.01/16039, MNCN/ADN 85647 | – | KX084817 | KX084913 | KX084968 |
| Trypanedenta gemmipara comb. nov. | Tgemmi 1 | 1 | Cavalli Islands, New Zealand | Rainbow Warrior wreck, 25 m | 1 February 2012 | −36.834167, 174.771389 | MNCN 16.01/16042, MNCN/ADN 85648 | – | – | KX084920 | – |
| Tgemmi 2 | 1 | Cavalli Islands, New Zealand | Rainbow Warrior wreck, 25 m | 1 February 2012 | −36.834167, 174.771389 | MNCN 16.01/16043 (SEM), MNCN/ADN 85649 | – | – | KX084921 | – | |
| Trypanedenta giganteacomb. nov. | Tgiga 1 | 1 | Elephant Island, Antarctica | Blake Trawl, 223–242 m | 22 October 2011 | −61.163333, −54.997222 | SIO A3211 | – | KX084842 | KX084918 | KX084946 |
| Tgiga 2 | 1 | Elephant Island, Antarctica | Blake Trawl, 223–242 m | 22 October 2011 | −61.163333, −54.997222 | SIO A2935 | – | – | KX084919 | KX084984 | |
| Tgiga 3 | 1 | Bransfield Strait, Antarctica | Blake Trawl, 150–247 m | 24 October 2011 | −63.080556, −59.156389 | SIO A2948 | – | – | – | KX084992 | |
| Tgiga 4 | 1 | Burdwood Bank East, Antarctica | Blake Trawl, 90–92 m | 24 April 2013 | −54.558889, −56.828889 | SIO A3565 | – | KX084843 | – | KX084947 | |
| Tgiga 5 | 1 | Burdwood Bank East, Antarctica | Blake Trawl, 122–123 m | 24 April 2013 | −54.540833, −56.626944 | SIO A3515 | – | KX084844 | – | KX084948 | |
| Trypanosyllis sp. 1 | – | 1 | ‘Sepok Point’, between Balayan Bay and Batangas Bay, Luzon Island, Philippines | Coral rubble, 6 m | 10 December 2010 | 13.683889, 120.895833 | MNCN 16.01/16056, MNCN/ADN 85650 | KX084809 | KX084818 | KX084914 | KX084935 |
| Trypanosyllis luzonensiscomb. nov. | Tluzo 1 | 1 | Hong Kong University of Science and Technology, Hong Kong, China | Aquarium system | 9 May 2014 | 22.338056, 114.2675 | SIO A6142 | – | KX084838 | KX084892 | KX084963 |
| Tluzo 2 | 1 | ‘Twin Rocks’, El Nido, Palawan Island, Philippines | Unidentified sponges, 6 m | 17 December 2010 | 11.297222, 119.318333 | MNCN 16.01/16053, MNCN/ADN 85652 | – | KX084839 | KX084893 | – | |
| Tluzo 3 | 1 | ‘Mainif Point’, between Balayan Bay and Batangas Bay, Luzon Island, Philippines | Coral rubble, 2 m | 8 December 2010 | 13.68, 120.855556 | MNCN 16.01/16054, MNCN/ADN 85653 | – | – | KX084895 | KX084978 | |
| Tluzo 4 | 1 | Sombrero Island, Balayan Bay, Luzon Island, Philippines | Coral rubble, 20 m | 9 December 2010 | 13.697778, 120.829722 | MNCN 16.01/16050, MNCN/ADN 85654 | – | – | KX084896 | KX084979 | |
| Tluzo 5 | 1 | Sombrero Island, Balayan Bay, Luzon Island, Philippines | Unidentified sponges, 2 m | 6 December 2010 | 13.697778, 120.829722 | MNCN 16.01/16055, MNCN/ADN 85655 | – | – | KX084894 | – | |
| Tluzo 6 | 1 | Montgomery Reef, WA, Australia | Acropora sp., 0 m | 23 October 2009 | −16.020556, 14.159167 | AM W.42432, MNCN/ADN 85656 | – | – | KX084897 | – | |
| Tluzo 7 | 1 | Sombrero Island, Balayan Bay, Luzon Island, Philippines | Coral rubble, 20 m | 9 December 2010 | 13.697778, 120.829722 | MNCN 16.01/16052, MNCN/ADN 85657 | – | – | KX084904 | KX084980 | |
| Tluzo 8 | 1 | Sombrero Island, Balayan Bay, Luzon Island, Philippines | Coral rubble, 20 m | 9 December 2010 | 13.697778, 120.829722 | MNCN 16.01/16044, MNCN/ADN 85658 | – | – | – | KX084986 | |
| Tluzo 9 | 1 | Port Jackson, NSW, Australia | Encrusting sponges and dead shells on subtidal rock ledge, 9 m | 6 October 2005 | −35.858333, 151.233333 | AM W.47275 MNCN/ADN 85710 | – | – | KX084898 | KX084977 | |
| Tluzo 10 | 1 | Port Jackson, NSW, Australia | Encrusting sponges and dead shells on subtidal rock ledge, 9 m | 6 October 2005 | −35.858333, 151.233333 | AM W.47276, MNCN/ADN 85659 | – | – | KX084907 | KX084981 | |
| Tluzo 11 | 1 | Adele Island, WA, Australia | Sublittoral fore-reef slope, 12.5 m | 18 October 2009 | −15.557778, 123.133889 | AM W.41723, MNCN/ADN 85660 | – | – | KX084899 | – | |
| Tluzo 12 | 1 | Ningaloo Reef, WA, Australia | Coarse coral rubble, 7 m | 20 May 2009 | −22.623611, 113.641111 | AM W.41647, MNCN/ADN 85661 | – | – | KX084900 | – | |
| Tluzo 13 | 1 | Ningaloo Reef, WA, Australia | Brown algae and coral rubble, 24 m | 17 May 2009 | −22.623611, 113.641111 | AM W.41648, MNCN/ADN 85662 | – | – | KX084901 | – | |
| Tluzo 14 | 1 | Ningaloo Reef, WA, Australia | Coarse coral rubble, 7 m | 20 May 2009 | −22.623611, 113.641111 | AM W.41646 (SEM), MNCN/ADN 85663 | – | – | KX084902 | – | |
| Tluzo 15 | 1 | Ningaloo Reef, WA, Australia | Coarse coral rubble, 7 m | 20 May 2009 | −22.623611, 113.641111 | AM W.41645, MNCN/ADN 85664 | – | – | KX084903 | – | |
| Tluzo 16 | 1 | Long Reef, WA, Australia | Sublitoral reef platform, 4 m | 21 October 2010 | −13.856667, 125.825 | AM W.41639 (SEM), MNCN/ADN 85665 | – | – | KX084905 | – | |
| Tluzo 17 | 1 | Prince of Wales Island, QLD, Australia | – | – | −10.681944, 142.188611 | – | – | – | JF903748 | – | |
| Tluzo 18 | 1 | Sombrero Island, Balayan Bay, Luzon Island, Philippines | Coral rubble, 20 m | 9 December 2010 | 13.697778, 120.829722 | MNCN 16.01/16046, MNCN/ADN 85666 | – | – | KX084906 | – | |
| Tluzo 19 | 1 | ‘Sepok Point’, between Balayan Bay and Batangas Bay, Luzon Island, Philippines | Coral rubble, 6 m | 10 December 2010 | 13.683889, 120.895833 | MNCN 16.01/16047, MNCN/ADN 85667 | – | KX084840 | KX084908 | KX084964 | |
| Tluzo 20 | 1 | ‘Koala Point’, Balayan Bay, Luzon Island, Philippines | Coral rubble, 16 m | 5 December 2010 | 13.798889, 120.869444 | MNCN 16.01/16045, MNCN/ADN 85668 | – | – | KX084909 | KX084982 | |
| Tluzo 21 | 1 | ‘Koala Point’, Balayan Bay, Luzon Island, Philippines | Thalyssias sp. and Acanthella sp. sponges, 16 m | 5 December 2010 | 13.798889, 120.869444 | MNCN 16.01/16048, MNCN/ADN 85669 | – | KX084841 | KX084910 | KX084965 | |
| Tluzo 22 | 1 | ‘Sepok Point’, between Balayan Bay and Batangas Bay, Luzon Island, Philippines | Coral rubble, 6 m | 10 December 2010 | 13.683889, 120.895833 | MNCN 16.01/16051, MNCN/ADN 85670 | – | – | KX084911 | KX084983 | |
| Tluzo 23 | 1 | Adele Island, edge of Frazer Inlet, WA, Australia | Sublittoral channel slope 0 m | 22 October 2009 | −15.444722, 123.170833 | AM W.41649, MNCN/ADN 85671 | – | – | KX084912 | – | |
| Trypanosyllis taboadaisp. nov. (lineage 5) | Tk 1 | Holotype 1 | Cavalli Islands, New Zealand | Calcareous algae, 17 m | 2 February 2012 | −34.984444, 173.944167 | MNCN 16.01/16087, MNCN/ADN 85672 | KX084810 | KX084819 | KX084866 | KX084936 |
| Tk 2 | 1 | Maitai Bay, Karikari Peninsule, New Zealand | Corallina sp. and unidentified brown algae, 3 m | 31 January 2012 | −34.831111, 173.409444 | MNCN 16.01/16086, MNCN/ADN 85673 | KX084811 | KX084832 | KX084868 | KX084937 | |
| Tk 3 | 1 | Cavalli Islands, New Zealand | Calcareous algae, 17 m | 2 February 2012 | −34.984444, 173.944167 | MNCN 16.01/16080, MNCN/ADN 85674 | – | KX084828 | KX084865 | KX084949 | |
| Tk 4 | 1 | Cavalli Islands, New Zealand | Unidentified sponges, algae 15 m | 31 January 2012 | −34.984444, 173.944167 | MNCN 16.01/16079, MNCN/ADN 85675 | – | KX084824 | KX084863 | KX084951 | |
| Tk5 | 1 | Maitai Bay, Karikari Peninsule, New Zealand | Kelp, 3 m | 31 January 2012 | −34.831111, 173.409444 | MNCN 16.01/16078, MNCN/ADN 85677 | – | KX084825 | KX084864 | KX084952 | |
| Trypanosyllis cf. krohnii (Australia) | Tk 6 | 1 | Port Jackson, NSW, Australia | Balanoid barnacles and sponges, 1 m | 19 February 2006 | −35.858333, 151.233333 | AM W.47278, MNCN/ADN 85677 | – | KX084831 | KX084878 | KX084954 |
| Tk 7 | 1 | Port Jackson, Vauclusse, Bottle&Glass Rock, Sydney, NSW, Australia | On Ecklonia sp. holdfast and under rocks, 8 m | 8 April 2010 | −35.858333, 151.233333 | AM W.42428, MNCN/ADN 85708 | – | – | KX084879 | – | |
| Trypanosyllis californiensissp. nov. (lineage 3) | Tk 8 | 1 | La Jolla, San Diego, California | Holdfast, 5 m | 18 April 2014 | 32.866944, −117.255833 | SIO A5007 (SEM) | KX084812 | KX084821 | KX084869 | KX084938 |
| Tk 9 | 1 | La Jolla, San Diego, California | Algae, 0 m | 18 April 2014 | 32.866944, −117.255833 | SIO A5009 | KX084813 | – | KX084870 | KX084939 | |
| Tk 10 | Holotype 1 | La Jolla, San Diego, California | Holdfast, 5 m | 18 April 2014 | 32.866944, −117.255833 | SIO A5008 | – | KX084820 | KX084871 | KX084950 | |
| Tk 11 | 1 | La Jolla, San Diego, California | Algae, 0 m | 18 April 2014 | 32.866944, −117.255833 | SIO A5006 | – | – | KX084872 | – | |
| Trypanosyllis cf. krohnii | Tk 12 | 1 | Itapua Beach, Bahgo, California&Glil | Halimeda opuntia, 0 m | June 2014 | −12.956944, −38.36 | MNCN/ADN 85680 | KX084814 | KX084823 | KX084881 | KX084940 |
| Trypanosyllis leivaisp. nov. (lineage 1) | Tk 13 | 1 | ‘Mainif Point’, between Balayan Bay and Batangas Bay, Luzon Island, Philippines | Coral rubble, 2 m | 8 December 2010 | 13.68, 120.855556 | MNCN 16.01/16081, MNCN/ADN 85679 | KX084816 | – | KX084874 | KX084942 |
| Tk 14 | 1 | ‘Beatrice Point’, Sombrero Island, Balayan Bay, Luzon Island, Philippines | Unidentified sponge, 2 m | 9 December 2010 | 13.697778, 120.829722 | MNCN/ADN 85680 | – | KX084827 | KX084876 | KX084953 | |
| Tk 15 | 1 | Sombrero Island, Balayan Bay, Luzon Island, Philippines | Unidentified sponges, 2 m | 6 December 2010 | 13.697778, 120.829722 | MNCN 16.01/16075, MNCN/ADN 85681 | – | KX084829 | KX084873 | – | |
| Tk 16 | 1 | ‘Koala Point’, Balayan Bay, Luzon Island, Philippines | Dead coral, 2 m | 5 December 2010 | 13.798889, 120.869444 | MNCN 16.01/16074, MNCN/ADN 85682 | – | KX084830 | – | KX084955 | |
| Tk 17 | 1 | El Nido, Palawan Island, Philippines | Unidentified sponge, 12 m | 18 December 2012 | 11.197222, 119.317222 | MNCN/ADN 85683 | – | KX084826 | – | KX084956 | |
| Tk 18 | Holotype 1 | ‘Mainif Point’, between Balayan Bay and Batangas Bay, Luzon Island, Philippines | Coral rubble, 2 m | 8 December 2010 | 13.68, 120.855556 | MNCN 16.01/16082, MNCN/ADN 85684 | – | KX084822 | KX084877 | – | |
| Tk 19 | 1 | ‘Mainif Point’, between Balayan Bay and Batangas Bay, Luzon Island, Philippines | Coral rubble, 2 m | 8 December 2010 | 13.68, 120.855556 | MNCN 16.01/16062, MNCN/ADN 85685 | – | – | KX084875 | KX084972 | |
| Tk 20 | 1 | Shark Bay, WA, Australia | – | – | −25.5, 113.5 | – | – | JF903677 | JF903751 | – | |
| Tk 21 | 1 | ‘Polpollcan’, El Nido, Palawan Island, Philippines | Coral rubble, 3 m | 15 December 2010 | 11.197222, 119.285 | MNCN 16.01/16061, MNCN/ADN 85686 | – | – | – | KX084987 | |
| Trypanosyllis cf. krohnii (lineage 10) | Tk 22 | 1 | Bahea de Aras 61, Philippinesangas Paulo, Brazil | Sponges, ascidians, 0 m | 10 May 2013 | 11.197222, 119.285 | SIO A6143 (SEM), MNCN/ADN 85687 | – | – | – | KX084985 |
| Tk 23 | 1 | Port Jackson, NSW, Australia | – | – | −35.858333, 151.233333 | – | – | JF903678 | JF903752 | JF903790 | |
| Trypanosyllis luqueisp. nov. (lineage 2) | Tk 24 | 1 | Anza Cove, San Diego, California | Bryozoans, 0 m | October 2013 | 32.795278, −117.212778 | SIO A5003 | – | – | – | KX084991 |
| Tk 25 | 1 | Anza Cove, San Diego, California | Bryozoans, 0 m | October 2013 | 32.795278, −117.212778 | SIO A5004 (SEM) | – | – | – | KX084994 | |
| Tk 26 | Holotype 1 | Anza Cove, San Diego, California | Bryozoans, 0 m | October 2013 | 32.795278, −117.212778 | SIO A5005 | – | – | – | KX084993 | |
| Trypanosyllis kalkinsp. nov. (lineage 4) | Tk 27 | 1 | Las Cruces, Valparan, Californiaas P | Unidentified sponge, 18 m | 16 January 2013 | −33.847778, −72.0575 | MNCN 16.01/16060, MNCN/ADN 85688 | – | – | KX084880 | – |
| Tk 28 | 1 | Las Cruces, Valpara, Californiaas P | Dendrymenia skottsbergii, 0 m | 15 January 2013 | −33.847778, −72.0575 | MNCN 16.01/16057, MNCN/ADN 85689 | – | – | – | KX084970 | |
| Tk 29 | Holotype 1 | Las Cruces, Valparaergiiliforniaas P | Dendrymenia skottsbergii, 0 m | 15 January 2013 | −33.847778, −72.0575 | MNCN 16.01/16058, MNCN/ADN 85690 | – | – | – | KX084990 | |
| Trypanosyllis krohnii (lineage 7) | Tk 30 | 1 | Matarge Barcelona Spain | Algae, 0 m | March 2014 | 41.5325, 2.453056 | MNCN 16.01/16187, MNCN/ADN 85691 | – | – | KX084867 | KX084971 |
| Tz 31 | 1 | Cap de Creus, Girona, Spain | Petrosia sp., 16 m | 16 September 2011 | 41.5325, 2.453056 | MNCN 16.01/16066, MNCN/ADN 85692 | – | – | KX084887 | KX084973 | |
| Tk 32 | Neotype 1 | Banyuls-sur-Mer, France | Shallow water | 19 April 2001 | 42.483333, 3.133333 | MNCN/ADN 9623 | – | JF903676 | EF123817 | EF123786 | |
| Trypanosyllis sp. 2 (lineage 6) | Tk 33 | Holotype 1 | Cap de Creus, Girona, Spain | Paramuricea clavata, 40 m | 16 September 2011 | 42.320278, 3.320556 | MNCN 16.01/16065, MNCN/ADN 85693 | – | – | KX084888 | KX084974 |
| Tk 34 | 1 | Cap de Creus, Girona, Spain | Calcareous algae, 36 m | 16 September 2011 | 42.320278, 3.320556 | MNCN 16.01/16064, MNCN/ADN 85694 | – | – | KX084889 | KX084975 | |
| Trypanosyllis cf. krohnii (Crete clade) | Tk35 | 1 | Alykes, Crete | Cystoseira barbata | 20 June 2008 | 35.415833, 24.9875 | MNCN 16.01/16069, MNCN/ADN 85695 | – | – | – | KX084988 |
| Tk 36 | 1 | Elounda, Crete | Calcareous red algae, 1 m | 20 June 2008 | 35.251667, 25.758333 | MNCN 16.01/16190, MNCN/ADN 85696 | – | – | – | KX084989 | |
| Trypanosyllis cf. krohnii | Tk 37 | 1 | Port Philip, Vic, Australia | Rock covered with epibionts, 1 m | 9 February 2010 | −38.103056, 144.4375 | AM W.42429, MNCN/ADN 85697 | – | – | KX084890 | KX084976 |
| Trypanosyllis cf. krohnii (lineage 9) | Tk 38 | 1 | Maunalaya Bay Beach Park, South shore of Oahu, Hawaii | Unidentified algae, 0 m | 25 September 2013 | 21.300556, −157.992778 | MNCN 16.01/16089 (SEM) | KX084815 | – | – | KX084941 |
| Tk 39 | 1 | ‘Mainif Point’, between Balayan Bay and Batangas Bay, Luzon Island, Philippines | Coral rubble, 2 m | 8 December 2010 | 13.68, 120.855556 | MNCN 16.01/16083, MNCN/ADN 85698 | – | KX084836 | KX084885 | KX084961 | |
| Tk 40 | 1 | ‘Mainif Point’, between Balayan Bay and Batangas Bay, Luzon Island, Philippines | Coral rubble, 2 m | 8 December 2010 | 13.68, 120.855556 | MNCN 16.01/16076, MNCN/ADN 85699 | – | KX084837 | KX084886 | KX084962 | |
| Trypanosyllis cf. krohnii (lineage 8) | Tk 41 | 1 | ‘Beatrice Point’, Sombrero Island, Balayan Bay, Luzon Island, Philippines | Sedimet, 3 m | 9 December 2010 | 13.68, 120.855556 | MNCN 16.01/16085, MNCN/ADN 85700 | – | KX084833 | KX084884 | KX084957 |
| Tk 42 | 1 | Balayan Bay, Luzon Island, Philippines | Coral rubble, 2–4 m | 7 December 2010 | 13.740556, 120.892778 | MNCN 16.01/16084, MNCN/ADN 85701 | – | KX084834 | KX084882 | KX084958 | |
| Tk 43 | 1 | Sombrero Island, Balayan Bay, Luzon Island, Philippines | Unidentified sponges, 2 m | 6 December 2010 | 13.697778, 120.829722 | MNCN 16.01/16088, MNCN/ADN 85702 | – | KX084835 | KX084883 | – | |
| Trypanosyllis cf. krohnii | Tk 44 | 1 | Port Jackson, Vauclusse, Bottle&Glass Rock, Sydney, NSW, Australia | On Ecklonia sp. holdfast and under rocks, 4 m | 8 April 2010 | −35.858333, 151.233333 | AM W.42431, MNCN/ADN 85709 | – | – | KX084891 | – |
| Tk 45 | 1 | Lizard island, QLD, Australia | – | – | −14.668889, 145.459444 | – | – | – | JF903750 | JF903793 | |
| Tk 46 | 1 | Port Jackson, NSW, Australia | Balanoid barnacles and sponges, 1 m | 19 February 2006 | −35.858333, 151.233333 | AM W.47277, MNCN/ADN 85703 | – | – | – | KX084995 | |
| Xenosyllis moloch | Xenosyllis moloch 1 | 1 | ‘Sepok Point’, between Balayan Bay and Batangas Bay, Luzon Island, Philippines | Coral rubble, 6 m | 10 December 2010 | 13.683889, 120.895833 | MCZ 251314, MNCN/ADN 85704 | – | KX084856 | KX084915 | – |
| Xenosyllis moloch 2 | 1 | ‘Koala Point’, Balayan Bay, Luzon Island, Philippines | Thalyssias sp. and Acanthella sp. sponges, 16 m | 5 December 2010 | 13.798889, 120.869444 | MCZ 251315, MNCN/ADN 85705 | – | KX084857 | KX084916 | KX084944 | |
| Xenosyllis scabroides | Xenosyllis scabroides | 1 | Lizard Island, QLD, Australia | – | – | −14.668889, 145.459444 | – | – | JF913974 | JF903753 | – |
Sequences generated for this study appear in bold.
For scanning electron microscopy, selected specimens were prepared on an Emitech K850 Critical Point Dryer, gold-coated with a Q150T-S Turbo-Pumper Sputter Coater, and examined with a Hitachi S-3000N scanning electron microscope (SEM) at the Servicio Interdepartamental de Investigación (SIDI) of the UAM.
Type material of Trypanosyllis luzonensis (Pillai, 1965) comb. nov. was loaned from The Natural History Museum, London (NHM). Comparative material from the type locality of Trypanosyllis ingensJohnson, 1902 and type material of Trypanosyllis intermediaMoore, 1909 was loaned from the Invertebrate Zoology Collection of the California Academy of Science (CAS-IZ). Comparative material of Trypanosyllis coeliaca and Trypanosyllis zebra from the Adriatic was loaned from the Museum für Naturkunde, Berlin (ZMB). Most of the Trypanosyllis luzonensis comb. nov. specimens were loaned from the Australian Museum (AM). All the newly collected specimens were deposited at the Museo Nacional de Ciencias Naturales, Madrid (MNCN), Museum of Comparative Zoology, Harvard University, Cambridge (MZC), and Scripps Institution of Oceanography, Benthic Invertebrate Collection, La Jolla (SIO). Catalogue numbers are listed in Table 1.
Molecular analyses
Genomic DNA were extracted from 92 individuals of Trypanosyllis, Eurysyllis, Plakosyllis, and Xenosyllis, and out-groups of Syllis and Parahaplosyllis (Table 1), using the DNeasy Blood & Tissue Kit (Qiagen), following manufacturer's protocols. Fragments of the nuclear genes 28S rRNA (531 bp) and 18S rRNA (∼1800 bp), and the mitochondrial 16S rRNA (470 bp) and cytochrome c oxidase subunit I (COI; ∼650 bp) were amplified by polymerase chain reaction (PCR). Primers 28Sa and 28Srd5b (Edgecombe & Giribet, 2006) were used to amplify the 28S rRNA fragment. For 18S rRNA, three overlapping pairs of primers were used: 18S1F-18S4R, 18S3F-18Sbi, and 18Sa2.0-18S9R (Giribet et al., 1996; Whiting et al., 1997). Primers 16SarL and 16SbrH (Palumbi, 1996) were used to amplify 16S rRNA, and the modified primers with inosine jgLCO1490 and jgHCO2198 (Geller et al., 2013) were employed to amplify COI in all specimens.
The PCR reactions consisted of 1 μL of DNA template in 25 μL reaction volumes containing 18 μL H2O; 5 μL of 5× USB buffer, 0.25 μL of each of 10 μM primers, 0.5 μL of 10 mM dNTPs, and 0.13 μL of 1.25 U/μL GOTaq DNA Polymerase (Promega). The temperature profile for the 18S and 28S rRNA nuclear markers was as follows: 95 °C/120 s; (95 °C/30 s; 47 °C/30 s; 72 °C/180 s) × 35 cycles; 72 °C/300 s; for 16S rRNA: 95 °C/5 min; (95 °C/30 s; 45 °C/30 s; 72 °C/60 s) × 35 cycles; 72 °C/10 min; and for COI: 95 °C/15 min; (94 °C/30 s; 45 °C/70 s; 72 °C/90 s) × 40 cycles; 72 °C/10 min. For cycle sequencing, 10 μL reactions were prepared using 1.5 μL of cleaned template DNA for each primer direction, using the following protocol: 4.3 μL ultrapure H20, 0.5 μL of 5× BigDye buffer, 3.2 μL 10 μM primer, 1.0 μL of BigDye Terminator v3.1 (Life Technologies, ABI). The sequencing reaction temperature profile was as follows: 94 °C/180 s; (94 °C/10 s; 50 °C/5 s; 60 °C/4 min) × 25 cycles. Sequencing products for each primer were precipitated using 3 g Sephadex beads in 45 mL of ultrapure distilled water. Then, samples were evaporated and resuspended in 20 μL of formamide. The sequencing reaction products were then analysed using an ABI Prism 3730xl Genetic Analyzer (Applied Biosystems).
Sequences were edited in GENEIOUS 6.1.6 (Kearse et al., 2012), where primers were removed and overlapping amplicons in 18S rRNA were merged into a consensus sequence. Multiple sequence alignments were run in MAFFT 7 under default parameters (Katoh & Standley, 2013). GBLOCKS 0.91b (Castresana, 2002) was used to remove highly variable positions in the nuclear ribosomal markers, with a maximum of eight contiguous non-conserved positions and five positions as the minimum length of blocks.
Phylogenetic analysis
In order to assess the monophyly of Trypanosyllis, we analysed the four molecular markers available for 81 specimens traditionally considered as belonging to Trypanosyllis and 13 specimens of the closely related genera Eurysyllis, Plakosyllis, and Xenosyllis. Sequences (seven) of Parahaplosyllis brevicirra Hartmann-Schröder, 1990 from GenBank and newly sequenced specimens of Parahaplosyllis kumpolÁlvarez-Campos, San Martín & Aguado, 2013, Syllis amica Quatrefages, 1866, S. bella Chamberlin, 1919, S. columbretensis (Campoy, 1982), and S. gerundensis (Alós & Campoy, 1981) were used as out-groups (Table 1). All mitochondrial and nuclear data sets were concatenated. The best-fitting model of sequence evolution was selected using the Akaike information criterion (AIC) in jModeltest 2 (Darriba et al., 2012). The best model for the data set of both mitochondrial and nuclear markers was a general time-reversible (GTR) model of sequence evolution with gamma-distributed rates across sites and a proportion of invariable sites (GTR + G + I). Partitions for each of the markers were used in all subsequent phylogenetic analyses: for ribosomal markers, non-codon-specific models were used; and for COI, we used codon-specific models. We also tried a third codon model for COI in the concatenated analysis, but the results did not differ from those using the codon-specific model so we are only presenting the first set of results.
Maximum-likelihood (ML) partitioned analyses of the concatenated data sets were run in RAxML 7.4.2 (Stamatakis, 2006) using a GTR + G + I model of sequence evolution. Bootstrap support values were estimated using 1000 replicates and ten starting trees (Stamatakis, Hoover & Rougemont, 2008). Bayesian inference (BI) partitioned analyses were also run with the concatenated data sets and the GTR + G + I model, implemented in MrBayes 3.2.1 (Ronquist et al., 2012), and run with four Markov chains that were started from a random tree and run simultaneously for 10 000 000 generations, with trees sampled every 500 generations (samplefreq = 500). The first 25% of trees were discarded as burn-in (burninfrac = 0.25) after assessing convergence using TRACER 1.6 (Rambaut et al., 2014) and AWTY (Wilgenbusch, Warren & Swofford, 2004).
Species delimitation analysis
In order to test whether Trypanosyllis krohnii was a species complex, we analysed our COI and 16S rRNA data set for all putative Trypanosyllis krohnii specimens with two different methods of species delimitation analysis: the Poisson tree processes (PTP) model (Zhang et al., 2013) and the generalized mixed Yule coalescent (GMYC) approach with both the single-threshold (S) and the multiple-threshold (M) variants of the method (Pons et al., 2006). Also, the PTP analysis was used to analyse our concatenated nuclear and mitochondrial genes. Both PTP and GMYC provide operational criteria of a gene-coalescent view of the phylogenetic species concept (Baum & Shaw, 1995). For species delimitation with PTP, we used the rooted BI phylogenetic tree for each data set (COI, 16S rRNA, and concatenated data), implemented in the PTP web server (Zhang et al., 2013) using 10 000 000 generations for the Markov chains with a thinning of 100 and a burn-in of 0.1. Convergence was checked using the log likelihood for the Markov chain Monte Carlo (MCMC) iterations after thinning in the same web server used for PTP. To explore the performance of GMYC, ultrametric trees were generated in BEAST 2.0 using the alignment for each data set and defining a relaxed lognormal clock, with a single partition, under a GTR + G evolutionary model, with the clock.rate parameter set to 1, and selecting a constant population size coalescent tree prior. The GMYC method was run with the packages APE 2.2-2 (Paradis, Claude & Strimmer, 2004) and SPLITS (Pons et al., 2006; Monaghan et al., 2009) using R 2.8.0 (R Development Core Team, 2008). Accounting for uncertainty in species delimitation and model-averaged estimates of diversity within individual samples were conducted following (Fernández & Giribet 2014).
Genetic distances between and within clades were calculated using Kimura's two-parameter (K2P) model (Kimura, 1980), including transitions and transversions, with uniform rates among sites, a pairwise deletion of gaps, and 500 bootstrap replicates to estimate the variance on the COI and 16S rRNA sequences using MEGA 5.2.2 (Tamura et al., 2011). In addition, we calculated the barcoding gap in the COI data set (Meyer & Paulay, 2005) within the clades defined by PTP and GMYC analyses using the furthest intraspecific distance among its own species and the closest, non-conspecific (interspecific) distance, calculated with the K2P model using the R package SPIDER (Brown et al., 2012).
Results
Phylogenetic reconstruction
Final alignments were composed of partial sequences of the nuclear genes 28S rRNA (371 bp) and 18S rRNA (1398 bp), and the mitochondrial 16S rRNA (430 bp) and COI (623 bp) from 101 specimens. Both ML and BI analyses of the four concatenated genes agreed on the paraphyly of Trypanosyllis, as Eurysyllis, Plakosyllis, and Xenosyllis appeared nested within Trypanosylliss.l. (Figs 1, 2). There were also differences between the topologies recovered in both analyses. In particular, two different well-supported clades (A and B) of species previously considered to belong to Trypanosylliss.l. appeared more related to Eurysyllis, Plakosyllis, and Xenosyllis than to other Trypanosyllis species in BI analyses, whereas only clade B was closely related to them in the ML analysis. Clade A was composed of Trypanosyllis coeliaca, whereas Trypanosyllis gemmipara and Trypanosyllis gigantea belong to clade B (Figs 1, 2). As a consequence, these two clades were named by resurrecting previously synonymized genera and re-delimiting Trypanosylliss.s. (bootstrap support, BS = 100%; posterior probability, PP = 1.00), which is here re-diagnosed (see morphological examination Section). Clade A is therefore referred to here as Pseudosyllis Grube, 1863 and clade B as TrypanedentaImajima & Hartman, 1964 (see below).
Maximum clade credibility tree from concatenated, partitioned Bayesian analysis of all genetic data (28S rRNA, 18S rRNA, 16S rRNA, and COI) showing the phylogenetic relationships between Eurysyllis, Xenosyllis, Plakosyllis, and Trypanosyllis s.l. Numbers over branches indicate posterior probability (PP) support values (only PP > 0.90 are indicated); red circles indicate a PP value of 1.00. The 23 specimens of Trypanosyllis luzonensiscomb. nov. are abbreviated as Tluzo1–Tluzo23. The 46 specimens of Trypanosyllis krohnii s.l. are abbreviated as Tk1–Tk46.
Phylogenetic relationships of Eurysyllis, Xenosyllis, Plakosyllis, and Trypanosyllis s.l. inferred from the maximum-likelihood analysis of the four concatenated genetic markers (18S rRNA, 28S rRNA, 16S rRNA, and COI). Numbers above branches indicate bootstrap support (BS) values (only BS > 50% are indicated); red circles indicate a BS value of 100%. The 23 specimens of Trypanosyllis luzonensiscomb. nov. are abbreviated as Tluzo1–Tluzo23. The 46 specimens of Trypanosyllis krohnii s.l. are abbreviated as Tk1–Tk46.
Major differences among topologies reconstructed by ML and BI were related to the relationships between Pseudosyllis, Trypanedenta, and Trypanosyllis (Figs 1, 2). The topology of the maximum clade credibility tree generated by the BI analysis showed two major, well-supported groups (Fig. 1): one clade including Eurysyllis, Plakosyllis, Xenosyllis, Pseudosyllis, and Trypanedenta, and the second major clade only containing the genus Trypanosylliss.s. In contrast, the ML results showed Pseudosyllis as the sister group to the other species in the in-group, which comprised a well-supported clade including the genera Eurysyllis, Plakosyllis, and Xenosyllis, and a clade including Trypanedenta as sister group to Trypanosyllis, although the relationships among them received low support values (Fig. 2).
Resolution within the newly delineated Trypanosyllis includes two main clades that are well supported both in the BI and ML topologies (Figs 1, 2). These two clades are Trypanosyllis luzonensis comb. nov. (previously considered to be within the striped complex) and the large clade containing the Trypanosyllis krohnii complex. The relationship between Trypanosyllis aeolisLangerhans, 1879 and the remaining Trypanosyllis differed between the BI and ML trees. An unidentified Trypanosyllis, labelled as Trypanosyllis sp. 1, was more closely related to Trypanosyllis luzonensis comb. nov. in both analyses. Within Trypanosyllis luzonensis comb. nov. we found several nodes that were strongly supported. With respect to the Trypanosyllis krohnii species complex, ML and BI topologies recovered slightly different internal topologies. We found 12 main geographical clades with PP > 0.95 within this species complex, namely: (a) a New Zealand clade; (b) an Australian clade (see Discussion section); (c, d) two clades from San Diego, California; (e) a clade containing specimens from the Philippines plus one specimen from Western Australia (see Discussion section); (f) a clade with individuals from Northern Brazil and Australia; (g) a Chilean (Las Cruces) clade; (h) a clade restricted to the Gulf of Lion (North-Western Mediterranean); (i) one clade appearing in the Gulf of Lion and the north-western coast of Spain; (j) a clade from Crete; and (k, l) two Pacific clades containing individuals from Hawaii and the Philippines (Figs 1, 2). The remaining specimens from Southern Brazil and Australia (Tk12, Tk44–Tk46) were not placed in any of the former clades (Figs 1, 2). The clade that appears restricted to the Gulf of Lion (Figs 1, 2) included specimens from the type locality of Trypanosyllis krohnii (Port–Vendres, France; Claparède, 1864), the type species of Trypanosyllis, and they agreed morphologically and ecologically with the original description of the species.
Species delimitation in the Trypanosyllis krohnii species complex
The PTP and GMYC analyses of the mitochondrial marker COI for 38 specimens of the Trypanosyllis krohnii species complex identified between seven and nine lineages (Fig. 3A), although only seven of them had a PP > 0.95 (Fig. 3A): one lineage from the Philippines (lineage 1); two lineages from San Diego, California, USA (lineages 2 and 3); one lineage from Chile (lineage 4); one lineage from New Zealand (lineage 5); one lineage from the Gulf of Lion (lineage 6); and one lineage from the Gulf of Lion and the north-western coast of Spain (lineage 7). The PTP and GMYC analyses of the 16S rRNA for 29 specimens of the complex identified between six and eight lineages (Fig. 3B), all with PP > 0.95 (Fig. 3B). Five lineages corresponded to the same lineages detected in the COI analyses (Fig. 3A, B: lineages 1, 3, and 5–7), and three more lineages that included specimens from the Philippines (lineage 8), Hawaii and Philippines (lineage 9), and Australia and Brazil (lineage 10). Lineage 2 from the COI analyses (Fig. 3A) was not detected in the 16S rRNA analyses as none of the specimens from the lineage were sequenced for 16S rRNA. Similarly, lineage 4 from the COI analyses (Fig. 3A) was not detected, as only one specimen was sequenced for 16S rRNA. We detected some incongruence between the PTP- and the GMYC-supported lineages, and also among the variants within the GMYC analyses for both mitochondrial genes (Fig. 3A, B). The S variant of GMYC used for the COI marker recognized nine lineages, although two of them are unsupported, with PP > 0.95 (Fig. 3A); however, the M variant of GMYC for the same gene presented seven well-supported lineages, in agreement with the PTP analysis (Fig. 3A). The S variant of GMYC for the 16S rRNA found eight well-supported lineages, whereas the M variant and the PTP results obtained seven and six well-supported lineages, respectively (Fig. 3B). In addition, there were also differences between the M variant of GMYC and the PTP analysis of the COI with respect to the number of specimens contained in some of the lineages. In the M variant of GMYC, lineage 1 contained only four specimens (Tk13, Tk16, Tk19, and Tk21), whereas PTP also included Tk14 (Fig. 3A). Likewise, in lineage 5 the GMYC analysis grouped three specimens (Tk1, Tk4, and Tk5), but only two were grouped by PTP (Tk4 and Tk5) (see Discussion section and Fig. 3A). With regards to the 16S rRNA analyses, all the lineages contained the same specimens for all of the analyses (Fig. 3B). Finally, the PTP analysis of the concatenated nuclear and mitochondrial data set for the 46 specimens identified ten lineages within the complex (Fig. 4). These ten lineages corresponded to ten of the 12 clades outlined by the phylogenetic analyses (Figs 1, 2). Our morphological examination of all of the specimens (see the remarks below) could only reconcile six of the lineages found by all of the species delimitation analyses as species, lineages 1–5 and lineage 7 (Figs 3, 4). Therefore, we only considered these six lineages as valid species (Fig. 4), although further molecular studies should be conducted in order to test the real diversity of the complex. One of these represents the type species of Trypanosyllis, Trypanosyllis krohnii, which is redescribed herein, and the other five are described as new species (see taxonomic results). The other four lineages identified within the complex (lineages 6 and 8–10) did not present any distinctive morphological features to be taxonomically named.
Pseudocryptic speciation within Trypanosyllis khronii s.l. obtained from mitochondrial genes. A, Bayesian COI ultrametric gene tree with posterior probabilities (>0.95) on each node. Vertical thick lines indicate the putative species delineated with GMYC and PTP methods. GMYC results include the single-threshold (S) and multiple-threshold (M) variants of the method. Grey thick lines indicate discordant results relative to the specimens included in each lineage by the different methods. B, Bayesian 16S rRNA ultrametric gene tree with posterior probabilities (>0.95) on each node. Vertical thick lines indicate the putative species delineated with GMYC and PTP methods for the mitochondrial gene. GMYC results include the single-threshold (S) and multiple-threshold (M) variants of the method. Note that red branches indicate the well-supported (>0.90) lineages found in the species delimitation analyses of Figure 3A, B, C, graphic representation of the barcoding gap existing between the infraspecific (red) and interspecific (green) genetic distances in the seven lineages obtained from the species delimitation analyses for COI (Fig. 3A) within the Trypanosyllis krohnii species complex.
Pseudocryptic speciation within Trypanosyllis s.l. obtained from the concatenated data set of nuclear and mitochondrial genes (28S rRNA, 18S rRNA, 16S rRNA, and COI). A, Bayesian gene tree with posterior probabilities (>0.90) on each node. Red branches indicate the well-supported (>0.90) lineages found in the species delimitation analyses. B, world map showing the location of the new pseudocryptic species (microscope pictures of ethanol-fixed material) recovered after species delimitation analysis. Neotype of Trypanosyllis krohnii (MNCN/ADN 9623), holotypes of Trypanosyllis taboadaisp. nov. (MNCN 16.01/16087), Trypanosyllis luqueisp. nov. (SIO A5005), Trypanosyllis leivaisp. nov. (MNCN 16.01/16082), Trypanosyllis californiensissp. nov. (SIO A5008), and Trypanosyllis kalkinsp. nov. (MNCN 16.01/16058), are shown.
The K2P genetic distances for the COI data set of all of the lineages identified by the species delimitation analyses were similar among sister species of Trypanosyllis s.s., ranging from 17.2 ± 1.8% to 27.5 ± 2.7% (lower left side of Table 2). In addition, a barcode gap was detected among the interspecific and intraspecific distances of the seven widely supported lineages of the Trypanosyllis krohnii species complex (Fig. 3C) found in the all-species delimitation analyses of COI (Fig. 3A). Species living in close proximity (lineages 2 and 3 from San Diego and lineages 6 and 7 from the Gulf of Lion) showed lower genetic distances for COI than the remaining species, however, ranging from 13.9 ± 1.7% to 14.9 ± 1.8% (lower left side of Table 2). The lowest genetic distance for COI appeared between lineages 8 and 9 (10.5 ± 1.6%), which also shared their distribution area (the Philippines) except for specimen Tk39 collected in Hawaii (Table 2). The K2P genetic distances of the 16S rRNA were also similar between the species within the complex, ranging from 15.7 ± 2.4% to 34.3 ± 4.1% (upper right side of Table 2). In this case, the lowest distances (Table 2) appeared again among lineages 8 and 9 (8.0 ± 1.5) and lineages 6 and 7 (7.2 ± 1.4). Although lineages 6 and 7 showed overlapping distributions, both were clearly distinguishable not only by morphological features (see Section 3.3), but also by ecological factors such as bathymetric range and substrate (Tables 1 and 3). Trypanosyllis krohnii (lineage 7) appeared on mussels from the infralittoral zone (0–16 m depth), whereas lineage 6 inhabits gorgonian species in deeper waters of the circalittoral zone, down to 36–40 m (see collection data below; Tables 1 and 3). In the case of the sympatric lineages from San Diego, the differences found between lineage 2 (Trypanosyllis luquei sp. nov.) and lineage 3 (Trypanosyllis californiensis sp. nov.) were both morphological (see Section 3.3) and ecological, including substrate type (Table 3). Whereas Trypanosyllis californiensis sp. nov. was collected from algae, Trypanosyllis luquei sp. nov. was only observed on bryozoans (see collection data below and Tables 1 and 3).
Upper right, interspecific uncorrected Kimura two-parameter (K2P) distances within Trypanosyllis species for the mitochondrial gene 16S rRNA; lower left, interspecific uncorrected K2P distances within Trypanosyllis species for the mitochondrial gene COI
| . | Trypanosyllis sp. 1 . | Trypanosyllis luzonensis . | Trypanosyllis aeolis . | Trypanosyllis leivai PHI (lineage 1) . | Trypanosyllis luquei SD, CA (lineage 2) . | Trypanosyllis californiensis LJ, CA (lineage 3) . | Trypanosyllis kalkin CL (lineage 4) . | Trypanosyllis taboadai NZ (lineage 5) . | Trypanosyllis krohnii PI-F (lineage 7) . | Lineage 6 (Trypanosyllis sp. 2) . | Lineage 8 (Trypanosyllis cf. krohnii) . | Lineage 9 (Trypanosyllis cf. krohnii) . | Lineage 10 (Trypanosyllis cf. krohnii) . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Trypanosyllis sp. 1 | n/a//n/a | 38.5 ± 4.9 | 34.6 ± 4.0 | 28.1 ± 3.9 | – | 31.8 ± 4.3 | 29.6 ± 3.9 | 38.5 ± 5.0 | 34.3 ± 4.5 | 30.6 ± 4.0 | 30.9 ± 3.9 | 30.6 ± 4.1 | 34.7 ± 4.2 |
| Trypanosyllis luzonensis | 24.2 ± 2.1 | 18.1 ± 1.6//10.1 ± 1.1 | 34.3 ± 4.1 | 25.1 ± 3.1 | – | 24.5 ± 3.1 | 28.6 ± 3.4 | 31.6 ± 3.8 | 30.4 ± 3.7 | 31.3 ± 3.7 | 29.2 ± 3.4 | 32.4 ± 3.7 | 27.2 ± 3.2 |
| Trypanosyllis aeolis | 24.9 ± 2.6 | 23. 5 ± 2.2 | n/a//n/a | 25.5 ± 3.3 | – | 26.3 ± 3.4 | 23.6 ± 3.2 | 30.1 ± 3.9 | 25.1 ± 3.4 | 24.9 ± 3.3 | 29.7 ± 3.9 | 31.9 ± 4.2 | 27.4 ± 3.5 |
| Trypanosyllis leivai PHI (lineage 1) | 26.6 ± 2.8 | 25.6 ± 2.3 | 25.0 ± 2.7 | 0.2 ± 0.1//0.3 ± 0.2 | – | 12.0 ± 1.9 | 13.7 ± 2.2 | 16.7 ± 2.3 | 20.7 ± 2.9 | 19.2 ± 2.8 | 15.7 ± 2.4 | 15.7 ± 2.4 | 15.8 ± 2.2 |
| Trypanosyllis luquei SD, CA (lineage 2) | 24.4 ± 2.6 | 23.8 ± 2.2 | 25.3 ± 2.8 | 18 ± 2 | 0.1 ± 0.1//− | – | – | – | – | – | – | – | – |
| Trypanosyllis californiensis LJ, CA (lineage 3) | 25.3 ± 2.5 | 25 ± 2.3 | 25 ± 2.6 | 18.5 ± 2.1 | 13.9 ± 1.7 | 0//0.3 ± 0.2 | 14.4 ± 2.1 | 17.2 ± 2.5 | 18.0 ± 2.6 | 18.0 ± 2.6 | 16.6 ± 2.5 | 15.9 ± 2.5 | 17.0 ± 2.3 |
| Trypanosyllis kalkin CL (lineage 4) | 25.9 ± 2.6 | 23.9 ± 2.1 | 22.7 ± 2.4 | 23.8 ± 2.3 | 17.2 ± 1.8 | 19.9 ± 2 | 0.1 ± 0.1//n/a | 17.9 ± 2.5 | 16.9 ± 2.5 | 15.7 ± 2.4 | 17.6 ± 2.5 | 17.3 ± 2.5 | 14.5 ± 2.2 |
| Trypanosyllis taboadai NZ (lineage 5) | 27.5 ± 2.7 | 24.5 ± 2.3 | 22.4 ± 2.4 | 23.5 ± 2.3 | 23.2 ± 2.2 | 23.8 ± 2.2 | 23 ± 2.2 | 0//3.9 ± 0.7 | 16.1 ± 2.4 | 16.1 ± 2.4 | 21.0 ± 2.8 | 20.7 ± 2.8 | 18.6 ± 2.6 |
| Trypanosyllis krohnii PI-F (lineage 7) | 25.5 ± 2.6 | 23.4 ± 2.1 | 23.1 ± 2.6 | 21.6 ± 2.4 | 20.3 ± 2.1 | 20.9 ± 2.1 | 21.5 ± 2.2 | 19.2 ± 2 | 0.1 ± 0.1//0.5 ± 0.3 | 7.2 ± 1.4 | 20.0 ± 2.8 | 23.2 ± 3.1 | 17.3 ± 2.5 |
| Lineage 6 (Trypanosyllis sp. 2) | 24. 9 ± 2.6 | 23.8 ± 2.2 | 21.7 ± 2.4 | 23.3 ± 2.4 | 24.3 ± 2.4 | 24.5 ± 1.7 | 27.4 ± 2.5 | 20.5 ± 2 | 14.9 ± 1.8 | 1 ± 0.4//0.5 ± 0.3 | 20.1 ± 2.7 | 23.2 ± 3.1 | 18.9 ± 2.6 |
| Lineage 8 (Trypanosyllis cf. krohnii) | 30.8 ± 3.7 | 29.3 ± 3.1 | 28.9 ± 3.7 | 30.1 ± 3.6 | 23.7 ± 3.0 | 26.3 ± 3.3 | 28.3 ± 3.5 | 28.0 ± 3.5 | 25.6 ± 3.1 | 27.2 ± 3.3 | 0.1 ± 0.1//1.2 ± 0.5 | 8.0 ± 1.5 | 20.2 ± 2.8 |
| Lineage 9 (Trypanosyllis cf. krohnii) | 29.9 ± 3.7 | 26.5 ± 2.8 | 26.8 ± 3.4 | 29.9 ± 3.9 | 21.0 ± 2.8 | 24.7 ± 3.1 | 26.3 ± 3.1 | 24.6 ± 3.1 | 23.3 ± 3.0 | 26.1 ± 3.2 | 10.5 ± 1.6 | 0.1 ± 0.1//0 ± 0 | 20.3 ± 2.7 |
| Lineage 10 (Trypanosyllis cf. krohnii) | 30.3 ± 3.3 | 27.3 ± 2.7 | 27.5 ± 3.2 | 27.0 ± 3.0 | 24.6 ± 2.8 | 25.1 ± 3.3 | 26.0 ± 3.0 | 25.6 ± 2.8 | 24.5 ± 2.9 | 25.7 ± 2.9 | 18.1 ± 2.0 | 20.2 ± 2.3 | 0.1 ± 0.1//2.5 ± 0.7 |
| . | Trypanosyllis sp. 1 . | Trypanosyllis luzonensis . | Trypanosyllis aeolis . | Trypanosyllis leivai PHI (lineage 1) . | Trypanosyllis luquei SD, CA (lineage 2) . | Trypanosyllis californiensis LJ, CA (lineage 3) . | Trypanosyllis kalkin CL (lineage 4) . | Trypanosyllis taboadai NZ (lineage 5) . | Trypanosyllis krohnii PI-F (lineage 7) . | Lineage 6 (Trypanosyllis sp. 2) . | Lineage 8 (Trypanosyllis cf. krohnii) . | Lineage 9 (Trypanosyllis cf. krohnii) . | Lineage 10 (Trypanosyllis cf. krohnii) . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Trypanosyllis sp. 1 | n/a//n/a | 38.5 ± 4.9 | 34.6 ± 4.0 | 28.1 ± 3.9 | – | 31.8 ± 4.3 | 29.6 ± 3.9 | 38.5 ± 5.0 | 34.3 ± 4.5 | 30.6 ± 4.0 | 30.9 ± 3.9 | 30.6 ± 4.1 | 34.7 ± 4.2 |
| Trypanosyllis luzonensis | 24.2 ± 2.1 | 18.1 ± 1.6//10.1 ± 1.1 | 34.3 ± 4.1 | 25.1 ± 3.1 | – | 24.5 ± 3.1 | 28.6 ± 3.4 | 31.6 ± 3.8 | 30.4 ± 3.7 | 31.3 ± 3.7 | 29.2 ± 3.4 | 32.4 ± 3.7 | 27.2 ± 3.2 |
| Trypanosyllis aeolis | 24.9 ± 2.6 | 23. 5 ± 2.2 | n/a//n/a | 25.5 ± 3.3 | – | 26.3 ± 3.4 | 23.6 ± 3.2 | 30.1 ± 3.9 | 25.1 ± 3.4 | 24.9 ± 3.3 | 29.7 ± 3.9 | 31.9 ± 4.2 | 27.4 ± 3.5 |
| Trypanosyllis leivai PHI (lineage 1) | 26.6 ± 2.8 | 25.6 ± 2.3 | 25.0 ± 2.7 | 0.2 ± 0.1//0.3 ± 0.2 | – | 12.0 ± 1.9 | 13.7 ± 2.2 | 16.7 ± 2.3 | 20.7 ± 2.9 | 19.2 ± 2.8 | 15.7 ± 2.4 | 15.7 ± 2.4 | 15.8 ± 2.2 |
| Trypanosyllis luquei SD, CA (lineage 2) | 24.4 ± 2.6 | 23.8 ± 2.2 | 25.3 ± 2.8 | 18 ± 2 | 0.1 ± 0.1//− | – | – | – | – | – | – | – | – |
| Trypanosyllis californiensis LJ, CA (lineage 3) | 25.3 ± 2.5 | 25 ± 2.3 | 25 ± 2.6 | 18.5 ± 2.1 | 13.9 ± 1.7 | 0//0.3 ± 0.2 | 14.4 ± 2.1 | 17.2 ± 2.5 | 18.0 ± 2.6 | 18.0 ± 2.6 | 16.6 ± 2.5 | 15.9 ± 2.5 | 17.0 ± 2.3 |
| Trypanosyllis kalkin CL (lineage 4) | 25.9 ± 2.6 | 23.9 ± 2.1 | 22.7 ± 2.4 | 23.8 ± 2.3 | 17.2 ± 1.8 | 19.9 ± 2 | 0.1 ± 0.1//n/a | 17.9 ± 2.5 | 16.9 ± 2.5 | 15.7 ± 2.4 | 17.6 ± 2.5 | 17.3 ± 2.5 | 14.5 ± 2.2 |
| Trypanosyllis taboadai NZ (lineage 5) | 27.5 ± 2.7 | 24.5 ± 2.3 | 22.4 ± 2.4 | 23.5 ± 2.3 | 23.2 ± 2.2 | 23.8 ± 2.2 | 23 ± 2.2 | 0//3.9 ± 0.7 | 16.1 ± 2.4 | 16.1 ± 2.4 | 21.0 ± 2.8 | 20.7 ± 2.8 | 18.6 ± 2.6 |
| Trypanosyllis krohnii PI-F (lineage 7) | 25.5 ± 2.6 | 23.4 ± 2.1 | 23.1 ± 2.6 | 21.6 ± 2.4 | 20.3 ± 2.1 | 20.9 ± 2.1 | 21.5 ± 2.2 | 19.2 ± 2 | 0.1 ± 0.1//0.5 ± 0.3 | 7.2 ± 1.4 | 20.0 ± 2.8 | 23.2 ± 3.1 | 17.3 ± 2.5 |
| Lineage 6 (Trypanosyllis sp. 2) | 24. 9 ± 2.6 | 23.8 ± 2.2 | 21.7 ± 2.4 | 23.3 ± 2.4 | 24.3 ± 2.4 | 24.5 ± 1.7 | 27.4 ± 2.5 | 20.5 ± 2 | 14.9 ± 1.8 | 1 ± 0.4//0.5 ± 0.3 | 20.1 ± 2.7 | 23.2 ± 3.1 | 18.9 ± 2.6 |
| Lineage 8 (Trypanosyllis cf. krohnii) | 30.8 ± 3.7 | 29.3 ± 3.1 | 28.9 ± 3.7 | 30.1 ± 3.6 | 23.7 ± 3.0 | 26.3 ± 3.3 | 28.3 ± 3.5 | 28.0 ± 3.5 | 25.6 ± 3.1 | 27.2 ± 3.3 | 0.1 ± 0.1//1.2 ± 0.5 | 8.0 ± 1.5 | 20.2 ± 2.8 |
| Lineage 9 (Trypanosyllis cf. krohnii) | 29.9 ± 3.7 | 26.5 ± 2.8 | 26.8 ± 3.4 | 29.9 ± 3.9 | 21.0 ± 2.8 | 24.7 ± 3.1 | 26.3 ± 3.1 | 24.6 ± 3.1 | 23.3 ± 3.0 | 26.1 ± 3.2 | 10.5 ± 1.6 | 0.1 ± 0.1//0 ± 0 | 20.3 ± 2.7 |
| Lineage 10 (Trypanosyllis cf. krohnii) | 30.3 ± 3.3 | 27.3 ± 2.7 | 27.5 ± 3.2 | 27.0 ± 3.0 | 24.6 ± 2.8 | 25.1 ± 3.3 | 26.0 ± 3.0 | 25.6 ± 2.8 | 24.5 ± 2.9 | 25.7 ± 2.9 | 18.1 ± 2.0 | 20.2 ± 2.3 | 0.1 ± 0.1//2.5 ± 0.7 |
Intraspecific distances in bold (left for COI and right for 16S rRNA).
Upper right, interspecific uncorrected Kimura two-parameter (K2P) distances within Trypanosyllis species for the mitochondrial gene 16S rRNA; lower left, interspecific uncorrected K2P distances within Trypanosyllis species for the mitochondrial gene COI
| . | Trypanosyllis sp. 1 . | Trypanosyllis luzonensis . | Trypanosyllis aeolis . | Trypanosyllis leivai PHI (lineage 1) . | Trypanosyllis luquei SD, CA (lineage 2) . | Trypanosyllis californiensis LJ, CA (lineage 3) . | Trypanosyllis kalkin CL (lineage 4) . | Trypanosyllis taboadai NZ (lineage 5) . | Trypanosyllis krohnii PI-F (lineage 7) . | Lineage 6 (Trypanosyllis sp. 2) . | Lineage 8 (Trypanosyllis cf. krohnii) . | Lineage 9 (Trypanosyllis cf. krohnii) . | Lineage 10 (Trypanosyllis cf. krohnii) . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Trypanosyllis sp. 1 | n/a//n/a | 38.5 ± 4.9 | 34.6 ± 4.0 | 28.1 ± 3.9 | – | 31.8 ± 4.3 | 29.6 ± 3.9 | 38.5 ± 5.0 | 34.3 ± 4.5 | 30.6 ± 4.0 | 30.9 ± 3.9 | 30.6 ± 4.1 | 34.7 ± 4.2 |
| Trypanosyllis luzonensis | 24.2 ± 2.1 | 18.1 ± 1.6//10.1 ± 1.1 | 34.3 ± 4.1 | 25.1 ± 3.1 | – | 24.5 ± 3.1 | 28.6 ± 3.4 | 31.6 ± 3.8 | 30.4 ± 3.7 | 31.3 ± 3.7 | 29.2 ± 3.4 | 32.4 ± 3.7 | 27.2 ± 3.2 |
| Trypanosyllis aeolis | 24.9 ± 2.6 | 23. 5 ± 2.2 | n/a//n/a | 25.5 ± 3.3 | – | 26.3 ± 3.4 | 23.6 ± 3.2 | 30.1 ± 3.9 | 25.1 ± 3.4 | 24.9 ± 3.3 | 29.7 ± 3.9 | 31.9 ± 4.2 | 27.4 ± 3.5 |
| Trypanosyllis leivai PHI (lineage 1) | 26.6 ± 2.8 | 25.6 ± 2.3 | 25.0 ± 2.7 | 0.2 ± 0.1//0.3 ± 0.2 | – | 12.0 ± 1.9 | 13.7 ± 2.2 | 16.7 ± 2.3 | 20.7 ± 2.9 | 19.2 ± 2.8 | 15.7 ± 2.4 | 15.7 ± 2.4 | 15.8 ± 2.2 |
| Trypanosyllis luquei SD, CA (lineage 2) | 24.4 ± 2.6 | 23.8 ± 2.2 | 25.3 ± 2.8 | 18 ± 2 | 0.1 ± 0.1//− | – | – | – | – | – | – | – | – |
| Trypanosyllis californiensis LJ, CA (lineage 3) | 25.3 ± 2.5 | 25 ± 2.3 | 25 ± 2.6 | 18.5 ± 2.1 | 13.9 ± 1.7 | 0//0.3 ± 0.2 | 14.4 ± 2.1 | 17.2 ± 2.5 | 18.0 ± 2.6 | 18.0 ± 2.6 | 16.6 ± 2.5 | 15.9 ± 2.5 | 17.0 ± 2.3 |
| Trypanosyllis kalkin CL (lineage 4) | 25.9 ± 2.6 | 23.9 ± 2.1 | 22.7 ± 2.4 | 23.8 ± 2.3 | 17.2 ± 1.8 | 19.9 ± 2 | 0.1 ± 0.1//n/a | 17.9 ± 2.5 | 16.9 ± 2.5 | 15.7 ± 2.4 | 17.6 ± 2.5 | 17.3 ± 2.5 | 14.5 ± 2.2 |
| Trypanosyllis taboadai NZ (lineage 5) | 27.5 ± 2.7 | 24.5 ± 2.3 | 22.4 ± 2.4 | 23.5 ± 2.3 | 23.2 ± 2.2 | 23.8 ± 2.2 | 23 ± 2.2 | 0//3.9 ± 0.7 | 16.1 ± 2.4 | 16.1 ± 2.4 | 21.0 ± 2.8 | 20.7 ± 2.8 | 18.6 ± 2.6 |
| Trypanosyllis krohnii PI-F (lineage 7) | 25.5 ± 2.6 | 23.4 ± 2.1 | 23.1 ± 2.6 | 21.6 ± 2.4 | 20.3 ± 2.1 | 20.9 ± 2.1 | 21.5 ± 2.2 | 19.2 ± 2 | 0.1 ± 0.1//0.5 ± 0.3 | 7.2 ± 1.4 | 20.0 ± 2.8 | 23.2 ± 3.1 | 17.3 ± 2.5 |
| Lineage 6 (Trypanosyllis sp. 2) | 24. 9 ± 2.6 | 23.8 ± 2.2 | 21.7 ± 2.4 | 23.3 ± 2.4 | 24.3 ± 2.4 | 24.5 ± 1.7 | 27.4 ± 2.5 | 20.5 ± 2 | 14.9 ± 1.8 | 1 ± 0.4//0.5 ± 0.3 | 20.1 ± 2.7 | 23.2 ± 3.1 | 18.9 ± 2.6 |
| Lineage 8 (Trypanosyllis cf. krohnii) | 30.8 ± 3.7 | 29.3 ± 3.1 | 28.9 ± 3.7 | 30.1 ± 3.6 | 23.7 ± 3.0 | 26.3 ± 3.3 | 28.3 ± 3.5 | 28.0 ± 3.5 | 25.6 ± 3.1 | 27.2 ± 3.3 | 0.1 ± 0.1//1.2 ± 0.5 | 8.0 ± 1.5 | 20.2 ± 2.8 |
| Lineage 9 (Trypanosyllis cf. krohnii) | 29.9 ± 3.7 | 26.5 ± 2.8 | 26.8 ± 3.4 | 29.9 ± 3.9 | 21.0 ± 2.8 | 24.7 ± 3.1 | 26.3 ± 3.1 | 24.6 ± 3.1 | 23.3 ± 3.0 | 26.1 ± 3.2 | 10.5 ± 1.6 | 0.1 ± 0.1//0 ± 0 | 20.3 ± 2.7 |
| Lineage 10 (Trypanosyllis cf. krohnii) | 30.3 ± 3.3 | 27.3 ± 2.7 | 27.5 ± 3.2 | 27.0 ± 3.0 | 24.6 ± 2.8 | 25.1 ± 3.3 | 26.0 ± 3.0 | 25.6 ± 2.8 | 24.5 ± 2.9 | 25.7 ± 2.9 | 18.1 ± 2.0 | 20.2 ± 2.3 | 0.1 ± 0.1//2.5 ± 0.7 |
| . | Trypanosyllis sp. 1 . | Trypanosyllis luzonensis . | Trypanosyllis aeolis . | Trypanosyllis leivai PHI (lineage 1) . | Trypanosyllis luquei SD, CA (lineage 2) . | Trypanosyllis californiensis LJ, CA (lineage 3) . | Trypanosyllis kalkin CL (lineage 4) . | Trypanosyllis taboadai NZ (lineage 5) . | Trypanosyllis krohnii PI-F (lineage 7) . | Lineage 6 (Trypanosyllis sp. 2) . | Lineage 8 (Trypanosyllis cf. krohnii) . | Lineage 9 (Trypanosyllis cf. krohnii) . | Lineage 10 (Trypanosyllis cf. krohnii) . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Trypanosyllis sp. 1 | n/a//n/a | 38.5 ± 4.9 | 34.6 ± 4.0 | 28.1 ± 3.9 | – | 31.8 ± 4.3 | 29.6 ± 3.9 | 38.5 ± 5.0 | 34.3 ± 4.5 | 30.6 ± 4.0 | 30.9 ± 3.9 | 30.6 ± 4.1 | 34.7 ± 4.2 |
| Trypanosyllis luzonensis | 24.2 ± 2.1 | 18.1 ± 1.6//10.1 ± 1.1 | 34.3 ± 4.1 | 25.1 ± 3.1 | – | 24.5 ± 3.1 | 28.6 ± 3.4 | 31.6 ± 3.8 | 30.4 ± 3.7 | 31.3 ± 3.7 | 29.2 ± 3.4 | 32.4 ± 3.7 | 27.2 ± 3.2 |
| Trypanosyllis aeolis | 24.9 ± 2.6 | 23. 5 ± 2.2 | n/a//n/a | 25.5 ± 3.3 | – | 26.3 ± 3.4 | 23.6 ± 3.2 | 30.1 ± 3.9 | 25.1 ± 3.4 | 24.9 ± 3.3 | 29.7 ± 3.9 | 31.9 ± 4.2 | 27.4 ± 3.5 |
| Trypanosyllis leivai PHI (lineage 1) | 26.6 ± 2.8 | 25.6 ± 2.3 | 25.0 ± 2.7 | 0.2 ± 0.1//0.3 ± 0.2 | – | 12.0 ± 1.9 | 13.7 ± 2.2 | 16.7 ± 2.3 | 20.7 ± 2.9 | 19.2 ± 2.8 | 15.7 ± 2.4 | 15.7 ± 2.4 | 15.8 ± 2.2 |
| Trypanosyllis luquei SD, CA (lineage 2) | 24.4 ± 2.6 | 23.8 ± 2.2 | 25.3 ± 2.8 | 18 ± 2 | 0.1 ± 0.1//− | – | – | – | – | – | – | – | – |
| Trypanosyllis californiensis LJ, CA (lineage 3) | 25.3 ± 2.5 | 25 ± 2.3 | 25 ± 2.6 | 18.5 ± 2.1 | 13.9 ± 1.7 | 0//0.3 ± 0.2 | 14.4 ± 2.1 | 17.2 ± 2.5 | 18.0 ± 2.6 | 18.0 ± 2.6 | 16.6 ± 2.5 | 15.9 ± 2.5 | 17.0 ± 2.3 |
| Trypanosyllis kalkin CL (lineage 4) | 25.9 ± 2.6 | 23.9 ± 2.1 | 22.7 ± 2.4 | 23.8 ± 2.3 | 17.2 ± 1.8 | 19.9 ± 2 | 0.1 ± 0.1//n/a | 17.9 ± 2.5 | 16.9 ± 2.5 | 15.7 ± 2.4 | 17.6 ± 2.5 | 17.3 ± 2.5 | 14.5 ± 2.2 |
| Trypanosyllis taboadai NZ (lineage 5) | 27.5 ± 2.7 | 24.5 ± 2.3 | 22.4 ± 2.4 | 23.5 ± 2.3 | 23.2 ± 2.2 | 23.8 ± 2.2 | 23 ± 2.2 | 0//3.9 ± 0.7 | 16.1 ± 2.4 | 16.1 ± 2.4 | 21.0 ± 2.8 | 20.7 ± 2.8 | 18.6 ± 2.6 |
| Trypanosyllis krohnii PI-F (lineage 7) | 25.5 ± 2.6 | 23.4 ± 2.1 | 23.1 ± 2.6 | 21.6 ± 2.4 | 20.3 ± 2.1 | 20.9 ± 2.1 | 21.5 ± 2.2 | 19.2 ± 2 | 0.1 ± 0.1//0.5 ± 0.3 | 7.2 ± 1.4 | 20.0 ± 2.8 | 23.2 ± 3.1 | 17.3 ± 2.5 |
| Lineage 6 (Trypanosyllis sp. 2) | 24. 9 ± 2.6 | 23.8 ± 2.2 | 21.7 ± 2.4 | 23.3 ± 2.4 | 24.3 ± 2.4 | 24.5 ± 1.7 | 27.4 ± 2.5 | 20.5 ± 2 | 14.9 ± 1.8 | 1 ± 0.4//0.5 ± 0.3 | 20.1 ± 2.7 | 23.2 ± 3.1 | 18.9 ± 2.6 |
| Lineage 8 (Trypanosyllis cf. krohnii) | 30.8 ± 3.7 | 29.3 ± 3.1 | 28.9 ± 3.7 | 30.1 ± 3.6 | 23.7 ± 3.0 | 26.3 ± 3.3 | 28.3 ± 3.5 | 28.0 ± 3.5 | 25.6 ± 3.1 | 27.2 ± 3.3 | 0.1 ± 0.1//1.2 ± 0.5 | 8.0 ± 1.5 | 20.2 ± 2.8 |
| Lineage 9 (Trypanosyllis cf. krohnii) | 29.9 ± 3.7 | 26.5 ± 2.8 | 26.8 ± 3.4 | 29.9 ± 3.9 | 21.0 ± 2.8 | 24.7 ± 3.1 | 26.3 ± 3.1 | 24.6 ± 3.1 | 23.3 ± 3.0 | 26.1 ± 3.2 | 10.5 ± 1.6 | 0.1 ± 0.1//0 ± 0 | 20.3 ± 2.7 |
| Lineage 10 (Trypanosyllis cf. krohnii) | 30.3 ± 3.3 | 27.3 ± 2.7 | 27.5 ± 3.2 | 27.0 ± 3.0 | 24.6 ± 2.8 | 25.1 ± 3.3 | 26.0 ± 3.0 | 25.6 ± 2.8 | 24.5 ± 2.9 | 25.7 ± 2.9 | 18.1 ± 2.0 | 20.2 ± 2.3 | 0.1 ± 0.1//2.5 ± 0.7 |
Intraspecific distances in bold (left for COI and right for 16S rRNA).
Summary of the morphological differences found between the striped Trypanosyllis, including the new species described here
| Species . | Body coloration and stripe pattern . | N articles in dorsal cirri (longest/shortest) . | Body length/width (mm) . | Substrate/depth . | Distribution . | References . |
|---|---|---|---|---|---|---|
| Trypanosyllis krohnii holotype | Two violet stripes* | 24/12 | 23/? | −/− | Port Vendres, France | Claparède, 1864; |
| Trypanosyllis krohnii neotype (lineage 7) | Two brown transverse stripes in anterior and midbody parts, in the middle and anterior end of segment. Purple dorsal cirri† | 30–32/20–22 | 8 (incomplete)/1.5 | Epifauna on mussels/intertidal | Banyuls-sur-Mer, Gulf of Lion, France | This paper |
| Trypanosyllis luzonensiscomb. nov. | Two bands almost fused into a single band. Dark-red spots on cirrophores. Red pigmentation on dorsal cirri† | 58–60/30–32 | 32/2 | Brackish water, fish pond/− | China, Philippines, Australia | Pillai, 1965; this paper |
| Trypanosyllis taboadaisp. nov. (lineage 5) | Two brown stripes delimiting segments, reaching parapodia† | 21–23/17–19 | 20/1.1 | Calcareous algae/17 m | Cavalli Islands, New Zealand | This paper |
| Trypanosyllis californiensissp. nov. (lineage 3) | Dark-brown transverse stripes across anterior segments. Fixed material red/pink with dark red–purple stripes. Stripes at each end of segment*, † | 30–32/20–22 | 12/0.8 | Kelp holdfast/5 m | La Jolla, San Diego, USA | This paper |
| Trypanosyllis leivaisp. nov. (lineage 1) | Two red–brown transverse stripes across anterior segments, one wider, not reaching end of segments† | 30–32/16–18 | 15/0.3–0.4 | Coral rubble/2 m | Balayan and Batangas Bay, Luzon Is.; El Nido, Palawan Is., Philippines | This paper |
| Trypanosyllis luqueisp. nov. (lineage 2) | Two brown transverse stripes across anterior segments, close to segmental ends. Brown appendages only coloured in the limit of each article*, † | 18–20/12–14 | 12/0.2–0.4 | Bryozoans/intertidal | San Diego Bay, California, USA | This paper |
| Trypanosyllis kalkinsp. nov. (lineage 4) | Two light-brown transverse stripes in anterior segments. Well-preserved specimens with two bands reaching eyes† | 20–22/14–16 | 4/0.3–0.4 | Dendrymenia skottsbergii / intertidal | Las Cruces, Valparaergii in anterior segment | This paper |
| Trypanosyllis sp. 2 (lineage 6) | Two red–brown transverse stripes across anterior segments, similar width and length, † anterior stripe reaching the end of parapodia | 40–42/23–24 | 6/0.7 | Paramuricea clavata/ 40 m | Cap de Creus, Girona, Catalonia (Gulf of Lion), Spain | This paper |
| Trypanosyllis richardi | Two transverse dark red–brown bands, as small dark spots arranged in parallel/transverse lines, thicker in the middle* | 40–50/− | 42/2 | −/− | Djibouti, Red Sea | Gravier, 1900 |
| Trypanosyllis parazebra | One thick brown band* | 9–11/6–7 | 6/0.7 | −/1 m | Arica, Chile | Hartmann-Schröder, 1965; |
| Trypanosyllis taeniaformis | Light red coloration, with numerous narrow transverse brown bands both in dorsum and between the joints of cirri and tentacles* | 46/− | −/− | −/shallow water | Port Jackson, Sydney, Australia | Haswell, 1886; Çinar, 2007; |
| Trypanosyllis prampramensis | Two stripes* | 8–10/6–8 | 6.6/0.7 | −/− | Greater Accra Regio, Ghana | Augener, 1918 |
| Trypanosyllis vittigera | Two brown bands* | 30/− | 13.5/2.5 | −/13 m | Key West, Florida, USA | Elhers, 1887 |
| Trypanosyllis zebra holotype | Two transverse dark-brown bands. White dorsal cirri | 20/− | 48.6/20.32‡ | −/− | Cherso, Adriatic Sea, Croatia | Grube 1860 |
| Trypanosyllis zebra (ZMB Q4428) | Two transverse brown bands. White dorsal cirri | 37–39/20–22 | 18/2 | −/− | Lessina, Adriatic Sea, Italy | This paper |
| Species . | Body coloration and stripe pattern . | N articles in dorsal cirri (longest/shortest) . | Body length/width (mm) . | Substrate/depth . | Distribution . | References . |
|---|---|---|---|---|---|---|
| Trypanosyllis krohnii holotype | Two violet stripes* | 24/12 | 23/? | −/− | Port Vendres, France | Claparède, 1864; |
| Trypanosyllis krohnii neotype (lineage 7) | Two brown transverse stripes in anterior and midbody parts, in the middle and anterior end of segment. Purple dorsal cirri† | 30–32/20–22 | 8 (incomplete)/1.5 | Epifauna on mussels/intertidal | Banyuls-sur-Mer, Gulf of Lion, France | This paper |
| Trypanosyllis luzonensiscomb. nov. | Two bands almost fused into a single band. Dark-red spots on cirrophores. Red pigmentation on dorsal cirri† | 58–60/30–32 | 32/2 | Brackish water, fish pond/− | China, Philippines, Australia | Pillai, 1965; this paper |
| Trypanosyllis taboadaisp. nov. (lineage 5) | Two brown stripes delimiting segments, reaching parapodia† | 21–23/17–19 | 20/1.1 | Calcareous algae/17 m | Cavalli Islands, New Zealand | This paper |
| Trypanosyllis californiensissp. nov. (lineage 3) | Dark-brown transverse stripes across anterior segments. Fixed material red/pink with dark red–purple stripes. Stripes at each end of segment*, † | 30–32/20–22 | 12/0.8 | Kelp holdfast/5 m | La Jolla, San Diego, USA | This paper |
| Trypanosyllis leivaisp. nov. (lineage 1) | Two red–brown transverse stripes across anterior segments, one wider, not reaching end of segments† | 30–32/16–18 | 15/0.3–0.4 | Coral rubble/2 m | Balayan and Batangas Bay, Luzon Is.; El Nido, Palawan Is., Philippines | This paper |
| Trypanosyllis luqueisp. nov. (lineage 2) | Two brown transverse stripes across anterior segments, close to segmental ends. Brown appendages only coloured in the limit of each article*, † | 18–20/12–14 | 12/0.2–0.4 | Bryozoans/intertidal | San Diego Bay, California, USA | This paper |
| Trypanosyllis kalkinsp. nov. (lineage 4) | Two light-brown transverse stripes in anterior segments. Well-preserved specimens with two bands reaching eyes† | 20–22/14–16 | 4/0.3–0.4 | Dendrymenia skottsbergii / intertidal | Las Cruces, Valparaergii in anterior segment | This paper |
| Trypanosyllis sp. 2 (lineage 6) | Two red–brown transverse stripes across anterior segments, similar width and length, † anterior stripe reaching the end of parapodia | 40–42/23–24 | 6/0.7 | Paramuricea clavata/ 40 m | Cap de Creus, Girona, Catalonia (Gulf of Lion), Spain | This paper |
| Trypanosyllis richardi | Two transverse dark red–brown bands, as small dark spots arranged in parallel/transverse lines, thicker in the middle* | 40–50/− | 42/2 | −/− | Djibouti, Red Sea | Gravier, 1900 |
| Trypanosyllis parazebra | One thick brown band* | 9–11/6–7 | 6/0.7 | −/1 m | Arica, Chile | Hartmann-Schröder, 1965; |
| Trypanosyllis taeniaformis | Light red coloration, with numerous narrow transverse brown bands both in dorsum and between the joints of cirri and tentacles* | 46/− | −/− | −/shallow water | Port Jackson, Sydney, Australia | Haswell, 1886; Çinar, 2007; |
| Trypanosyllis prampramensis | Two stripes* | 8–10/6–8 | 6.6/0.7 | −/− | Greater Accra Regio, Ghana | Augener, 1918 |
| Trypanosyllis vittigera | Two brown bands* | 30/− | 13.5/2.5 | −/13 m | Key West, Florida, USA | Elhers, 1887 |
| Trypanosyllis zebra holotype | Two transverse dark-brown bands. White dorsal cirri | 20/− | 48.6/20.32‡ | −/− | Cherso, Adriatic Sea, Croatia | Grube 1860 |
| Trypanosyllis zebra (ZMB Q4428) | Two transverse brown bands. White dorsal cirri | 37–39/20–22 | 18/2 | −/− | Lessina, Adriatic Sea, Italy | This paper |
Here ‘stripe’ refers to lines that are thinner than bands.
Coloration observed in live specimens.
Coloration observed in ethanol-fixed specimens.
Original measurements given in the description of Grube (1860), which might be mistaken given the huge numbers reported that were never seen before in any other Trypanosyllis species.
Summary of the morphological differences found between the striped Trypanosyllis, including the new species described here
| Species . | Body coloration and stripe pattern . | N articles in dorsal cirri (longest/shortest) . | Body length/width (mm) . | Substrate/depth . | Distribution . | References . |
|---|---|---|---|---|---|---|
| Trypanosyllis krohnii holotype | Two violet stripes* | 24/12 | 23/? | −/− | Port Vendres, France | Claparède, 1864; |
| Trypanosyllis krohnii neotype (lineage 7) | Two brown transverse stripes in anterior and midbody parts, in the middle and anterior end of segment. Purple dorsal cirri† | 30–32/20–22 | 8 (incomplete)/1.5 | Epifauna on mussels/intertidal | Banyuls-sur-Mer, Gulf of Lion, France | This paper |
| Trypanosyllis luzonensiscomb. nov. | Two bands almost fused into a single band. Dark-red spots on cirrophores. Red pigmentation on dorsal cirri† | 58–60/30–32 | 32/2 | Brackish water, fish pond/− | China, Philippines, Australia | Pillai, 1965; this paper |
| Trypanosyllis taboadaisp. nov. (lineage 5) | Two brown stripes delimiting segments, reaching parapodia† | 21–23/17–19 | 20/1.1 | Calcareous algae/17 m | Cavalli Islands, New Zealand | This paper |
| Trypanosyllis californiensissp. nov. (lineage 3) | Dark-brown transverse stripes across anterior segments. Fixed material red/pink with dark red–purple stripes. Stripes at each end of segment*, † | 30–32/20–22 | 12/0.8 | Kelp holdfast/5 m | La Jolla, San Diego, USA | This paper |
| Trypanosyllis leivaisp. nov. (lineage 1) | Two red–brown transverse stripes across anterior segments, one wider, not reaching end of segments† | 30–32/16–18 | 15/0.3–0.4 | Coral rubble/2 m | Balayan and Batangas Bay, Luzon Is.; El Nido, Palawan Is., Philippines | This paper |
| Trypanosyllis luqueisp. nov. (lineage 2) | Two brown transverse stripes across anterior segments, close to segmental ends. Brown appendages only coloured in the limit of each article*, † | 18–20/12–14 | 12/0.2–0.4 | Bryozoans/intertidal | San Diego Bay, California, USA | This paper |
| Trypanosyllis kalkinsp. nov. (lineage 4) | Two light-brown transverse stripes in anterior segments. Well-preserved specimens with two bands reaching eyes† | 20–22/14–16 | 4/0.3–0.4 | Dendrymenia skottsbergii / intertidal | Las Cruces, Valparaergii in anterior segment | This paper |
| Trypanosyllis sp. 2 (lineage 6) | Two red–brown transverse stripes across anterior segments, similar width and length, † anterior stripe reaching the end of parapodia | 40–42/23–24 | 6/0.7 | Paramuricea clavata/ 40 m | Cap de Creus, Girona, Catalonia (Gulf of Lion), Spain | This paper |
| Trypanosyllis richardi | Two transverse dark red–brown bands, as small dark spots arranged in parallel/transverse lines, thicker in the middle* | 40–50/− | 42/2 | −/− | Djibouti, Red Sea | Gravier, 1900 |
| Trypanosyllis parazebra | One thick brown band* | 9–11/6–7 | 6/0.7 | −/1 m | Arica, Chile | Hartmann-Schröder, 1965; |
| Trypanosyllis taeniaformis | Light red coloration, with numerous narrow transverse brown bands both in dorsum and between the joints of cirri and tentacles* | 46/− | −/− | −/shallow water | Port Jackson, Sydney, Australia | Haswell, 1886; Çinar, 2007; |
| Trypanosyllis prampramensis | Two stripes* | 8–10/6–8 | 6.6/0.7 | −/− | Greater Accra Regio, Ghana | Augener, 1918 |
| Trypanosyllis vittigera | Two brown bands* | 30/− | 13.5/2.5 | −/13 m | Key West, Florida, USA | Elhers, 1887 |
| Trypanosyllis zebra holotype | Two transverse dark-brown bands. White dorsal cirri | 20/− | 48.6/20.32‡ | −/− | Cherso, Adriatic Sea, Croatia | Grube 1860 |
| Trypanosyllis zebra (ZMB Q4428) | Two transverse brown bands. White dorsal cirri | 37–39/20–22 | 18/2 | −/− | Lessina, Adriatic Sea, Italy | This paper |
| Species . | Body coloration and stripe pattern . | N articles in dorsal cirri (longest/shortest) . | Body length/width (mm) . | Substrate/depth . | Distribution . | References . |
|---|---|---|---|---|---|---|
| Trypanosyllis krohnii holotype | Two violet stripes* | 24/12 | 23/? | −/− | Port Vendres, France | Claparède, 1864; |
| Trypanosyllis krohnii neotype (lineage 7) | Two brown transverse stripes in anterior and midbody parts, in the middle and anterior end of segment. Purple dorsal cirri† | 30–32/20–22 | 8 (incomplete)/1.5 | Epifauna on mussels/intertidal | Banyuls-sur-Mer, Gulf of Lion, France | This paper |
| Trypanosyllis luzonensiscomb. nov. | Two bands almost fused into a single band. Dark-red spots on cirrophores. Red pigmentation on dorsal cirri† | 58–60/30–32 | 32/2 | Brackish water, fish pond/− | China, Philippines, Australia | Pillai, 1965; this paper |
| Trypanosyllis taboadaisp. nov. (lineage 5) | Two brown stripes delimiting segments, reaching parapodia† | 21–23/17–19 | 20/1.1 | Calcareous algae/17 m | Cavalli Islands, New Zealand | This paper |
| Trypanosyllis californiensissp. nov. (lineage 3) | Dark-brown transverse stripes across anterior segments. Fixed material red/pink with dark red–purple stripes. Stripes at each end of segment*, † | 30–32/20–22 | 12/0.8 | Kelp holdfast/5 m | La Jolla, San Diego, USA | This paper |
| Trypanosyllis leivaisp. nov. (lineage 1) | Two red–brown transverse stripes across anterior segments, one wider, not reaching end of segments† | 30–32/16–18 | 15/0.3–0.4 | Coral rubble/2 m | Balayan and Batangas Bay, Luzon Is.; El Nido, Palawan Is., Philippines | This paper |
| Trypanosyllis luqueisp. nov. (lineage 2) | Two brown transverse stripes across anterior segments, close to segmental ends. Brown appendages only coloured in the limit of each article*, † | 18–20/12–14 | 12/0.2–0.4 | Bryozoans/intertidal | San Diego Bay, California, USA | This paper |
| Trypanosyllis kalkinsp. nov. (lineage 4) | Two light-brown transverse stripes in anterior segments. Well-preserved specimens with two bands reaching eyes† | 20–22/14–16 | 4/0.3–0.4 | Dendrymenia skottsbergii / intertidal | Las Cruces, Valparaergii in anterior segment | This paper |
| Trypanosyllis sp. 2 (lineage 6) | Two red–brown transverse stripes across anterior segments, similar width and length, † anterior stripe reaching the end of parapodia | 40–42/23–24 | 6/0.7 | Paramuricea clavata/ 40 m | Cap de Creus, Girona, Catalonia (Gulf of Lion), Spain | This paper |
| Trypanosyllis richardi | Two transverse dark red–brown bands, as small dark spots arranged in parallel/transverse lines, thicker in the middle* | 40–50/− | 42/2 | −/− | Djibouti, Red Sea | Gravier, 1900 |
| Trypanosyllis parazebra | One thick brown band* | 9–11/6–7 | 6/0.7 | −/1 m | Arica, Chile | Hartmann-Schröder, 1965; |
| Trypanosyllis taeniaformis | Light red coloration, with numerous narrow transverse brown bands both in dorsum and between the joints of cirri and tentacles* | 46/− | −/− | −/shallow water | Port Jackson, Sydney, Australia | Haswell, 1886; Çinar, 2007; |
| Trypanosyllis prampramensis | Two stripes* | 8–10/6–8 | 6.6/0.7 | −/− | Greater Accra Regio, Ghana | Augener, 1918 |
| Trypanosyllis vittigera | Two brown bands* | 30/− | 13.5/2.5 | −/13 m | Key West, Florida, USA | Elhers, 1887 |
| Trypanosyllis zebra holotype | Two transverse dark-brown bands. White dorsal cirri | 20/− | 48.6/20.32‡ | −/− | Cherso, Adriatic Sea, Croatia | Grube 1860 |
| Trypanosyllis zebra (ZMB Q4428) | Two transverse brown bands. White dorsal cirri | 37–39/20–22 | 18/2 | −/− | Lessina, Adriatic Sea, Italy | This paper |
Here ‘stripe’ refers to lines that are thinner than bands.
Coloration observed in live specimens.
Coloration observed in ethanol-fixed specimens.
Original measurements given in the description of Grube (1860), which might be mistaken given the huge numbers reported that were never seen before in any other Trypanosyllis species.
Taxonomy
Our molecular analyses (Figs 1, 2) combined with morphological examination (see below) suggested six different clades that were assigned the status of genera: the previously described genera Eurysyllis, Xenosyllis, and Plakosyllis, the newly reorganized Trypanosyllis, and the resurrected Pseudosyllis Grube, 1863 and TrypanedentaImajima & Hartman, 1964 (Fig. 5). Pseudosyllis was originally described as a monotypic genus containing Pseudosyllis brevipennis Grube 1863, later synonymised with Trypanosyllis coeliaca Claparède, 1868 by San Martín (2003). Our results now show that the former T. coeliaca does not belong to Trypanosyllis and it is here considered a junior synonym of P. brevipennis following the priority rules of the International Code of Zoological Nomenclature (ICZN). Pseudosyllis brevipennis is characterized by its small body size and short unpigmented cirri (Figs 6A–D, 7). Interestingly, Pseudosyllis brevipennis was found to be morphologically more similar to species of Eurysyllis, Plakosyllis, and Xenosyllis than to those of Trypanosyllis. As the type material of Pseudosyllis brevipennis is lost, a neotype from the type locality is designated herein (ZMB 7978), although the specimen could not be sequenced as it was originally preserved in formalin. In addition, the newly assigned genus Trypanedenta contained two species that share a reproductive strategy based on producing several stolons, as well as the presence of chaetae with few or no spines on the margin (Fig. 6E–I), as opposed to other genera with many spines. Trypanosyllis gemmiparaJohnson, 1901 and Trypanosyllis gigantea (McIntosh, 1885) are now included in TrypanedentaImajima & Hartman, 1964 as Trypanedenta gemmipara comb. nov. and Trypanedenta gigantea comb. nov. (Figs 6E–I, 8, 9).
Maximum clade credibility tree from concatenated, partitioned Bayesian analysis of all genetic data (28S rRNA, 18S rRNA, 16S rRNA, and COI) showing the phylogenetic relationships between Eurysyllis, Xenosyllis, Plakosyllis, the newly assigned Pseudosyllis, Trypanedenta, and the newly organized Trypanosyllis s.s. Numbers above branches indicate posterior probabilities support values (only PP > 0.9 are indicated); red circles indicate a PP value of 1.00. The 23 specimens of Trypanosyllis luzonensiscomb. nov. are abbreviated as Tluzo1–Tluzo23. The 46 specimens of T. krohnii sensu lato are abbreviated as Tk1–Tk46. Drawings showing the morphology of the different genera have been included with Fauna Iberica permission (San Martín, 2003), except the one of the genus Trypanedenta which was originally created by the authors.
Morphological features of species from the genera Pseudosyllis and Trypanedenta. A, light microscopy image of Pseudosyllis brevipennis neotype (ZMB 7978) anterior part, arrows point to the trepan (t) and the proventricle (p). B, light microscopy image of Pseudosyllis brevipennis neotype midbody chaetae showing the distal spines (arrow). C, light microscopy image of Pseudosyllis brevipennis (MNCN 16.01/16177) posterior chaetae showing the distal spines (arrow). D, scanning electron microscopy (SEM) micrographs of Pseudosyllis brevipennis midbody chaetae (MNCN 16.01/16176). E, light microscopy image of Trypanedenta gemmiparacomb. nov. anterior part (MNCN 16.01/16042). F, SEM micrographs of the posterior chaetae of Trypanedenta gemmiparacomb. nov. (MNCN 16.01/16043). G, light microscopy image of a complete specimen (SIO A2948) Trypanedenta giganteacomb. nov. showing a chain of stolon (sc) in posterior part. H, detached mature stolon of Trypanedenta giganteacomb. nov. (SIO A3211). I, SEM micrographs of the posterior chaetae of Trypanedenta giganteacomb. nov. (MNCN 16.01/16508).
Scanning electron micrographs of Pseudosyllis brevipennis (MNCN 16.01/16177): A, anterior part; B, midbody chaetae; C, midbody aciculae. The arrows point to the nuchal organs (no) and ciliary rows (cr).
Scanning electron micrographs of Trypanedenta gemmiparacomb. nov. (MNCN 16.01/16043): A, anterior part; B, prostomium; C, detail of nuchal organs (no); D, detail of ciliary rows on anterior segments (cr); E, anterior chaetae; F, posterior chaetae, arrow points to papillae (pp) of bilobed parapodia.
Scanning electron micrographs of Trypanedenta giganteacomb. nov.: A, anterior part (MNCN 16.01/16505); B, prostomium (MNCN 16.01/16505), arrow points to the nuchal organs (no); C, detail of dorsal cirri (MNCN 16.01/16506); D, detail of the trepan pharynx in apical view, arrow points to the trepan (t) (MNCN 16.01/16501); E, pharynx and proventricle (MNCN 16.01/165052); F, anterior chaetae (MNCN 16.01/16505); G, midbody chaetae, arrow points to four protruding aciculae (a; MNCN 16.01/16506); H, posterior chaetae, arrow points to two protruding aciculae (a; MNCN 16.01/16508).
Based on the results from both phylogenetic and species delimitation analyses, and assessing differences in morphological and ecological features, we conclude that Trypanosyllis krohnii is not a cosmopolitan species, but is instead a complex of pseudocryptic species. Here we illustrate the recognized pseudocryptic species found within the Trypanosyllis krohnii complex based on specimens collected around the world (Figs 3–5, 10). Furthermore, a neotype and a re-description of the type species of the genus are provided in order to resolve the taxonomic confusion related to its status (see below). Although chaetae have been considered one of the key characters to identify syllid species, variations in the morphology of chaetae among these seven Trypanosyllis species did not match the clades defined by our molecular data. Despite the lack of differences in chaetal morphology, the presence of colour polymorphism (only observable in well-preserved specimens), as well as differences in body width, length of cirri, and habitat choice, have been observed in these seven lineages (Figs 4B, 10).
Light microscopy images of the anterior of Trypanosyllis s.s. species: A, Trypanosyllis krohnii neotype (MNCN/ADN 9623); B, Trypanosyllis aeolis (MNCN 16.01/16039); C, Trypanosyllis zebra (ZMB Q4428); D, Trypanosyllis luzonensiscomb. nov. (AM W.47275); E, Trypanosyllis taboadaisp. nov. holotype (MNCN 16.01/16087); F, Trypanosyllis luqueisp. nov. holotype (SIO A5005); G, Trypanosyllis leivaisp. nov. holotype (MNCN 16.01/16082); H, Trypanosyllis californiensissp. nov. holotype, live specimen (SIO A5008); I, Trypanosyllis sp. 2 (MNCN 16.01/16065); J, Trypanosyllis kalkinsp. nov. holotype (MNCN 16.01/16058).
The five new species are named and described here, and Trypanosyllis krohnii s.s. is re-described and a type designated following the requirements of the ICZN. It is important to note that the holotype of each of the five newly described species and the neotype of the newly collected specimen of Trypanosyllis krohnii s.s. are incomplete: some segments were used for DNA sequencing. Following advice from a member of the International Commission on Zoological Nomenclature (Dr M.A. Alonso-Zarazaga), we used specimens for which molecular data are available as the primary types. A summary of the morphological differences found between our new species and all other striped Trypanosyllis species described so far is provided in Table 3.
Pseudosyllis Grube, 1863
Pseudosyllis Grube, 1863: 44.
TrypanosyllisClaparède, 1864 (partim).
Type species
Pseudosyllis brevipennis Grube, 1863.
Pseudosyllis brevipennis Grube, 1863 Figs 6A–D, 7
Trypanosyllis coeliaca Claparède, 1868, 19(2): 203; San Martín, 2003: 308–311, Figs 169, 170.
Type material examined
Neotype: Rovigno, Croatia (45.066667, 13.616667): one specimen mounted on a slide (ZMB 797), 1912, coll. Heider (Figs 6A, B).
Other material examined
Rovigno, Croatia: two specimens mounted on slides (ZMB 7978a, ZMB 7978b), collection details as for the neotype. Spain, Catalonia: one specimen in 96% EtOH (MNCN/ADN 9622), Port de la Selva (42.3375, 3.203333), Posidonia oceanica, 10 m, 21 September 2004, no collector data; two specimens in 96% EtOH (MNCN 16.01/16041, 16176), Barcelona, Mataró (41.5325, 2.453056), intertidal algae, March 2014, leg. M. Ballesteros; one specimen mounted for SEM (MNCN 16.01/16177) Girona, Cap Falcó (42.433333, 3.174722), calcareous algae, 15 September 2011, leg. G. San Martín (Figs 5C–D, 8). Spain, Alborán Sea: one specimen in 96% EtOH (MNCN 16.01/16040), (42.433333, 3.174722), algae, 42–48 m, 21 September 2011, leg. A. Luque and Alborán-INDEMARES Oceanographic Campaign collecting team; one specimen in 10% formalin buffered in seawater (MNCN 16.01/16178), (35.983333, −2.916667), calcareous algae, 68–70 m, 24 September 2011, leg. A. Luque Alborán-INDEMARES Oceanographic Campaign collecting team; one specimen in 10% formalin buffered in seawater (MNCN 16.01/16079), (35.878667, −3.077833), calcareous algae, 96–100 m, Alborán-INDEMARES Oceanographic Campaign collecting team.
Diagnosis
Body of medium to small size (up to about 12 mm long) without colour pattern. Short appendages, relatively thick, with few articles (Figs 6A, 7A). Bidentate chaetae with similar teeth; with spines along margin, distalmost spines reaching the distalmost tooth (Figs 6B–D, 7B). Proventricle shorter than pharynx, occupying two or three segments (Fig. 6A). Reproduction by single tetraglene stolon San Martín, (2003).
Description
Neotype, complete specimen: 5 mm long, 0.5 mm wide, 51 chaetigers, without distinct colour pattern (Fig. 6A). Oval prostomium with two pairs of eyes in trapezoidal arrangement, anterior eyes slightly larger (Figs 6A, 7A); antennae originating on anterior margin of prostomium; long median antenna with about eight articles; lateral antennae shorter, with about six articles. Oval palps shorter, completely separate. Nuchal organs as two densely ciliated semicircular areas, surrounding prostomial lobes (Fig. 7A). Segment 1 slightly smaller than subsequent segments; dorsal enlarged anterior cirri similar in length to antennae, with eight or nine articles; ventral cirri shorter with between four and six articles. Dorsal cirri similar in length to median antenna, with seven or eight articles (Figs 6A, 7A). Anterior segments with two rows of cilia, reaching cirrophores (Fig. 7A). Between ten and 12 compound bidentate, heterogomph falcigers in anterior and midbody segments, and seven or eight in posterior parapodia. All compound chaetae morphologically similar throughout body, bidentate, with distal tooth slightly larger than proximal one, and spines along margin, most distal ones larger and reaching distal tooth (Figs 6B–D, 7B). All parapodia with two protruding, acuminate, and thick aciculae (Fig. 7C). Dorsal and ventral simple chaetae not seen. Pharynx through about six segments, with an anterior tooth and a trepan with between eight and ten small teeth (Fig. 6A). Proventricle through three segments, with about 22 muscle cell rows (Fig. 6A). Reproduction by a single cephalous stolon with two pairs of eyes.
Remarks
San Martín (2003) considered that Trypanosyllis coeliaca Claparède, 1868 (type-locality, Naples, W Mediterranean) and Pseudosyllis brevipennis Grube, 1863 (type-locality: Adriatic Sea) were synonymous species. Although the author noted that Pseudosyllis Grube, 1863 had priority over TrypanosyllisClaparède, 1864; he considered Pseudosyllis as a nomen dubium, and therefore the species was named as Trypanosyllis coeliaca for stability purposes (see remarks in San Martín, 2003). Our study shows that Trypanosyllis coeliaca, as traditionally considered, belongs to a different genus, more closely related to the genera Xenosyllis, Eurysyllis, and Plakosyllis (Figs 1, 2, 5). These results agree with the morphological features: all species included in these four genera share a small body size, short cirri and proventricle, and a trepan with minute teeth, except for Xenosyllis, the trepan of which was secondarily lost. In addition, the species presented a large spine on the chaetae margin that is unique compared with other species of Trypanosyllis. Therefore, we reinstate Pseudosyllis (following ICZN requirements) to include the former Trypanosyllis coeliaca, which should now be considered Pseudosyllis brevipennis, the type and unique species of the genus. One of the specimens examined from the Museum für Naturkunde (Berlin) from the type locality (Croatia) agrees with the characters described by Grube (1863) and also in the morphological features examined in our specimens. Since the type material is lost, we have designated a neotype for the species (ZMB 7978), but further molecular studies are required to test if the specimens collected in the Spanish coasts are the same species as the specimens from the type locality. The other two specimens examined from the ZMB, identified as Trypanosyllis coeliaca (slides ZMB 7978a and ZMB 7978b), belonging to the same locality, might represent a different species because they have shorter chaetae with a minute proximal tooth and without spines on the margin. Nevertheless, they also share some of the morphological characters defined for Pseudosyllis species, so it will be necessary to sequence and compare specimens of these two morphotypes from Croatia to assess if they are different species. In addition, in order to check the status of Trypanosyllis coeliaca, further molecular studies are required including material from Naples (the type locality). Although Pseudosyllis is currently monotypic, there are also other species traditionally considered within Trypanosyllis that could also belong to Pseudosyllis, as they share the synapomorphies identified for the group. This is the case for Trypanosyllis parvidentataPerkins, 1981 from Florida (Perkins, 1981) and Trypanosyllis microdenticulataSalcedo-Oropeza, San Martín & Solis-Weiss, 2011, from the southern Mexican Pacific coast (Salcedo-Oropeza et al., 2011). Further molecular analyses are still needed in order to test whether Trypanosyllis parvidentata and Trypanosyllis microdenticulata are more closely related to Pseudosyllis brevipennis than to species within Trypanosyllis, and therefore to establish the status of Pseudosyllis brevipennis s.s.
Type locality
Rovigno, Croatia (Adriatic Sea).
Distribution
Mediterranean Sea.
Trypanedenta Imajima & Hartman, 1964; Figs 6E–I, 8, 9
Trypanosyllis (Trypanedenta) Imajima & Hartman: 125.
TrypanosyllisClaparède, 1864 (partim).
Type species
Trypanosyllis gemmiparaJohnson, 1901.
Diagnosis
Body variable in size, from 7 mm to more than 80 mm in length, and from 1 mm to 5 mm wide, with uniform yellowish coloration (Figs 6E, G). Unidentate or bidentate chaetae, without serration in margin or with a few, minute spines (Fig. 6F, I). Reproduction by clusters of several simultaneous tetraglene stolons (Fig. 6G, H).
Remarks
Trypanedenta was considered a subgenus of Trypanosyllis, because of the lack of a middorsal tooth in the pharynx (Imajima & Hartman, 1964). Our study concluded that TrypanedentaImajima & Hartman, 1964 should be raised to genus level, although the synapomorphies for the group require a re-evaluation of pharyngeal teeth during ontogeny (San Martín, 2003). Indeed, the type of reproduction, which differs from that in the other genera analysed, seems to be one of the most important features to differentiate this genus from the remaining genera. Both species included in the genus, Trypanedenta gemmipara comb. nov. and Trypanedenta gigantea comb. nov., present more than one stolon during their reproductive stages. Trypanedenta gemmipara comb. nov. develops a cluster of stolons (Johnson, 1901; Imajima & Hartman, 1964; Imajima, 1966), whereas Trypanedenta gigantea comb. nov. forms a chain of stolons (first report, this paper; Fig. 6G). Another character that seems to be important for the diagnosis of the genus is the presence of chaetae with just a few small spines on the margin (Trypanedenta gigantea comb. nov.) or none (Trypanedenta gemmipara comb. nov.) (Figs 6F, I, 8E–F, 9F–H). Further molecular studies are required to assess if other species currently included within Trypanosyllis, presenting similar chaetae and more than one stolon during their reproductive stage, actually belong to Trypanedenta. That may be the case for Trypanosyllis ingensJohnson, 1902, Trypanosyllis sanmartiniÇinar, 2007, and Trypanosyllis auranticusNogueira & Fukuda, 2008 (Johnson, 1902; Çinar, 2007; Nogueira & Fukuda, 2008). In addition, the budding of numerous simultaneous stolons has also been observed in Trypanobia asterobiaOkada, 1933, but the relationship between the recently resurrected genus TrypanobiaImajima & Hartman, 1964 (Aguado et al., 2015) and Trypanedenta needs further study.
Distribution
Cosmopolitan.
Trypanedenta gemmipara (Johnson, 1901) comb. nov. Figs 6E, F, 8
Trypanosyllis gemmiparaJohnson, 1901: 405; pl. 7, figs 72–76.
Trypanosyllis (Trypanedenta) gemmiparaImajima & Hartman (1964): 126; pl. 30, figs f, g. Imajima (1966): 237; figs 44a–m.
Trypanosyllis misakiensis Izuka, 1906: 283; figs 1–4. Izuka (1912): 185; pl. 20, figs 2–6. Fauvel (1932): 80. Fauvel (1953): 158. Uschakov (1955): 183; fig. 52a–d.
Material examined
New Zealand, Cavalli Islands (−36.834167, 174.771389): two specimens in 96% EtOH (MNCN 16.01/16042–16043), one of them mounted for SEM (MNCN 16.01/16043), Rainbow Warrior wreck, 25 m, 1 February 2012, leg. G. San Martín Project CGL2009-12292 BOS collecting team.
Trypanosyllis intermedia. Holotype (CAS-IZ 20550), California, Monterey County, Monterey Bay (36.616667, −121.951111), intertidal, 12 July 1904, leg. H.P. Johnson.
Diagnosis
Uniform yellow colour in well-preserved specimens (Fig. 6E), compound bidentate chaetae, all with short blades, without serrations (Figs 6F, 8E, F), stolon clusters during reproductive stage (Johnson, 1901; Imajima & Hartman, 1964; Imajima, 1966).
Description
Complete longest specimen: 10 mm long, 1 mm wide, 90 chaetigers, with uniform yellow coloration (Fig. 6E). Oval posteriorly bilobed prostomium with four large eyes in trapezoidal arrangement (Figs 6E, 8A, B). Antennae originating on anterior margin (Figs 6E, 8A, B); median antenna slightly longer than combined length of prostomium and palps, with about 20–22 articles, slightly longer than lateral antennae, with 19 or 20 articles. Oval palps shorter than prostomium, completely separated, ventrally folded. Nuchal organs surrounding prostomial lobes (Fig. 8C). Segment 1 slightly smaller than subsequent segments; dorsal enlarged anterior cirri similar in length to the antennae, with 21 articles, slightly longer than ventral cirri, with 18 articles. Dorsal cirri shorter than body width, alternately long (20–22 articles) and short (12–14 articles) (Figs 6E, 8A). Anterior segments with two rows of cilia (Fig. 8D). Parapodial lobes distinctly bilobed, presenting a rounded papilla situated ventrally to the lobes and dorsally to the ventral cirri (Fig. 8F). Ventral digitiform cirri, shorter than parapodial lobes (Figs 6F, 8F). About six compound bidentate, heterogomph falciger chaetae per parapodium, with distal tooth slightly larger than proximal tooth and smooth margins (Figs 6F, 8E, F). Three anterior aciculae, straight, acute, protruding from parapodia (Fig. 8E); two aciculae in midbody and a single acicula in posterior parapodia. Dorsal and ventral simple chaetae not seen. Pharynx slender and long, through 16 segments (Fig. 6E); trepan with ten teeth, surrounded by a crown of ten papillae. Proventricle half shorter than pharynx, through eight segments, with about 40 muscle cell rows.
Remarks
Although Imajima & Hartman (1964) and San Martín (2003) synonymized Trypanedenta gemmipara with Trypanosyllis aeolis, our results concluded that it is a different and well-delineated species that in fact belongs to a different genus (Figs 1, 2, 5). The morphological examination agrees with the molecular results, as Trypanedenta gemmipara largely differs from Trypanosyllis aeolis in coloration pattern and type of chaetae. Trypanedenta gemmipara presents uniform yellow coloration (Fig. 6E), with all chaetae bidentate, short, and smooth (Figs 6F, 8E, F), whereas the later show pink–pale coloration in body and yellow–orange cirri (Fig. 10B), with bidentate chaetae with spines on the margin (Fig. 12B–D). In addition, the specimens examined from New Zealand agree with the descriptions of the species from other areas in the Pacific Ocean. In spite of this apparent similarity, further studies are needed to test the putative wide distribution of the species, as the analysis in other syllid species pointed to a restricted geographical distribution rather than a wide distribution. The type material examined of Trypanosyllis intermedia from California has the same kind of chaetae observed in Trypanedenta gemmipara comb. nov., but only on its midbody and posterior parapodia. As Trypanosyllis intermedia could not be sequenced, and the kind of reproduction has not been reported, we cannot be sure if it is a Trypanosyllis species, or another species of Trypanedenta.
Type locality
Washington, USA (Pacific Ocean).
Distribution
Pacific Ocean: from Alaska to Mexico; Japan, India. First report for New Zealand.
Trypanedenta gigantea (Mcintosh, 1885) comb. nov. Figs 6G–I, 9
Syllis gigantea McIntosh, 1885:193–195, pl. 30, figs 1–3.
Material examined
Antarctica: two specimens in 96% EtOH (SIO A3211, A2935), Elephant Island (−61.163333, −54.997222), Blake Trawl, 223–242 m, 22 October 2011, leg. G.W. Rouse; one specimen in 96% EtOH (SIO A2948), Bransfield Strait (−63.080556, −59.156389), Blake Trawl, 150–247 m, 24 October 2011; one specimen in 96% EtOH (SIO A3565), Burdwood Bank East (−54.558889, −56.828889), Blake Trawl, 90–92 m, 24 April 2013, leg. G.W. Rouse; one specimen in 96% EtOH (SIO A3515), Burdwood Bank East (−54.540833, −56.626944), Blake Trawl, 122–123 m, 24 April 2013, leg. G.W. Rouse. Weddell Sea: two specimens in 10% formalin buffered in seawater (MNCN 16.01/16504), (−70.933333, −10.533333), Agassiz Trawl, 288 m, 9 December 2003, leg. C. Avila and M. Ballesteros, i.d. by S. Taboada; one specimen in four different parts, mounted for SEM (MNCN 16.01/16505–16508), collection details as for MNCN 16.01/16504; four specimens in 10% formalin buffered in seawater (MNCN 16.01/16502), (−71.066667, −1.566667), Bottom Trawl, 308 m, 23 December 2003, leg. C. Avila and M. Ballesteros, i.d. by S. Taboada; pharynx and trepan of one specimen mounted for SEM (MNCN 16.01/16503), collection details as for MNCN 16.01/16502; pharynx and trepan of one specimen mounted for SEM (MNCN 16.01/16501) and six specimens in 10% formalin buffered in seawater (MNCN 16.01/8875), (−70.866667, −10.85), Bottom Trawl, 294 m, 27 December 2003, leg. C. Avila and M. Ballesteros, i.d. by S. Taboada; one specimen in 10% formalin buffered in seawater (MNCN 16.01/3561), (−71.1, −11.45), Agassiz Trawl, 277 m, 28 December 2003, leg. C. Avila and M. Ballesteros, i.d. by S. Taboada; four specimens in 10% formalin buffered in seawater (MNCN 16.01/8769), (−70.95, −10.55), Bottom Trawl, 332 m, 24 December 2003, leg. C. Avila and M. Ballesteros, i.d. by S. Taboada.
Trypanosyllis ingens. California, Marin County: one specimen (CAS-IZ 181627) and one stolon (CAS-IZ 181664), Cordell Bank National Marine Sanctuary, Cordell Bank, north-west pinacle sea mount (38.03, −123.426667), 40 m, 15 December 1981, leg. R. Schmieder; six stolons (CAS-IZ 181663), Cordell Bank National Marine Sanctuary, Cordell Bank, north-west pinacle sea mount (38.031667, −123.418333), 40 m, 10 October 1981, leg. R. Schmieder.
Diagnosis
Compound unidentate posterior chaetae, ventral chaetae with short blades, with few spines on margin (Figs 6I, 9F–H), stolon chains during the reproductive stage (first report in this paper, Fig. 6G, H).
Description
Complete longest specimen: 55 mm long, 30 mm wide, 304 chaetigers, with yellow pale coloration on body and cirri, but stolons with a strong brown pigmentation (Fig. 6G, H). Bilobed prostomium with two pairs of eyes, anterior eyes larger than posterior eyes (Fig. 6G); antennae originating on the anterior margin of prostomium, median antenna long, with about 21–23 articles; lateral antennae slightly shorter, with about 18–20 articles. Oval palps shorter than prostomium, completely separate. Nuchal organs as a row of cilia on anterior margin of segment 1 (Fig. 9B). Segment 1 slightly smaller than subsequent segments; dorsal enlarged anterior cirri longer than antennae, with about 28–30 articles, longer than ventral ones, with about 22–24 articles. Dorsal cirri alternating long (20–22 articles) and short (15–17 articles). All articles on appendages joined only by the central part (Fig. 9C). Ventral cirri digitiform, reaching edge of parapodia. Between eight and ten compound heterogomph falciger chaetae per parapodium, similar throughout body. Most part of chaetae unidentate with a few spines on margin (Figs 6I, 9F, G). Some chaetae of posterior parapodia slightly bidentate with a minute proximal tooth (Fig. 9H). All aciculae thick and distally pointed, protruding from parapodia (Fig. 9G, H), four on anterior and midbody parapodia (Fig. 9G), and two on posterior parapodia (Fig. 9H). Dorsal and ventral simple chaetae not seen. Pharynx through about 19 segments (Fig. 6G); trepan as a corneous disc surrounded by ten papillae (Fig. 9D). Proventricle through 17 segments, with about 41 muscle cell rows (Fig. 9E).
Reproduction
Scissiparity (gemmiparity). Development of a chain of stolons during the reproductive stages, brown in colour, first stolons acephalous (Fig. 6G), and last two stolons better developed, presenting pair of ventral eyes. One detached cephalous female stolon found, 40 mm long, 20 mm wide, with 60 segments, and with two pairs of brown eyes (Fig. 6H).
Remarks
The most similar species to Trypanedenta gigantea comb. nov. is Trypanosyllis ingensJohnson, 1902; described from Pacific Grove (California, USA). Johnson (1902) described not only the same kind of unidentate chaetae seen in Trypanedenta gigantea comb. nov., but also the development of numerous budding stolons. The specimens examined from Marin County (California, USA) agree with the original description, but also present some smooth bidentate chaetae, similar to those that appear in Trypanedenta gemmipara comb. nov., which were not described by Johnson (1902), and are the main difference between these two large-bodied species. As the specimens of Trypanosyllis ingens were not able to sequenced, we cannot be sure whether it also belongs to Trypanedenta.
Type locality
Ross Sea (Southern Ocean).
Distribution
Southern Ocean, Strait of Magellan; south-western Pacific (New Zealand); north-eastern Atlantic Ocean; eastern Mediterranean Sea (Adriatic and Aegean seas).
Trypanosyllis Claparède, 1864
TrypanosyllisClaparède, 1864: 558.
Parautolytus Pillai, 1965: 123.
Type species
Trypanosyllis krohniiClaparède, 1864.
Diagnosis
As in (San Martín et al. 2008).
Remarks
Although Imajima & Hartman (1964) divided the genus into four subgenera, based on body shape, type of chaetae, and the presence of a pharyngeal tooth, most authors did not follow this classification, arguing that these features depended on the ontogeny (e.g., San Martín, 2003; Çinar, 2007; Nogueira & Fukuda, 2008; San Martín et al., 2008). Our results show that Trypanosyllis s.l. contains at least two clades, which we have designated as genera: TrypanedentaImajima & Hartman, 1964 and Pseudosyllis Grube, 1863. These genera differ from Trypanosyllis s.s. in the type of chaetae, body size, length of cirri, and reproductive mode (see remarks above).
Trypanosyllis krohnii Claparède, 1864; Figs 4B, 10A, 11
Scanning electron micrographs of Trypanosyllis krohnii (MNCN 16.01/16186): A, anterior part; B, prostomiun; C, detail of ciliary bands in midbody dorsal segments and parapodia; D, anterior dorsal chaeta; E, anterior ventral chaetae; F, posterior dorsal chaeta; G, posterior ventral chaetae.
Type material examined
Neotype. France, Banyuls-sur-Mer (42.483333, 3.133333): one specimen in 96% EtOH (MNCN ADN/9623), Harbour on docks, snorkelling, epifauna on mussels (i.e. hydroids, sponges), 19 April 2001, no collector data (Fig. 9A).
Other material examined
Spain, Catalonia: one specimen in 96% EtOH (MNCN 16.01/16187), Barcelona, Mataró (41.5325, 2.453056), intertidal algae, March 2014, leg. M. Ballesteros; one specimen in 96% EtOH (MNCN 16.01/16066), Girona, Cap de Creus (42.320278, 3.320556), Petrosia sp., 16 m, 16 September 2011, leg. G. San Martín; one specimen mounted for SEM (MNCN 16.01/16186), collection data as for MNCN 16.01/16066; four midbody parts (MNCN 16.01/16188), collection data as for MNCN 16.01/16066.
Morphologically similar species examined
Italy: Trypanosyllis zebra, one specimen (ZMB Q4428), Lesina (41.766667, 15.433333), 1874, leg. Grube. Lessina coll. Grube.
Diagnosis
Colour pattern in preserved specimens as brown transverse stripes across anterior and midbody parts, one close to anterior end of segment and the other in middle of segment (Figs 4B, 10A). All stripes similar in length, reaching parapodia (Figs 4B, 10A). Large anterior cirri with about 42 articles (n = 5); dorsal cirri alternating long (30–32 articles) and short (20–22 articles).
Description
Neotype incomplete, 8 mm long, 1.5 mm wide, 83 chaetigers. Long dorsoventrally flattened body (Figs 10A, 11A). Colour pattern as two thin, brown, transverse stripes across limit of anterior and midbody segments (Fig. 10A). Some specimens with purple pigmentation remaining on anterior and dorsal cirri (Fig. 4B). Oval prostomium with two pairs of eyes in trapezoidal arrangement (Fig. 10A); antennae originating on anterior margin of prostomium, long median antenna with about 30 articles; lateral antennae slightly shorter, with about 25 articles (Fig. 10A, 11A, B). Oval palps shorter than prostomium, completely separated. Nuchal organs as two densely ciliated semicircular areas, extending on prostomium and surrounding prostomial lobes. In addition, ciliary bands on dorsum of anterior and midbody segments and parapodia (Fig. 11C). Segment 1 slightly smaller than subsequent segments; dorsal enlarged anterior cirri longer than antennae, with about 42 articles, longer than ventral cirri, with about 35 articles Dorsal cirri alternating long (30–32 articles) and short (20–22 articles). Ventral digitiform cirri, reaching edge of parapodia. Parapodia with two anterior, digitiform lobes. Compound bidentate, heterogomph falciger chaetae, about 15–17 on anterior parapodia, 13–15 on midbody and 12–14 on posterior ones. All chaetae similar throughout body, with both teeth similar in length and serrated margin (Fig. 11D–G); all parapodia with dorsal chaetae with longer blades and very short spines on the margin (Fig. 11D, F), and ventral bidentate chaetae, shorter than ventral chaetae, with minute spines on the margin of anterior and midbody chaetae (Fig. 11E), and almost smooth on posterior parapodia (Fig. 11G). Three or four anterior straight aciculae, all distally blunt; two or three midbody and posterior straight, distally pointed aciculae thicker than anterior aciculae. Dorsal and ventral simple chaetae not seen. Pharynx running through 11 segments. Proventricle running through 13 segments, with about 40 muscle cell rows.
Remarks
Trypanosyllis, together with the species Trypanosyllis krohnii, were described by Claparède (1864) from Port-Vendres (France). Four years earlier, Syllis zebraGrube, 1860 was described from the Adriatic Sea, and Marenzeller (1874) later placed it in Trypanosyllis because it presented a trepan. In 1879, Langerhans synonymized Trypanosyllis krohnii with Trypanosyllis zebra on the basis of the original descriptions, without studying the type specimens (subjective synonymy, ICZN, article 61.3). All subsequent authors followed this synonymy, probably because of the similarity in the striped coloration found in both species. Many years later Hartman (1959) designed Trypanosyllis zebra as the type species of the genus by subsequent designation (ICZN, article 69.1), and since then all the authors except Day (1967) considered this designation as valid (e.g., Imajima, 1966; Uebelacker, 1984; San Martín, 2003). Our study found that Hartman's designation is in fact not valid, because Claparède (1864) described the genus Trypanosyllis and its monotypic type species Trypanosyllis krohnii (ICZN, art 68.3), which is a valid species. Lineage 6, which inhabits the same geographical areas as the type species, presented enough distinctive morphological features to separate them, such as the colour pattern, the length of the dorsal cirri, and the body width (Fig. 10A, I; Table 3). In addition, the comparative material from the Adriatic deposited in the ZMB, which was collected in 1874 and identified by Grube as Syllis zebra, also differed from lineage 6 in the same characters (Fig. 10C, I; Table 3), but was slightly more similar to Trypanosyllis krohnii than lineage 6 (Fig. 10A, C; Table 3). The ZMB specimen did not fully agree with the original description of Trypanosyllis zebraGrube (1860), mainly because of the huge size of the holotype described by Grube (Table 3), and therefore we prefer not to designate it as the neotype of Trypanosyllis zebra. Likewise, even though this ZMB specimen is similar to Trypanosyllis krohnii, we cannot synonymize them because we have not sequenced this material. Therefore, until new material of striped Trypanosyllis from the Adriatic is collected for further sequencing, we will regard our specimens in lineage 6 as Trypanosyllis sp. 2.
Nevertheless, here we conclude that Trypanosyllis krohnii is not a synonym of Trypanosyllis zebra, but a valid and well-defined species distributed at least in the Gulf of Lion and along the north-west coast of Spain (Tables 1 and 3). In the present study we re-describe the type species of the genus, designating a neotype (as the type material is lost) following the ICZN requirements. The studied specimens agree with the original description, except for the colouration, which Claparède (1864) described as violet. As we have observed remnants of violet pigmentation in the anterior and midbody cirri of some specimens (Fig. 4B), we conclude that the coloration differences may be the result of preservation methods. Our results also suggest that the species is distributed in the same area (Gulf of Lion) as Trypanosyllis sp. 2, but inhabiting different bathymetric ranges, with Trypanosyllis krohnii occurring in more shallow waters (Table 3). Marion & Bobretzky (1875) reported Trypanosyllis krohnii from the Gulf of Marseille, noting that specimens collected in shallow waters presented body coloration and length of cirri that differed considerably from those of specimens collected in deeper waters, although they considered both as the same species. Our examination agrees with their observation, and thus we conclude that there is enough molecular, morphological, and ecological evidence to consider them as two different species.
Type locality
Banyuls-sur-Mer, France (North-Western Mediterranean Sea).
Distribution
Gulf of Lion (Mediterranean Sea), including the Cap de Creus, in the north-eastern coast of Spain (Girona).
Trypanosyllis aeolis Langerhans, 1879; Figs 10B, 12A, B
Scanning electron micrographs of Trypanosyllis aeolis (MNCN 16.01/16181): A, anterior part; B, anterior chaetae; C, posterior chaetae.
Trypanosyllis aeolisLangerhans, 1879: 558, figs 18a, b; Núñez et al., 1992: 114; San Martín, 2003: 315, figs 174–176.
Material examined
Spain, Mallorca, El Toro Island (39.4891, 2.4809): three specimens in 96% EtOH (MNCN 16.01/16039, 16180, 16181), Cladocora cespitosa and Miriapora sp., 12 m, 18 June 2012, leg. P. Álvarez-Campos and M. Capa. Australia, QLD, Lizard Island, Watsons Bay (−14.657222, 145.450833): one specimen in 96% EtOH (AM W.41717), coral rubble, 4.5 m, 28 August 2010, leg. P. Hutchings and M. Capa.
Diagnosis
Uniform pale pink coloration, with yellow dorsal cirri (Fig. 10B); compound bidentate chaetae, with small proximal tooth; sometimes blades unidentate on posterior parapodia (Fig. 12B).
Description
Longest complete specimen (MNCN 16.01/16039), 10 mm long, 1.2 mm wide, 65 chaetigers. Pale pink body coloration with yellow pigmentation in some cirri (Fig. 10B). Oval posteriorly bilobed prostomium (Figs 10B, 12A); two prostomial lobes, with two pairs of red eyes in trapezoidal arrangement (Fig. 10B). Oval palps slightly shorter than prostomium, completely separate. Nuchal organs as two densely ciliated semicircular areas, surrounding the eyes (Fig. 12A). Antennae originating on anterior margin of prostomium, median antenna long, with about 23 articles; lateral antennae distinctly shorter, with about 15 articles (Figs 10B, 12A). Segment 1 slightly shorter than subsequent segments; dorsal enlarged anterior cirri slightly longer than antennae, with about 25 articles, longer than ventral cirri, with about 20 articles. Anterior dorsal cirri as long as median antenna (25 articles); midbody and posterior dorsal cirri shorter, with 20 and 15 articles, respectively. Ventral cirri digitiform, shorter than parapodia. Compound bidentate heterogomph falciger chaetae with both teeth similar in length and with a few and minute spines on margin (Fig. 12B, C). About 13 or 14 chaetae on anterior parapodia (Fig. 12B), and with between eight and ten chaetae on midbody and posterior parapodia (Fig. 12C). One unique, thick, and straight aciculae protruding from each parapodia (Fig. 12B). Pharynx through about ten segments; trepan with eight teeth. Proventricle similar in length to pharynx, through nine segments, with about 22 muscle cell rows.
Remarks
See remarks for Trypanedenta gemmipara comb. nov. One of the specimens of Trypanosyllis aeolis analysed is from Australia, and even though we have not been able to find morphological differences with the specimens from the Iberian Peninsula, because of their disjunct distribution this may represent another case of cryptic speciation.
Type locality
Madeira (Atlantic Ocean).
Distribution
Pacific Ocean (Washington, USA); north-eastern Atlantic Ocean (UK, Portugal, and Spain); Mediterranean Sea, including Adriatic and Aegean seas.
Trypanosyllis luzonensis (Pillai, 1965) comb. nov. Figs 10D, 13, 14
Scanning electron micrographs and light microscopy images of Trypanosyllis luzonensiscomb. nov.: A, prostomium, arrows point to nuchal organs (no; AM W.41639); B, midbody segments showing the ciliation (AM W.41639); C, chaetae from anterior parapodia (AM W.41713); D, chaetae from midbody parapodia (AM W.41713); E, chaetae from posterior parapodia (AM W.41646); F, dorsal (dsc) and ventral (vsc) simple chaetae from posterior parapodia (AM W.41713); G, female detached stolon (MNCN 16.01/16054).
Trypanosyllis luzonensiscomb. nov. line drawing (AM W.47275): A, anterior part; B, trepan; C, anterior ventral cirri; D, midbody ventral cirri; E, midbody chaetae; F, posterior dorsal simple chaetae; G, posterior ventral simple chaetae; H, anterior and midbody aciculum; I, posterior aciculae. Scale bars: A, 1 mm; B, 0.375 mm; C, 0.18 mm; D, 0.375 mm; E–I, 20 μm.
Parautolytus luzonensis Pillai, 1965: 123, figs 5E–I, 6A–C.
Trypanosyllis zebraSan Martín et al., 2008 partim: 43–48, figs 32–34.
Type material examined
Paratype. Philippines: one specimen (BMNH 1965.33.29, The Natural History Museum, London, UK), Luzon Island, Bonva, Dagupan City (16.033056, 120.333056), brackish water, fish pond, 1965, leg. G. Pillai.
Other material examined
China: Hong Kong (22.338056, 114.2675): one specimen in 96% EtOH (SIO A6142), Hong Kong University of Science and Technology, aquarium system, 9 May 2014, leg. G.W. Rouse. Philippines: one specimen in 96% EtOH (MNCN 16.01/16053), Palawan Island, El Nido, ‘Twin Rocks’ (11.297222, 119.318333), unidentified sponges, 6 m, 17 December 2010, leg. G. San Martín Project CGL2009-12292 BOS collecting team; one specimen in 96% EtOH (MNCN 16.01/16054), Luzon Island, between Balayan Bay and Batangas Bay, ‘Mainif Point’ (13.68, 120.855556), coral rubble, 2 m, 8 December 2010, leg. G. San Martín Project CGL2009-12292 BOS collecting team; six specimens in 96% EtOH (MNCN 16.01/16045), Luzon Island, between Balayan Bay and Batangas Bay, ‘Mainif Point’ (13.68, 120.855556), coral rubble, 20 m, 8 December 2010, leg. G. San Martín Project CGL2009-12292 BOS collecting team; six specimens in 96% EtOH (MNCN 16.01/16047, 16049, 16051), Luzon Island, between Balayan Bay and Batangas Bay, ‘Sepok Point’ (13.68, 120.855556), coral rubble, 6 m, 10 December 2010, leg. G. San Martín Project CGL2009-12292 BOS collecting team; four complete specimens and four midbody parts in 96% EtOH (MNCN 16.01/16035, 16044, 16046, 16050, 16052), Luzon Island, Balayan Bay, Sombrero Island (13.697778, 120.829722), coral rubble, 20 m, 9 December 2010, leg. G. San Martín Project CGL2009-12292 BOS collecting team; two specimens in 96% EtOH (MNCN 16.01/16055, 16183), Luzon Island, Balayan Bay, Sombrero Island (13.697778, 120.829722), unidentified sponges, 2 m, 6 December 2010, leg. G. San Martín Project CGL2009-12292 BOS collecting team; one specimen in 96% EtOH (MNCN 16.01/16045), Luzon Island, Balayan Bay, ‘Koala Point’ (13.798889, 120.869444), coral rubble, 16 m, 5 December 2010, leg. G. San Martín Project CGL2009-12292 BOS collecting team; two specimens in 96% EtOH (MNCN 16.01/16045, 16082), Luzon Island, Balayan Bay, ‘Koala Point’ (13.798889, 120.869444), Thalyssias sp. and Acanthella sp. sponges, 16 m, 5 December 2010, leg. G. San Martín Project CGL2009-12292 BOS collecting team. Australia: one specimen in 80% EtOH (AM W.42432), WA, Montgomery Reef (−16.020556, 124.159167), mid-littoral lower terrace, Acropora sp., 0 m, 23 October 2009, leg. Western Australian Museum Kimberley Islands Collecting Team; one specimen in 80% EtOH (AM W.41723), WA, Adele Island (−15.557778, 123.133889), sublittoral fore-reef slope, 12.5 m, 18 October 2009, leg. Western Australian Museum Kimberley Islands Collecting Team; one specimen in 80% EtOH (AM W.41649), WA, Adele Island, edge of Frazer Inlet (−15.444722, 123.170833), sublittoral channel slope, 0 m, 22 October 2009, leg. Western Australian Museum Kimberley Islands Collecting Team; three specimens in 95% EtOH (AM W.41645, W.41646 mounted for SEM, W.41647), WA, Ningaloo Reef (−22.623611, 113.641111), coarse coral rubble, 7 m, 20 May 2009, leg. P. Hutchings, M. Capa, R.S. Wilson and L. Avery; one specimen in 95% EtOH (AM W.41648), WA, Ningaloo Reef (−22.769722, 113.645556), brown algae and coral rubble, 24 m, 17 May 2009, leg. M. Capa and L. Avery; two specimens in 80% EtOH (AM W.41638, W.41639 mounted for SEM), WA, Long Reef (−13.856667, 125.825), sublittoral reef platform, 4 m, 21 October 2009, leg. Western Australian Museum Kimberley Islands Collecting Team; one specimen in 80% EtOH (AM W.41635), WA, Long Reef (−13.832222, 125.811944), sublittoral reef platform, 2 m, 23 October 2009, leg. Western Australian Museum Kimberley Islands Collecting Team; two specimens in 95% EtOH (AM W.42860), WA, Cassini Island, (−13.932222, 125.618333), 12 m, 16 October 2010, leg. Western Australian Museum Kimberley Islands Collecting Team; two specimens in 95% EtOH (AM W.41636, 41637), WA, Cassini Island, (−13.942222, 125.643333), 10–12 m, 24 October 2010, leg. Western Australian Museum Kimberley Islands Collecting Team; four specimens in 80% EtOH (AM W.32041, W.47275, 47276), NSW, Port Jackson (−35.858333, 151.233333), encrusting sponges and dead shells on subtidal rock ledge, 9 m, 6 October 2005, leg. P. Hutchings; one specimen in 95% EtOH (AM W.42430), NSW, Nielsen Park, Bottle & Glass Rock, Sydney (−35.858333, 151.233333), kelp holdfasts, 6 m, 13 April 2000, leg. P. Hutchings; three specimens in 95% EtOH (AM W.41713, W.42420, 42421), QLD, Lizard Island, Watsons Bay (−14.657222, 145.450833), coral rubble, 4.5 m, 28 August 2010, leg. P. Hutchings and M. Capa; one specimen in 95% EtOH (AM W.41640), QLD, Lizard Island, Watsons Bay (−14.658333, 145.448889), sand, 9 and 30 August 2010, leg. P. Hutchings and M. Capa; one specimen in 95% EtOH (AM W.41714), QLD, Lizard Island, Bommie Bay (−14.659722, −145.471111), 7.4 m, 9 September 2010, leg. L. Avery.
Diagnosis
Colour pattern with two large bands on each segment, almost fused as a single band (Fig. 10D). Posterior ventral compound chaetae unidentate or with a very small proximal tooth (Figs 13E, F, 14E).
Description
Paratype (BMNH 1965.33.29): 32 mm long, complete but broken in three pieces, 2 mm wide, with 145 chaetigers; longest specimen examined 30 mm long, 0.5 mm wide, with 140 chaetigers. Body long, dorsoventrally flattened, ribbon-like, with numerous segments (Figs 10D, 14A). Colour pattern as two wide, dorsal, dark red transversal bands on each segment, almost fused (Fig. 10D); sometimes dark to red spots on cirrophores. Antennae, dorsal enlarged anterior cirri, and anterior long dorsal cirri with remains of red pigmentation; remaining cirri yellowish, pale. Some specimens without pigmentation (e.g. paratype). Oval posteriorly bilobed prostomium, with two pairs of red eyes in trapezoidal arrangement (Figs 10D, 13A, 14A). Oval palps slightly shorter than prostomium, completely separate. Nuchal organs as two densely ciliated semicircular areas, extending on prostomium, surrounding prostomial lobes (Fig. 13A). Some specimens with one transversal row of cilia on each segment, extending also on parapodia (Fig. 13B). Segment 1 smaller than subsequent segments, sometimes partially covered by the first chaetiger. Antennae originating on the margin of prostomium; median antenna short, with about 40 articles, longer than lateral ones, with about 25 articles (Figs 10D, 14A). Dorsal enlarged anterior cirri longer than antennae, with about 50 articles, distinctly longer than ventral cirri, with about 20 articles (Figs 10D, 14A). Dorsal cirri alternating in anterior and midbody parapodia between long (58. Dorsal cirri alternating in anterior a, except those of chaetigers 3 and 4, both long with about 60 articles (Figs 10D, 14A). Parapodia with one anterior digitform lobe and one shorter bilobed posterior lobe (Fig. 14C, D). Ventral digitiform cirri, longer than parapodial lobes on anterior parapodia (Fig. 14C), shorter in midbody and posterior parapodia (Fig. 14D). Compound bidentate, heterogomph falciger chaetae, about 15–17 per parapodium on anterior segments, diminishing to eight on midbody and five on posterior parapodia. Anterior and midbody compound chaetae with both teeth similar, and short spines on margin (Fig. 13C, D). Compound chaetae on posterior parapodia with thicker shafts and shorter blades, most ventral chaetae with a small proximal tooth, sometimes most ventral chaetae unidentate (Figs 13E, F, 14E). Some specimens with slender and bidentate dorsal and ventral simple chaetae on posterior parapodia (Figs 13F, 14F, G). Aciculae thick, acutely pointed (Fig. 14H, I), three anteriorly, one or two posteriorly. Pharynx long, through about 12 or 13 segments; trepan with ten large teeth, surrounded by a crown of about 20 soft papillae (Fig. 14B). Proventricle long and slender, slightly shorter that pharynx, with about 35 muscle cell rows. Pygidium with two anal cirri, shorter than dorsal cirri.
Reproduction
Scissiparity (schizogamy); cephalous stolons with two pairs of red eyes (Fig. 13G). One detached female stolon (from Philippines), yellowish in colour, 13 mm long, 2.0–2.5 mm wide, 50 segments, oocytes 60 μm in diameter (Fig. 13G). Male stolons not seen.
Remarks
Trypanosyllis luzonensis comb. nov. is very similar to Trypanosyllis krohnii. In fact, specimens of Trypanosyllis luzonensis comb. nov. from Australia were considered erroneously as Trypan by San Martín et al. (2008); however, our morphological and molecular analyses showed that Trypanosyllis luzonensis comb. nov. is a valid species. The most apparent features distinguishing Trypanosyllis luzonensis comb. nov. from Trypanosyllis krohnii are the colour pattern, which in Trypanosyllis luzonensis comb. nov. comprises two large bands on each segment, almost fused into a single band (Fig. 10D), and posterior ventral compound chaetae that are unidentate or with a very small proximal tooth. The unique detached stolon examined of Trypanosyllis luzonensis comb. nov. (identified by means of molecular techniques) is larger than any other examined of Trypanosyllis krohnii s.l. (Fig. 13G).
Type locality
Dagupan City, Bonva, Luzon Island, Philippines (Indo-Pacific Ocean).
Distribution
China (Hong Kong), Philippines (Luzon and Palawan Islands), and Australia.
Trypanosyllis taboadai Álvarez-Campos, Riesgo & San Martín sp.nov Figs 10E, 15A–C
Scanning electron micrographs of new species from the Trypanosyllis krohnii species complex: A–C, Trypanosyllis taboadaisp. nov. (MNCN 16.01/16078); D–F, Trypanosyllis californiensissp. nov. (SIO 5007); G–I, Trypanosyllis leivaisp. nov. (MNCN 16.01/16073). A, D, G, anterior chaetae. B, E, H, midbody chaetae. C, F, I, posterior chaetae.
Type material examined
Holotype. New Zealand: specimen in 96% EtOH (MNCN 16.01/16087), Cavalli Islands (−34.984444, 173.944167), calcareous algae, 17 m, 2 February 2012, leg. G. San Martín Project CGL2009-12292 BOS collecting team.
Paratypes. New Zealand: one complete specimen, two anterior and two posterior parts in 96% EtOH (MNCN 16.01/16080, 16097), collection data as for the holotype; one specimen in 96% EtOH (MNCN 16.01/16086), Karikari Peninsula, Maitai Bay (−34.831111, 173.409444), Corallina sp. and unidentified brown algae, 3 m, 31 January 2012, leg. G. San Martín Project CGL2009-12292 BOS collecting team; one specimen in 96% EtOH (MNCN 16.01/16079), Cavalli Islands (−34.984444, 173.944167), unidentified sponges and algae 15 m, 31 January 2012, leg. G. San Martín Project CGL2009-12292 BOS collecting team; two specimens in 96% EtOH (MNCN 16.01/16078, 16099), Karikari Peninsula, Maitai Bay (−34.831111, 173.409444), kelp, 3 m, 31 January 2012, leg. G. San Martín Project CGL2009-12292 BOS collecting team; one specimen in 96% EtOH (MNCN 16.01/16098), Karikari Peninsula, Maitai Bay (−34.831111, 173.409444), brown algae and Corallina sp., 3 m, 31 January 2012, leg. G. San Martín Project CGL2009-12292 BOS collecting team.
Diagnosis
Similar to Trypanosyllis krohnii except for pigmentation and length of appendages (Fig. 10E). Each segment with two brown stripes, one anterior and one posterior, reaching parapodia. Alternation of long and short dorsal cirri less marked than in T. krohnii, long cirri with 21–23 articles, short with 17–19 articles.
Description
Holotype complete specimen in two parts: 20 mm long, 1.1 mm wide, 165 chaetigers. Colour pattern defined by two brown stripes per segment, one on anterior margin and one on posterior margin, reaching parapodia (Fig. 10E). Oval, posteriorly bilobed prostomium, with two pairs of eyes in trapezoidal arrangement (Fig. 10E). Oval palps shorter than prostomium, completely separate. Nuchal organs not seen. Segment 1 slightly smaller than subsequent segments; antennae originating on anterior margin of prostomium, median antenna with about 12 articles, shorter than lateral antennae, with about 16 or 17 articles. Dorsal enlarged anterior cirri longer than antennae, with about 30 articles, longer than ventral cirri, with about 20 articles. Dorsal cirri alternating long (23 articles) and short (18 articles). Ventral digitiform cirri, shorter than parapodium. Compound bidentate, heterogomph falciger chaetae with spines on the margin; ventral chaetae with few spines, almost smooth margins (Fig. 15A–C). Anterior dorsal chaetae with blades longer than those in posterior parapodia (Fig. 15A–C). Between nine and 12 chaetae on anterior and midbody chaetigers, and between six and eight chaetae on posterior chaetigers. Two or three aciculae, thick and acutely pointed in all segments. Dorsal and ventral simple chaetae not seen. Pharynx through about 15 segments, shorter than proventricle through 18 segments, with about 50 muscle cell rows.
Remarks
The most similar species to Trypanosyllis taboadai sp. nov. are Trypanosyllis krohnii and Trypanosyllis taeniaformis (Haswell, 1886). Trypanosyllis krohnii is similar in body size and coloration to Trypanosyllis taboadai sp. nov., but it presents longer appendages and a shorter pharynx and proventricle. Trypanosyllis taeniaformis, originally described from Sydney (Australia), and reported later from New Zealand by Day & Hutchings (1979), also differs in the length of the appendages (longer than those of the new species) and in coloration, which is light red with numerous narrow bands instead of white with two brown stripes per segment, as in Trypanosyllis taboadai sp. nov. (Table 3).
Type locality
Cavalli Islands, New Zealand (Pacific Ocean).
Distribution
Only known from the type locality.
Etymology
Named after Dr Sergi Taboada, close colleague and dear friend, for his help collecting and sorting part of the analysed material, and for his encouragement and insight during the development of this research project.
Trypanosyllis californiensis Állvarez-Campos & Rouse sp. nov. Figs 10H, 15D–F
Type material examined
Holotype. California: specimen in 96% EtOH (SIO A5008), La Jolla, San Diego (32.866944, −117.255833), kelp holdfast, 5 m, 18 April 2014, leg. G.W. Rouse.
Paratypes. California: one specimen mounted for SEM (SIO A5007), collection data as for the holotype; two specimens in 96% EtOH (SIO A5006, 5009), La Jolla, San Diego (32.866944, −117.255833), algae, 0 m, 18 April 2014, leg. G.W. Rouse.
Diagnosis
Similar to Trypanosyllis krohnii except for pigmentation and body size (Fig. 10H). Dark-brown transverse stripes across anterior segments of living specimens. Fixed material appears red/pink with dark red–purple stripes: anterior stripe is situated close to anterior end of segment, whereas posterior stripe appears slightly before end of segment (anterior and posterior stripes appear as two close lines per segment, but, in fact, are on different segments; Fig. 10H). Appendages also with pigmentation only in limit of each article.
Description
Holotype incomplete: 12 mm long, 0.8 mm wide, 76 chaetigers. Body coloration in live specimens white/pale, with two dark-brown thin transverse stripes across anterior and midbody segments of live specimens (Fig. 10H). Fixed material appeared red pigmented with dark red–purple stripes. Anterior stripe situated only in anterior end of segment, whereas posterior stripe slightly precedes posterior end of segment (Fig. 9H). Appendages also with pigmentation in limit of each article. Oval prostomium with two pairs of red eyes in trapezoidal arrangement (Fig. 10H). Oval palps shorter than prostomium, completely separate. Nuchal organs not seen. Segment 1 slightly smaller than subsequent segments; antennae originating on anterior margin of prostomium, median antenna with about 20 articles; lateral antennae slightly shorter, with 15–17 articles. Dorsal enlarged anterior cirri longer than antennae, with about 32 articles, much longer than ventral cirri, with about 17 articles. Dorsal cirri alternating between long (30–32 articles) and short (20–22 articles). Dorsal cirri of the anteriormost segments slightly longer than the rest, with 34–36 articles. Ventral digitiform cirri, shorter than parapodia. Compound bidentate heterogomph falciger chaetae with spines on margin, with between eight and ten spines per parapodium (Fig. 15D–F), similar throughout body, except in most posterior parapodia, that are shorter (Fig. 15F). Two or three thick, acutely pointed aciculae in all segments. Dorsal and ventral simple chaetae not seen. Pharynx through about 14 segments, proventricle through 11 segments, with about 30 muscle cell rows.
Remarks
The examined specimens of Trypanosyllis californiensis sp. nov. collected in La Jolla differ markedly from those of Trypanosyllis luquei sp. nov., collected from San Diego Bay, in length of dorsal cirri (32 and 20 articles in the former, 20 and 14 articles the latter; four and three specimens from each species) and in coloration (Trypanosyllis californiensis sp. nov. has stripes on each segment, whereas Trypanosyllis luquei sp. nov. has stripes in the anterior and posterior end of the segment). In addition, there are also ecological differences: Trypanosyllis californiensis sp. nov. occurs in association with algae, whereas Trypanosyllis luquei sp. nov. lives in association with bryozoans. Another congeneric species described from California (from Monterey Bay), Trypanosyllis intermediaMoore, 1909; differs from Trypanosyllis californiensis sp. nov. in colour pattern, which is reddish brown in the dorsum with pale in anterior and posterior ends of segments (instead of the two brown transverse stripes of Trypanosyllis californiensis sp. nov.) and also in the form of midbody and posterior chaetae, which are much shorter, bidentate, and without spines, i.e. more similar to those in Trypanedenta gemmipara comb. nov. (Fig. 7E).
Type locality
La Jolla, San Diego (Pacific Ocean).
Distribution
Only known from the type locality.
Etymology
Named after the state of California, where the type locality of the species is located.
Trypanosyllis leivai Állvarez-Campos, Riesgo & San Martín sp.nov. Figs 10G, 15G–I
Type material examined
Holotype. Philippines: specimen in 96% EtOH (MNCN 16.01/16082), Luzon Island, between Balayan Bay and Batangas Bay, ‘Mainif Point’, (13.68, 120.855556), coral rubble, 2 m, 8 December 2010, leg. G. San Martín Project CGL2009-12292 BOS collecting team.
Paratypes. Philippines: three specimens in 96% EtOH (MNCN 16.01/16062, 16063, 16081), collection data as for the holotype; one specimen mounted for SEM (MNCN 16.01/16073) and one anterior part in 96% EtOH (MNCN 16.01/16077), Luzon Island, Balayan Bay (13.740556, 120.892778), coral rubble, 2–4 m, 7 December 2010, leg. G. San Martín Project CGL2009-12292 BOS collecting team; one specimen in 96% EtOH (MNCN/ADN 85680), Sombrero Island (Luzon), Balayan Bay, ‘Beatrice Point’, (13.697778, 120.829722), unidentified sponge, 2 m, 9 December 2010, leg. G. San Martín Project CGL2009-12292 BOS collecting team; one specimen in 96% EtOH (MNCN 16.01/16075), Sombrero Island (Luzon), Balayan Bay (13.697778, 120.829722), unidentified sponges, 2 m, 6 December 2010, leg. G. San Martín Project CGL2009-12292 BOS collecting team; one specimen in 96% EtOH (MNCN 16.01/16074), Luzon Island, Balayan Bay, ‘Koala Point’ (13.798889, 120.869444), coral rubble, 2 m, 5 December 2010, leg. G. San Martín Project CGL2009-12292 BOS collecting team; 1 specimen in 96% EtOH (MNCN ADN 85683, 16.01/16061), Palawan Island, El Nido (11.197222 119.317222), unidentified sponge, 12 m, 18 December 2010, leg. G. San Martín Project CGL2009-12292 BOS collecting team.
Diagnosis
Similar to Trypanosyllis krohnii except for pigmentation, body size, and length of appendages (Fig. 9G). Red-brown transverse stripes across anterior segments, two per segment, one wider than the other. Stripes do not reach end of segments (Fig. 10G). Specimens considerably (0.3–0.4 mm; n = 10) with slender and shorter anterior cirri (14–19 articles). Dorsal cirri alternating in anterior and midbody segments: Segments 1 and 3 presenting longer cirri (30–32 articles) and segments 2 and 4 with shorter ones (16–18 articles). Midbody segments with an alternation pattern less evident: long cirri with 16–18 articles and short cirri with 12–14 articles; posterior dorsal cirri all short (7–9 articles).
Description
Holotype incomplete: 15 mm long, 0.4 mm wide, 144 chaetigers. Well-preserved specimens white in colour with brown transverse stripes across anterior and midbody segments; one stripe slightly thicker, in middle of each segment, and the other two stripes thinner, in both anterior and posterior ends of segment (Fig. 10G). Only these two lines are similar in length, reaching the parapodia (Fig. 10G). Appendages also white. Oval prostomium with two pairs of red eyes in trapezoidal arrangement (Fig. 10G). Oval palps shorter than prostomium, completely separate. Nuchal organs not seen. Segment 1 slightly smaller than subsequent segments; antennae originating on anterior margin of prostomium, median antenna with about 22 articles; lateral antennae slightly shorter, with 15–17 articles. Dorsal enlarged anterior cirri longer than antennae, with about 19 articles, longer than ventral cirri, with about 14 articles. Dorsal cirri alternating long (16–18 articles) and short (12–14 articles). Dorsal cirri of the first and third segment longer than remaining cirri, with 32 articles; those from the second and fourth segment much shorter with 14 articles. Ventral digitiform cirri, shorter than parapodia. Compound bidentate heterogomph falciger chaetae with spines on margin. Between ten and 12 chaetae on anterior parapodia (Fig. 15G), between nine and 11 chaetae on midbody parapodia (Fig. 15H), and between six and eight chaetae in posterior parapodia (Fig. 15I). Blades similar throughout body, with ventral blades shorter both in midbody and posterior chaetae (Fig. 15H, I). Three aciculae in anterior parapodia, two thick, straight, acutely pointed and one slightly thinner, distally blended; only two thick, straight, acutely pointed aciculae in midbody parapodia and one in posterior parapodia. Dorsal and ventral simple chaetae not seen. Pharynx through about 14 segments, proventricle through 15 segments, with about 45 muscle cell rows.
Remarks
The general aspect of specimens from the Philippines is more slender and smaller than specimens of the type species of the genus, although they both share a similar coloration pattern (Fig. 10G). Trypanosyllis leivai sp. nov. is the second species of the genus described in the Philippines, together with Trypanosyllis luzonensis comb. nov., although our results showed that there are probably two other species inhabiting these waters (see Fig. 1, Trypanosyllis sp. 1 and specimens Tz39–Tz43). Within the Philippines, the most similar species that also presents stripes on the body is Trypanosyllis luzonensis comb. nov.; however, it clearly differs from Trypanosyllis leivai sp.nov in coloration pattern, size of body, and length of the dorsal cirri (see description above).
Type locality
‘Mainif Point’, between Balayan Bay and Batangas Bay, Luzon Island, Philippines (Indo-Pacific Ocean).
Distribution
Known from the type locality and from El Nido (Palawan Island), in the Philippines.
Etymology
Named after Carlos Leiva, colleague and friend of P.A.-C. and A.R., for his help collecting and sorting some of the analysed material.
Trypanosyllis luquei Álvarez-Campos & Verdes sp. nov. Figs 10F, 16A–C
Scanning electron micrographs of new species from the Trypanosyllis krohnii species complex: A–C, Trypanosyllis luqueisp. nov. (SIO A5004); D–F, Trypanosyllis kalkinsp. nov. (MNCN 16.01/16058).
Type material examined
Holotype. California: specimen in 96% EtOH (SIO A5005), San Diego, Anza Cove (32.795278, −117.212778), bryozoans, intertidal, October 2013, leg. A. Verdes.
Paratypes. California: one specimen mounted for SEM (SIO A5004) and one specimen in 96% EtOH (SIO A5006), collection data as for the holotype.
Diagnosis
Similar to Trypanosyllis krohnii except for pigmentation and length of appendages (Figs 3C, 10F). Two brown transverse stripes across anterior segments, situated next to anterior and posterior ends of segment. Appendages also with brown pigmentation only in limit of each article. Width 0.2–0.4 mm with slender and shorter anterior cirri (32–35 articles). Dorsal cirri alternating long (18–20 articles) and short (12–14 articles).
Description
Holotype incomplete, 12 mm long, 0.4 mm wide, 105 chaetigers. Body white with brown transverse stripes across anterior and midbody segments, situated in the anterior and posterior ends of the segment (Fig. 10G). Appendages also with brown pigmentation just in the end of each article (Fig. 10G). Oval prostomium with two pairs of red eyes in trapezoidal arrangement. Oval palps shorter than prostomium, completely separate. Nuchal organs not seen. Segment 1 slightly smaller than subsequent segments; antennae originating on anterior margin of prostomium, median antenna with about 14 articles; lateral antennae slightly shorter, with 11 articles. Dorsal enlarged anterior cirri much longer than antennae, with about 35 articles, longer than ventral cirri, with about 24 articles. Dorsal cirri alternating between long (20 articles) and short (14 articles). Dorsal cirri from the first to the fourth segment longer than those of remaining segments, with 28 articles. Ventral cirri digitiform, shorter than parapodia. Compound bidentate heterogomph falciger chaetae with spines on margin, with between ten and 12 on anterior parapodia (Fig. 16A), seven or eight on midbody parapodia (Fig. 16B), and five or six on posterior parapodia (Fig. 16C). Blades decreasing in length throughout the body, ventral blades always shorter than dorsal blades (Fig. 16A–C). Two or three aciculae in all parapodia, thick, straight, acutely pointed. Dorsal and ventral simple chaetae not seen. Pharynx through about 12 segments, proventricle also through 12 segments, with about 30 muscle cell rows.
Remarks
Same as those in Trypanosyllis californiensis sp. nov.
Type locality
San Diego Bay, California (Pacific Ocean).
Distribution
Only known from the type locality.
Etymology
Named after Dr. Ángel A. Luque, colleague, friend, and mentor in our malacological endeavours.
Trypanosyllis kalkin Álvarez-Campos & Verdes sp. nov. Figs 10J, 16D–F
Type material examined
Holotype. Chile: specimen in 96% EtOH (MNCN 16.01/16058), Valparaiso region, Las Cruces (−33.847778, −72.0575), Dendrymenia skottsbergii, intertidal, 15 January 2013, leg. P. Álvarez-Campos and A. Verdes.
Paratypes. Chile: one specimen mounted for SEM (MNCN 16.01/16059) and one specimen in 96% EtOH (MNCN 16.01/16057), collection data as for the holotype; one specimen in 96% EtOH (MNCN 16.01/16060), Valparaiso region, Las Cruces (−33.847778, −72.0575), unidentified sponge, 18 m, 16 January 2013, P. Álvarez-Campos and A. Verdes; one specimen in 96% EtOH (MNCN 16.01/160185), Valparaiso region, Las Cruces (−33.847778, −72.0575), unidentified sponge, 10 m, 18 January 2013, P. Álvarez-Campos and A. Verdes.
Diagnosis
Similar to Trypanosyllis krohnii except for pigmentation, body size, and length of appendages (Fig. 10J). Light-brown transverse stripes in anterior segments, two per segment. In addition, well-preserved specimens present pigmentation on prostomium as two bands that reach the eyes. Body width 0.3–0.4 mm (n = 5). Dorsal cirri shorter than those from the type species, alternating long cirri (20–22 articles) and short cirri (14–16 articles) (Fig. 10J).
Description
Holotype incomplete: 4 mm long, 0.6 mm wide, 28 chaetigers. Body white in colour with light-brown transverse stripes in anterior segments, two per segment. In addition, well-preserved specimens present pigmentation in prostomium as two bands reaching the eyes (Fig. 10J). Some appendages also with brown pigmentation just in the limit of each article (Fig. 10J). Oval prostomium with two pairs of red eyes in trapezoidal arrangement. Oval palps shorter than prostomium, completely separate. Nuchal organs not seen. Segment 1 slightly smaller than subsequent segments; antennae originating on anterior margin of prostomium, median antenna with about 12 articles; lateral antennae slightly shorter, with between eight and ten articles. Dorsal enlarged anterior cirri much longer than antennae, with about 22 articles, longer than ventral cirri, with about 18 articles. Dorsal cirri alternating between long (14 articles) and short (10 articles). Dorsal cirri from the first segments slightly longer than remaining cirri, with 16 articles. Ventral cirri digitiform, shorter than parapodia. Compound bidentate, heterogomph falciger chaetae with spines on margin, about 7–9 chaetae in all parapodia (Fig. 16D–F). Blades similar in length throughout body, ventral ones always shorter than dorsal (Fig. 16D–F). Two or three aciculae in anterior and midbody parapodia, thick, straight, acutely pointed, only one thicker in posterior parapodia (Fig. 16F). Dorsal and ventral simple chaetae not seen. Pharynx and proventricle similar in length through about seven segments, proventricle with about 30 muscle cell rows.
Remarks
There is no other species in the complex similar in coloration to Trypanosyllis kalkin sp. nov., as it is the only species presenting coloration on the prostomium (Fig. 10J). In addition, together with Trypanosyllis luquei sp. nov. it is the smallest species in the complex. The other two species described in the Pacific coast, Trypanosyllis luquei sp. nov. (Fig. 10F) and Trypanosyllis californiensis sp. nov. (Fig. 10H), from San Diego, present shorter and wider chaetae. There is another congeneric species described from Chile, Trypanosyllis parazebraHartmann-Schröder, 1965; from Arica, but it shows a single wide brown stripe on each anterior segment, instead of the two stripes of Trypanosyllis kalkin sp. nov. In addition, the compound chaetae have a less marked proximal tooth (Hartmann-Schröder, 1965).
Type locality
Las Cruces, Valparaiso region, Central Chile (Pacific Ocean).
Distribution
Only known from the type locality.
Etymology
From the Mapuche language of the indigenous population of Chile, referring to the stripes that the animals present.
Discussion
Our study provides molecular evidence for the paraphyly of Trypanosyllis, which we now divide into three genera: the resurrected Pseudosyllis stat. satd and Trypanedenta stat. nov., and an amended Trypanosyllis. In particular, traditional morphological characters used to recognize Trypanosyllis (a flattened body and the presence of a trepan) were not useful at the generic level. Thus, morphological diagnosis had to be aided by molecular species delimitation in order to recognize these particular genera of Syllidae, as has been performed previously for other organisms (e.g. Knowlton, 2000; Bickford et al., 2007; Carr et al., 2011; Nygren, 2014). Our new arrangement with Pseudosyllis, Trypanedenta, and Trypanosyllis has important implications for the evolution of the reproductive mode in syllids. The subfamily Syllinae reproduces using schizogamy (by means of stolons filled with gametes, at the end of the main animal, which swim to the surface and spawn; Garwood, 1991), which could imply either scissiparity (one stolon) or gemmiparity (multiple stolons). Both scissiparity and gemmiparity were independently acquired more than once within Syllidae (Nygren, 1999). Whereas gemmiparity was previously known within Syllinae as occurring exclusively in Trypanosyllis (Johnson, 1902; Okada, 1933; Çinar, 2007; Nogueira & Fukuda, 2008), our results and the recent study of Aguado et al. (2015) proved that gemmiparity is also present in other genera outside Trypanosyllis, Trypanobia, and Trypanedenta. Interestingly, Aguado et al. (2012) found seven clades within Syllinae characterized by a different stolon morphotype. Thus, they concluded that the different clades within Syllinae might be defined by a different stolon morphotype. This assumption was not adequate at least for some of the clades that they obtained, given the small taxon sampling considered. In particular, Aguado et al. (2012) suggested that the clade containing Xenosyllis, Eurysyllis, and Trypanosyllis was defined by the presence of acerous stolons (two pairs of eyes), which occurs in three of the four species included in their analysis. We have increased the taxon sampling in such a clade to 19 species, but only seven of them have information on the stolon morphology (Eurysyllis tuberculata, Pseudosyllis brevipennis, Trypanedenta gigantea comb. nov., Trypanedenta gemmipara comb. nov., Trypanosyllis krohnii, Trypanosyllis sp. 2, and Trypanosyllis luzonensis). Our results demonstrate that at least one of the species within Trypanosyllis (Trypanosyllis luzonensis comb. nov.) presents a stolon that cannot be classified as acerous (see remarks and Fig. 13G). Furthermore, Trypanedenta gigantea comb. nov., also in the same clade, showed two different stolon stages: one with a chain of non-mature acephalous stolons (Fig. 6G), whereas another specimen had a mature, detached, cephalous stolon (Fig. 6H), like the one appearing in Trypanosyllis ingens (Johnson, 1902). In addition, in the clade formed by Haplosyllis and Branchiosyllis, acephalous stolons were coded as the only apomorphy (Aguado et al., 2012), although Haplosyllis was previously reported to have several species with cephalous stolons (Lattig, Martin & San Martín, 2010; Lattig & Martin, 2011). The same case can be found within the genus AlcyonosyllisGlasby & Watson, 2001, which presents species with different kinds of stolons (i.e. Glasby & Watson, 2001; Álvarez-Campos et al., 2013). In the light of our results, we suggest that the stolon morphotype may respond to the ontogenetic stage of the specimens captured rather than a taxon-specific fixed character. Consequently, the evolutionary hypothesis provided for Syllinae (Aguado et al., 2012) may need to be further reviewed by largely increasing the taxon sampling of species with known reproductive strategy.
Within Trypanosyllis, the striped species have been traditionally considered as cosmopolitan, given that the morphological differences reported in purported specimens from different localities were considered insufficient to delimit new species (San Martín, 2003; Nogueira & Fukuda, 2008; San Martín et al., 2008). In our case, both the PTP and the GMYC species delimitation analyses, and also the combined phylogenetic analysis of the COI, 16S, 18S, and 28S rRNA genes, agreed in the presence of a complex of species within Trypanosyllis krohnii, comprising at least six pseudocryptic species. In addition, given the clades recovered within Trypanosyllis luzonensis comb. nov. (Figs 1, 2, 5), it may also constitute another case of pseudocryptic speciation. Similarly, the Mediterranean species Trypanosyllis zebra could represent another case of cryptic speciation (see species delimitation and taxonomic results). Both fall far from the scope of the present study, however, and future molecular and morphological analyses within these two species are required to solve this question. As Knowlton (1993, 2000) defined, pseudocryptic species are those that have been morphologically recognized as such only after other methods have unveiled their existence. Pseudocryptic speciation is common in many marine groups (e.g., Knowlton, 1993, 2000; Sáez et al., 2003; Mertens et al., 2012; Cornils & Held, 2014; Kawauchi & Giribet, 2014), including polychaetes (Luttikhuizen & Dekker, 2010), but this is the first reported case of pseudocryptic speciation within Syllidae.
In addition to the previously discussed analyses for accurate species delimitation, a large difference between the highest intraspecific genetic distance and the lowest interspecific genetic divergence using COI sequences is usually required (i.e. there is a DNA barcoding gap; see Hebert, Ratnasingham & de Waard, 2003a; Stoeckle, 2003; DeSalle, Egan & Siddall, 2005; Meyer & Paulay, 2005). The interspecific genetic distances in COI between our pseudocryptic lineages varied from 10.5 to 27.4%, and were always larger than the intraspecific divergences within the Trypanosyllis krohnii complex (Table 2). Our results fall in the range of those of other studies conducted in polychaetes, with reported K2P distances of 6.5–18.5% for COI in Phyllodocidae (Nygren & Pleijel, 2011) and 14.8–20.9% in Polynoidae (Neal et al., 2014). Given that the mtDNA evolves faster compared with most nuclear genes, the use of this marker has been extensively criticized (Ferguson, 2002; Ballard & Whitlock, 2004; Galtier et al., 2009). Furthermore, a recent study has demonstrated that a barcoding gap does not generally exist within Annelida (Kvist, 2014), as it is often found in other taxa (e.g. Hebert, Cywinska & Ball, 2003b). We, in turn, report a large barcoding gap among the seven pseudocryptic lineages of Trypanosyllis krohnii s.l. considered in the analyses of the COI data. In addition, we have obtained a similar trend for the genetic distances using another mitochondrial gene, the 16S rRNA (Table 2), which are also considerably higher than those found in other polychaete groups (2.2–5.3% in Spionidae; Bastrop, Jürss & Sturmbauer, 1998; 2.2–9.6% in Siboglinidae; Miglietta et al., 2010). Despite the agreement between all our molecular analyses in the definition of distinct species within Trypanosyllis, a combined approach using multiple genetic markers (mtDNA and nuclear) and morphological, ecological, physiological, or reproductive data has been proposed as essential to infer species boundaries in polychaetes (e.g. Lewis & Karageorgopoulos, 2008; Rice, Karl & Rice, 2008; Halt et al., 2009; Nygren, Eklöf & Pleijel, 2010; Nygren, 2014). Therefore, we have complemented our molecular analyses here with morphological and ecological information about the species, which provided robustness to the clarification of the taxonomic status of the new species and genera in this particular case within Syllidae.
In our phylogenetic analyses, Trypanosyllis krohnii and Trypanosyllis sp. 2 (lineage 6) were closely related clades occurring in sympatry, but both species presented differences in their bathymetric range together with slight morphological differences. Genetic divergence for this species pair found in COI (14.9%; Table 2) and 16S rRNA sequences (7.2%; Table 2), together with their ecological specialization, suggest that they are separate species. Therefore, we have revealed the presence of pseudocryptic species in a depth range of only a few metres for the first time in syllids, a pattern that has also been reported in other polychaete groups (e.g. Kruse & Reise, 2003; Nygren, Eklöf & Pleijel, 2009; Nygren et al., 2010; Luttikhuizen et al., 2011; Schüller, 2011). Likewise, Trypanosyllis californiensis sp. nov. and Trypanosyllis luquei sp. nov., separated by a distance of just a few kilometres, were highly divergent genetically, although belonging to closely related clades, and also exhibit slight morphological differences. In this case it is possible that further sampling at intermediate localities may unveil a different phylogenetic pattern regarding these two clades, as the lowest divergence in COI (13.9%, Table 2) was detected between these species; however, as both lineages possess diagnostic morphological and ecological traits and are reciprocally monophyletic, we consider them different species. Remaining species within the complex were found in allopatry and the pattern of isolation by distance may explain the high rates in their COI divergences (17.2–27.5%; Table 2).
Interestingly, the Australian lineage (represented by individuals Tk6 and Tk7), which appeared in a well-supported clade in the S variant of GMYC for the 16S rRNA, may correspond to Trypanosyllis taeniaiformis (Haswell, 1886), a species described from Port Jackson, which was also considered part of the Trypanosyllis krohnii complex (San Martín et al., 2008). The whereabouts of the type material of this species are unknown (Anna Murray, Australian Museum, pers.ncommun.), making direct comparisons with our material impossible. As we could not amplify COI for more than one specimen from this locality, and given that they are not considered as a supported lineage in the remaining species delimitation analyses, we adopted a conservative approach and decided to wait to take any taxonomic action until new material becomes available. In the case of Trypanosyllis leivai sp. nov., it is possible that this species is not only distributed in Philippines but also in Australia, as there is a specimen from Shark Bay (Western Australia) that appeared nested in the same clade in both phylogenetic analyses (see Tz20 in Figs 1, 2, 4). Specimen Tz20 was studied by Aguado et al. (2012), but morphological data or voucher information are not available for comparison with Trypanosyllis leivai sp. nov. Regarding Trypanosyllis taboadai sp. nov. (lineage 5), the species delimitation analysis of COI only considered specimens Tk1, Tk4, and Tk5 within the supported clade, but the same clade in both S and M variants of GMYC for the 16S rRNA also included specimen Tk3. Furthermore, the PTP analysis of all of the genes, as well as both ML and BI analyses of the whole data set, also included all of the New Zealand specimens. Given that all specimens (Tk1–Tk5) present the same morphological characters, we have considered Tk2 and Tk3 to also belong in Trypanosyllis taboadai sp. nov. (Figs 4, 5). The discordance between the species delimitation analyses may arise form the existence of more than one species in New Zealand, but further studies including more specimens are needed to test this hypothesis.
Even though cryptic and pseudocryptic species of polychaetes are routinely inferred in genetic surveys, few of them are formally described (see discussion in Westheide & Haß-Cordes, 2001). Such failure in providing taxonomic descriptions can result in an underestimation of the true biodiversity in the sea (Appeltans et al., 2012). We therefore describe here all the well-supported pseudocryptic species and clarify the taxonomy of Trypanosyllis. Our approach was conservative, however, because we only considered species that presented clear morphological, ecological, and molecular evidence as being distinct. Such an approach reduced our initial 12 supported clades obtained by BI and ML analyses (Figs 1, 2) to the seven lineages found in the species delimitation analyses (Figs 3, 4), which presented enough distinctive morphological and ecological features.
Conclusion
The present study constitutes yet another example indicating that poorly interpreted morphology may seriously misinform the taxonomy of a given group, and may also contribute to underestimating biodiversity. In light of our current results, we suggest that the traditional morphological characters used to recognize genera and species within Syllidae should be revised. Trypanosyllis was found to be paraphyletic as formerly construed and some species had to be assigned to two genera, Pseudosyllis and Trypanedenta to rectify this problem. Furthermore, our results also show an underestimation of species numbers within the genus, as all analyses agreed in the existence of at least seven species within the formerly cosmopolitan Trypanosyllis krohnii. Our study denotes that, as has happened in other polychaete clades (e.g. Nygren & Pleijel, 2011), intraspecific colour polymorphism may indicate the existence of cryptic species, as seen also in other groups of marine invertebrates (e.g. Sundberg et al., 2009). We therefore corroborate previous work using combined approaches to infer species boundaries. Taking into account that Syllidae has been reported as one of the most abundant and diverse families of Annelida, and considering the results here obtained within just a small clade, we anticipate the presence of a large amount of hidden diversity within the clade.
Acknowledgements
The authors are indebted to all of the colleagues and friends that have helped with fieldwork: Christos Arvanaditis, Giorgos Chatzigeorgiou, Luigi Musco, Barbara Micka, Marcelo V. Fukuda, Wagner Magalhães, Patricia Lattig, Fernando Fernández Álvarez, Charlotte Watson, Ángel A. Luque and the researchers participating in the cruise Alborán-INDEMARES Oceanographic Campaign (supported by the European Project LIFE+INDEMARES) and in the cruise from the Project CGL2009-12292 BOS (Spanish Ministry of Economy and Knowledge) to G.S.M. Special thanks are offered to Sergi Taboada and Aida Verdes, for help not only in the sampling, sorting, and laboratory work, but also for their useful comments and advice throughout the research. Many thanks to Miquel Arnedo (UB) for additional comments and discussion, and to the reviewers and associate editor for their insightful comments and editing, which helped to improve previous versions of this article. Thanks also to João Gil (Centre d'Estudis Avançats de Blanes, CEAB-CSIC) and M.A Alonso-Zarazaga (MNCN) for their help with taxonomic questions. The first author acknowledges all of the members of the Giribet laboratory and the MCZ Invertebrate Zoology Department for their help and support during her time at the MCZ (Harvard University), and especially to Rosa Fernández who assisted with analyses. Many thanks go to José Ignacio Carvajal (SIO) and Gemma Blasco who greatly helped with the molecular procedures. Thanks also to Pat Hutchings (AM) and Daniel Martin (CEAB-CSIC) who hosted the first author to study some of the material. We are grateful to the curators of the different museums for sending the comparative material on loan. Many thanks also to Enrique Rodríguez for his help with SEM observations at SIDI, UAM. This research received funding from the European Union's (European Atomic Energy Community's) Seventh Framework Program (FP7/2007–2013; FP7/2007–2011) under grant agreement 227799 to P.A.-C. and from the Systematics Research Fund 2013/2014 (Systematic Association of the Linnean Society of London) to P.A.-C. Sequencing was conducted with internal MCZ funds to G.G., funds from the Marie Curie IOF grant 237219 to A.R., and SIO funds and award #1043749 from the National Science Foundation Office of Polar Programs to G.W.R. A.R. was supported by a Juan de la Cierva Research contract from the Spanish Ministry of Science and Innovation.
References
Author notes
*Corresponding author. E-mail: patricia.alvarez@uam.es